Autologous hGMSC-Derived iPS: A New Proposal for Tissue Regeneration
Abstract
:1. Introduction
2. Results
2.1. hGMSCs Characterization
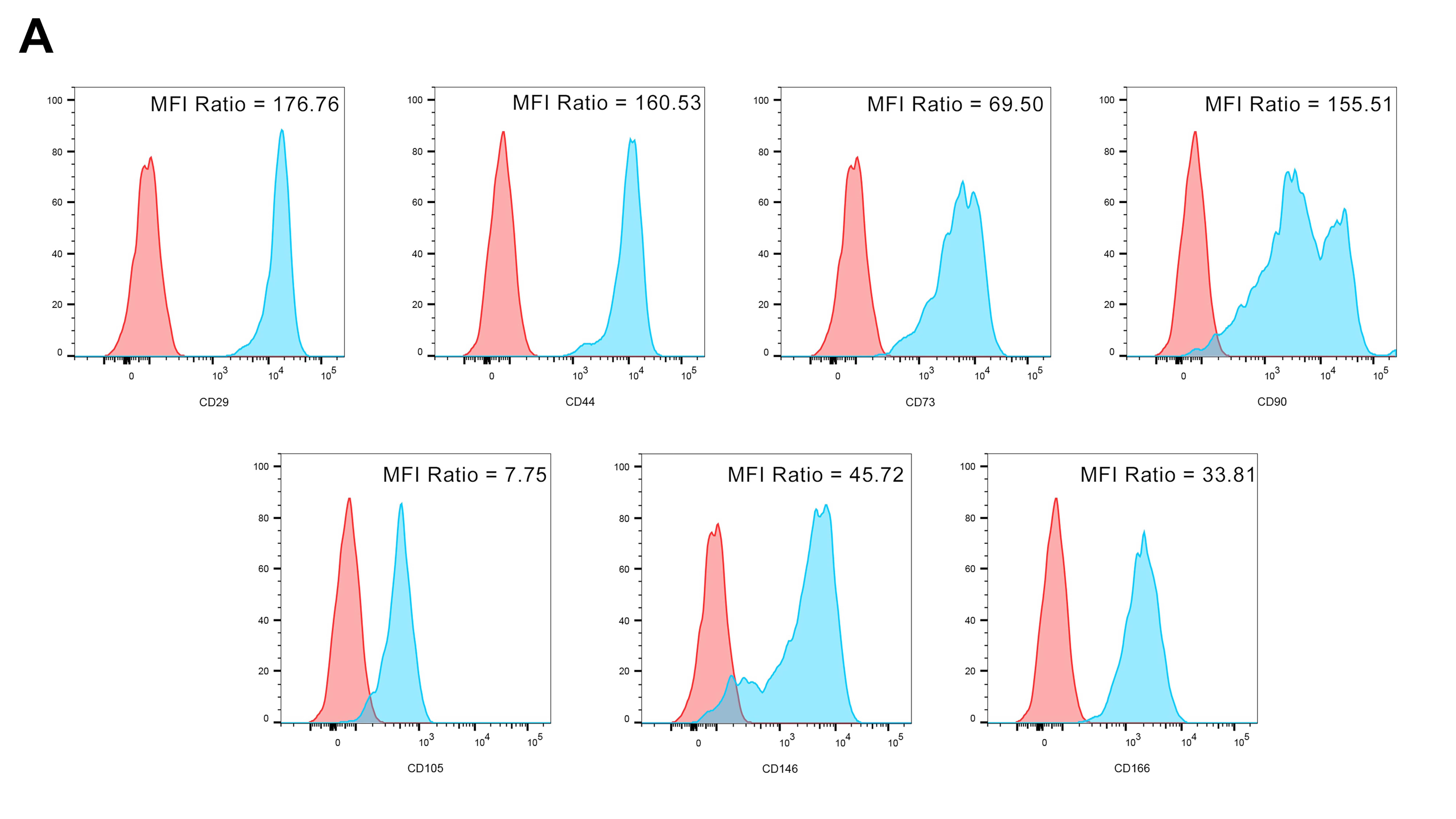
2.2. hGMSCs and hGMSC-Derived iPS Characterization by Flow Cytometry
2.3. hGMSC-Derived iPS Colonies Formation
2.4. Karyotype Reconstruction
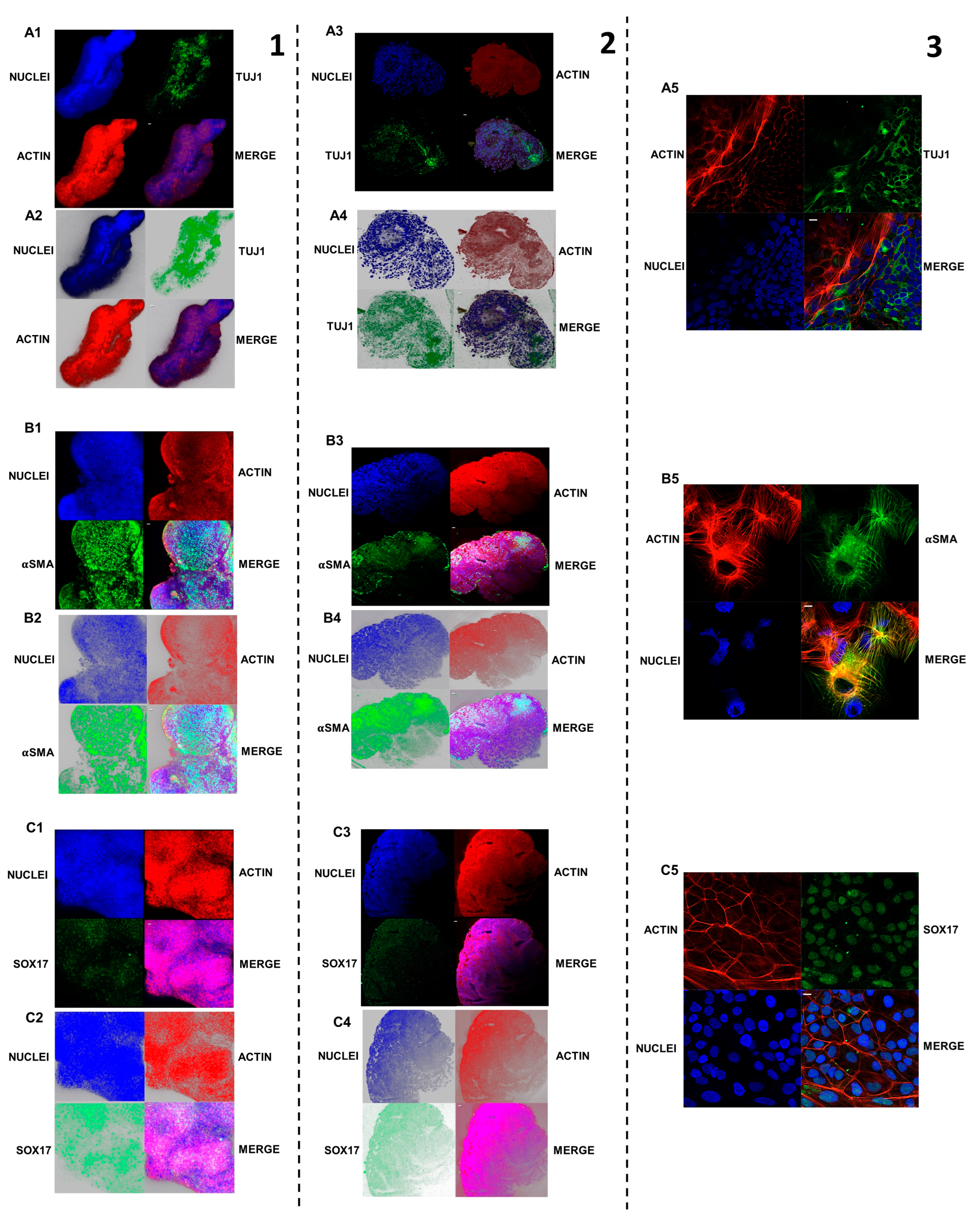
2.5. Pluripotency Markers Expression Analysis with Confocal Microscopy
2.6. DNA Finger Printing
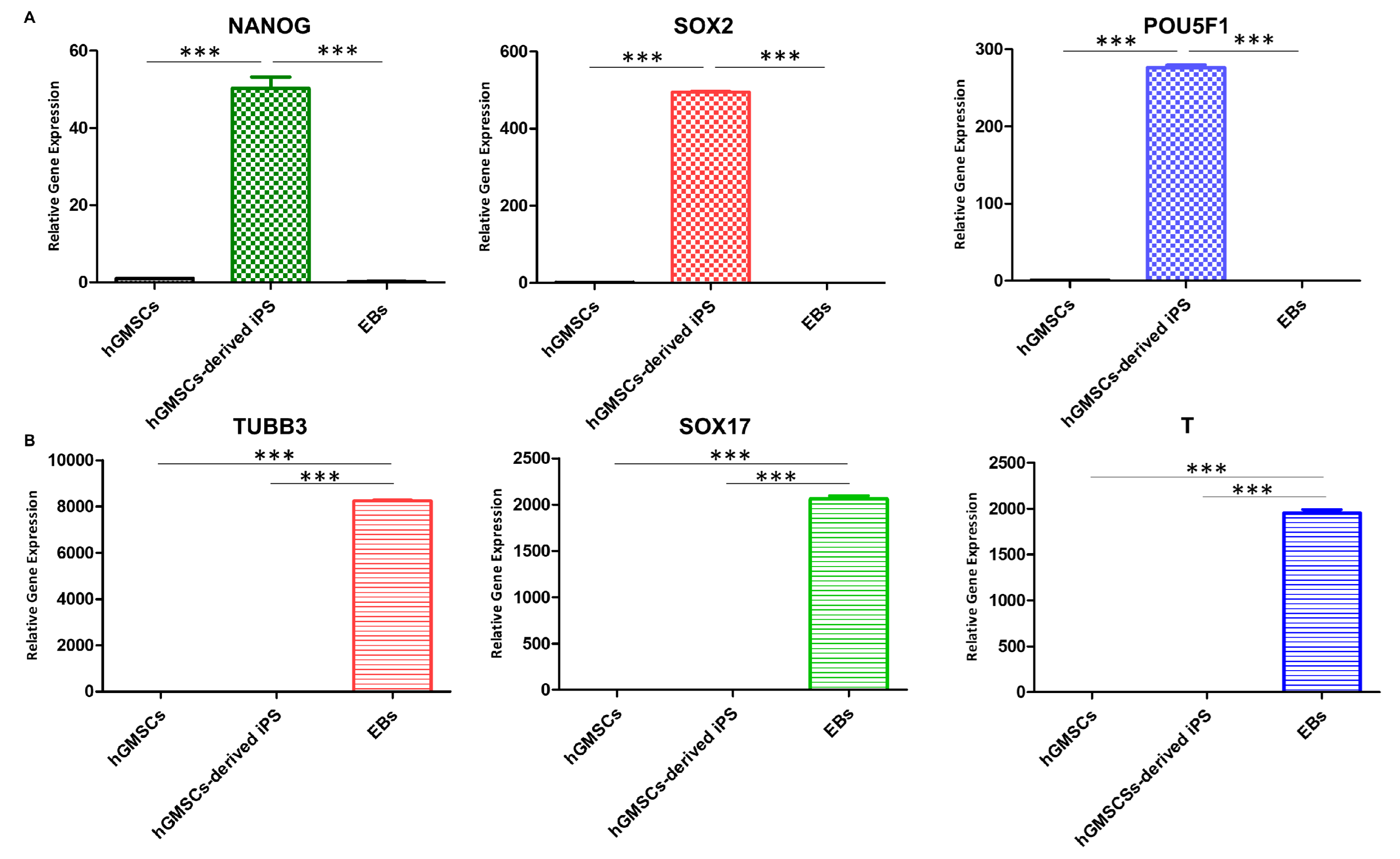
2.7. qPCR
2.8. Analysis of EBs Formation with Optical Microscopy
2.9. Histochemistry Analysis of EBs in Toluidine Blue
2.10. Three Embryonic Layers’ Markers’ Expression Analysis with Confocal Microscopy
2.11. hGMSCs and hGMSC-Derived iPS EVs Flow Cytometry
2.12. miRNA Characterization and Analysis
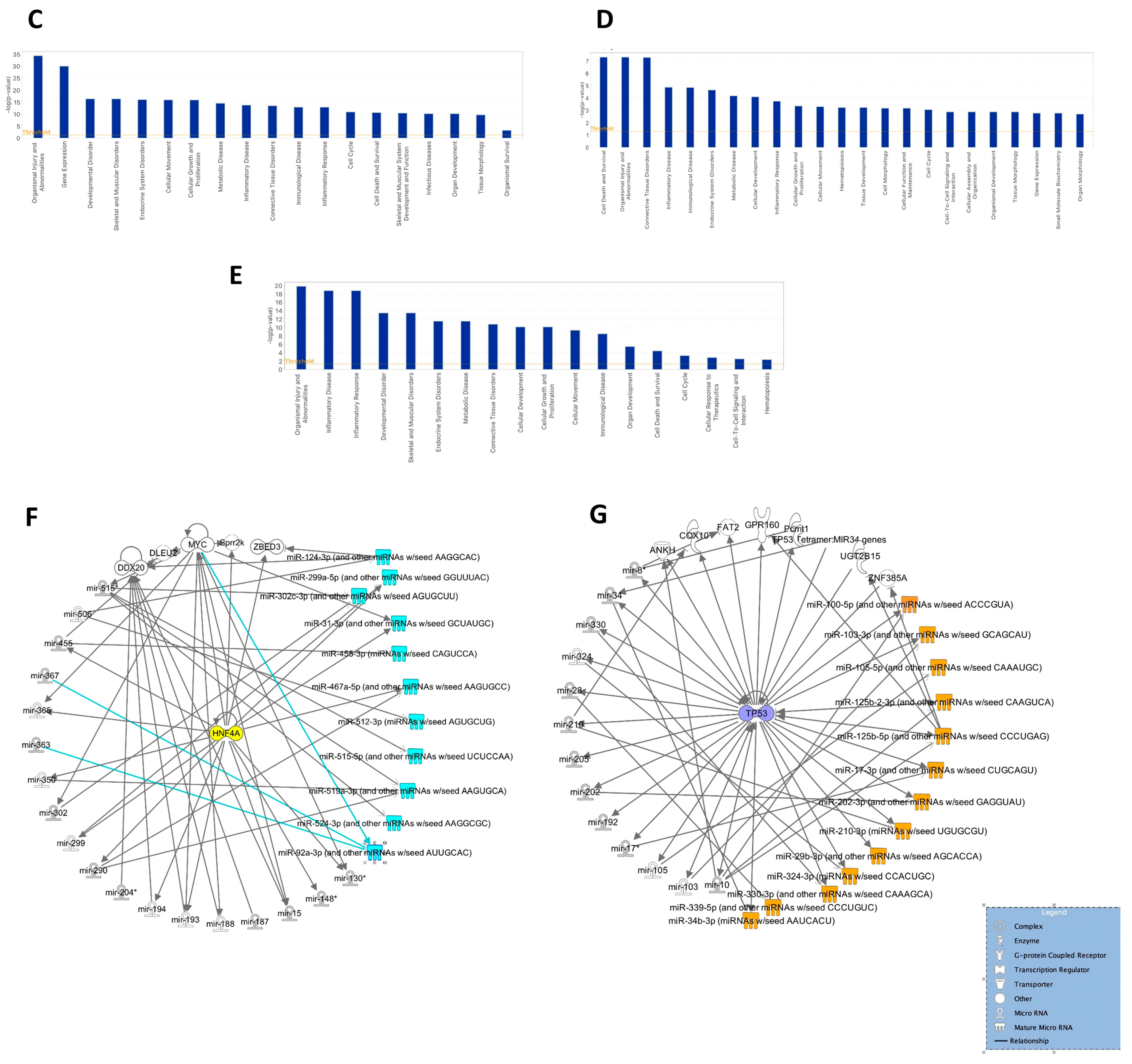
2.13. Ingenuity Pathway Functional Analysis
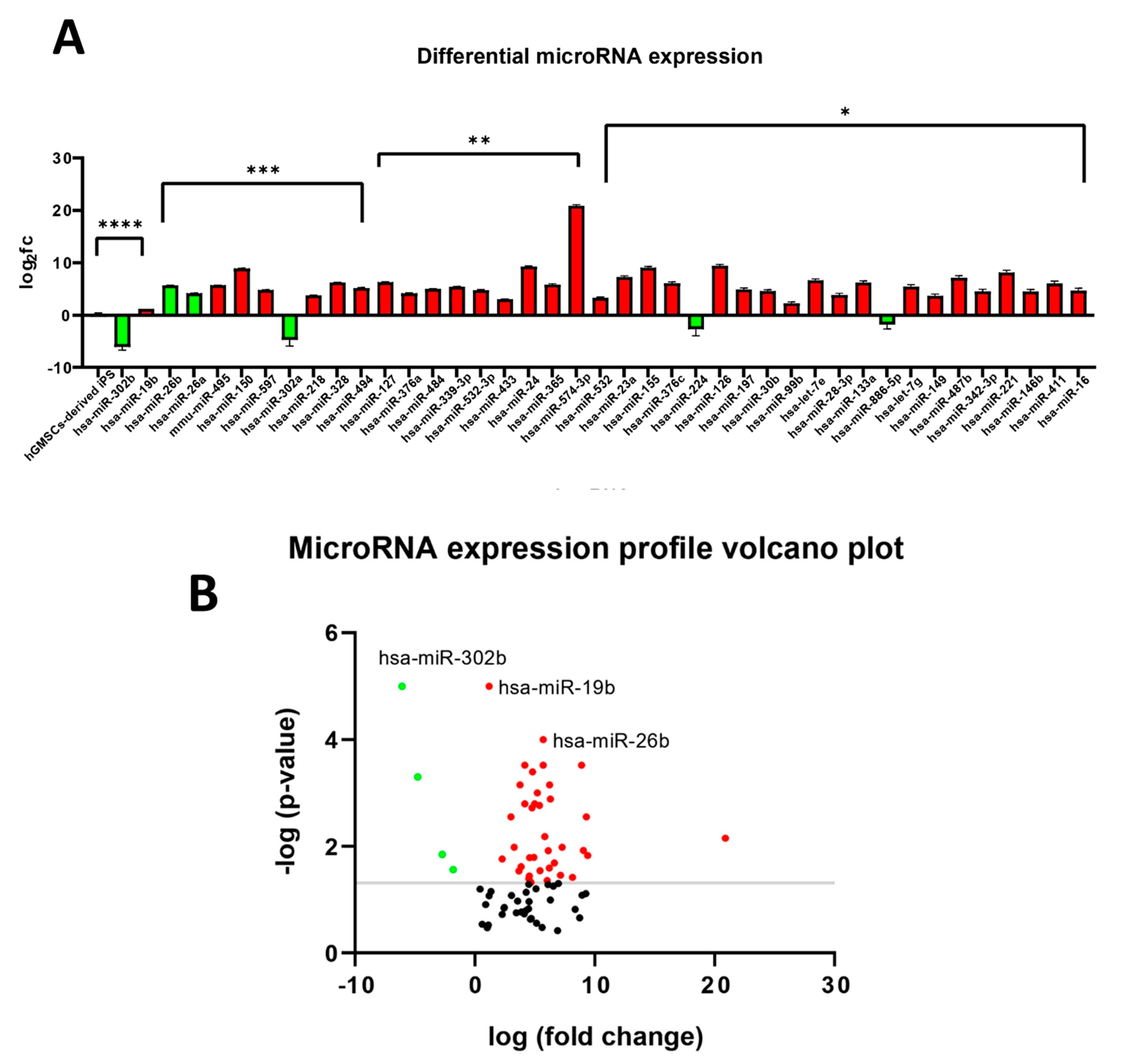
2.14. miRNAs Expression Profile
2.15. Ingenuity Pathway Network Analysis
3. Discussion
4. Materials and Methods
4.1. hGMSCs’ Culture Establishment
4.2. Flow Cytometry Analysis of hGMSC and hGMSC-Derived iPS Phenotypes
4.3. Colorimetric Detection of hGMSCs’ Mesenchymal Differentiation
4.4. RT-PCR Analysis of hGMSCs’ Mesenchymal Differentiation
4.5. Reprogramming of hGMSCs
4.6. hGMSC-Derived iPS Colonies Picking and Expansion
4.7. hGMSC-Derived iPS Optical Microscopy and Scanning Electron Microscope
4.8. Karyotype Reconstruction
4.9. hGMSC-Derived iPS Colonies Immunofluorescence Analysis
4.10. DNA Finger Printing
4.11. Formation of Embryoid Bodies (EBs)
4.12. qPCR
4.13. EB Sections’ Histochemistry and Immunofluorescence Staining
4.14. Immunofluorescence Analysis of 3D EBs and EBs Migrating Population
4.15. EVs Isolation and Quantification
4.16. EVs Characterization by FACS
4.17. EVs miRNAs Extraction
4.18. miRNAs’ Characterization Profile
4.19. IPA Functional and Biological Network Analysis
4.20. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christ, G.J.; Saul, J.M.; Furth, M.E.; Andersson, K.E. The pharmacology of regenerative medicine. Pharmacol. Rev. 2013, 65, 1091–1133. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.; Dunnill, P. A brief definition of regenerative medicine. Regen. Med. 2008, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Uccelli, A.; Moretta, L.; Pistoia, V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008, 8, 726–736. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Thirumala, S.; Goebel, W.S.; Woods, E.J. Clinical grade adult stem cell banking. Organogenesis 2009, 5, 143–154. [Google Scholar] [CrossRef]
- Martin, I.; Galipeau, J.; Kessler, C.; Le Blanc, K.; Dazzi, F. Challenges for mesenchymal stromal cell therapies. Sci. Transl. Med. 2019, 11, eaat2189. [Google Scholar] [CrossRef]
- Venkatesh, D.; Kumar, K.P.M.; Alur, J.B. Gingival mesenchymal stem cells. J. Oral Maxillofac. Pathol. 2017, 21, 296–298. [Google Scholar] [CrossRef]
- Aboul-Soud, M.A.M.; Alzahrani, A.J.; Mahmoud, A. Induced Pluripotent Stem Cells (iPSCs)-roles in regenerative therapies, disease modelling and drug screening. Cells 2021, 10, 2319. [Google Scholar] [CrossRef]
- Bindhya, S.; Sidhanth, C.; Shabna, A.; Krishnapriya, S.; Garg, M.; Ganesan, T.S. Induced pluripotent stem cells: A new strategy to model human cancer. Int. J. Biochem. Cell Biol. 2019, 107, 62–68. [Google Scholar] [CrossRef]
- Pan, G.; Li, J.; Zhou, Y.; Zheng, H.; Pei, D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. Faseb J. 2006, 20, 1730–1732. [Google Scholar] [CrossRef]
- Jaenisch, R.; Young, R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008, 132, 567–582. [Google Scholar] [CrossRef]
- Miyazaki, S.; Yamamoto, H.; Miyoshi, N.; Takahashi, H.; Suzuki, Y.; Haraguchi, N.; Ishii, H.; Doki, Y.; Mori, M. Emerging methods for preparing iPS cells. Jpn. J. Clin. Oncol. 2012, 42, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Della Rocca, Y.; Mazzone, A.; Pizzicannella, J.; Marconi, G.D.; Trubiani, O.; Diomede, F. Reprogramming methods for induced pluripotent stem cells generation. Ital. J. Anat. Embryol. 2023, 127, 89–93. [Google Scholar] [CrossRef]
- Panda, A.; Gurusamy, N.; Rajasingh, S.; Carter, H.K.; Thomas, E.L.; Rajasingh, J. Non-viral reprogramming and induced pluripotent stem cells for cardiovascular therapy. Differentiation 2020, 112, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Taranger, C.K.; Noer, A.; Sorensen, A.L.; Hakelien, A.M.; Boquest, A.C.; Collas, P. Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol. Biol. Cell 2005, 16, 5719–5735. [Google Scholar] [CrossRef]
- Jiang, Z.; Han, Y.; Cao, X. Induced pluripotent stem cell (iPSCs) and their application in immunotherapy. Cell. Mol. Immunol. 2014, 11, 17–24. [Google Scholar] [CrossRef]
- Harrell, C.R.; Fellabaum, C.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Molecular mechanisms responsible for therapeutic potential of mesenchymal stem cell-derived secretome. Cells 2019, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.F. Extracellular vesicles and neurodegenerative diseases. J. Neurosci. 2019, 39, 9269–9273. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, G.; Zhang, K.; Cao, Q.; Liu, T.; Li, J. Mesenchymal stem cells-derived exosomes for drug delivery. Stem Cell Res. Ther. 2021, 12, 561. [Google Scholar] [CrossRef]
- Diomede, F.; Fonticoli, L.; Marconi, G.D.; Della Rocca, Y.; Rajan, T.S.; Trubiani, O.; Murmura, G.; Pizzicannella, J. Decellularized dental pulp, extracellular vesicles, and 5-azacytidine: A new tool for endodontic regeneration. Biomedicines 2022, 10, 403. [Google Scholar] [CrossRef]
- Marchisio, M.; Simeone, P.; Bologna, G.; Ercolino, E.; Pierdomenico, L.; Pieragostino, D.; Ventrella, A.; Antonini, F.; Del Zotto, G.; Vergara, D.; et al. Flow Cytometry analysis of circulating extracellular vesicle subtypes from fresh peripheral blood samples. Int. J. Mol. Sci. 2020, 22, 48. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrugger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Van Pham, P.; Tran, N.Y.; Phan, N.L.; Vu, N.B.; Phan, N.K. Vitamin C stimulates human gingival stem cell proliferation and expression of pluripotent markers. Vitr. Cell. Dev. Biol. Anim. 2016, 52, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Paganelli, A.; Trubiani, O.; Diomede, F.; Pisciotta, A.; Paganelli, R. Immunomodulating profile of dental mesenchymal stromal cells: A comprehensive overview. Front. Oral Health 2021, 2, 635055. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Structure and regulation of Kit protein-tyrosine kinase—The stem cell factor receptor. Biochem. Biophys. Res. Commun. 2005, 338, 1307–1315. [Google Scholar] [CrossRef]
- De Toledo, M.A.S.; Fu, X.; Kluge, F.; Gotz, K.; Schmitz, S.; Wanek, P.; Schuler, H.M.; Pannen, K.; Chatain, N.; Koschmieder, S.; et al. CRISPR/Cas9-engineered human ES cells harboring heterozygous and homozygous c-KIT knockout. Stem Cell Res. 2022, 60, 102732. [Google Scholar] [CrossRef] [PubMed]
- Loukogeorgakis, S.P.; De Coppi, P. Concise review: Amniotic fluid stem cells: The known, the unknown, and potential regenerative medicine applications. Stem Cells 2017, 35, 1663–1673. [Google Scholar] [CrossRef]
- Sheridan, S.D.; Surampudi, V.; Rao, R.R. Analysis of embryoid bodies derived from human induced pluripotent stem cells as a means to assess pluripotency. Stem Cells Int. 2012, 2012, 738910. [Google Scholar] [CrossRef]
- Verma, M.; Lam, T.K.; Hebert, E.; Divi, R.L. Extracellular vesicles: Potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin. Pathol. 2015, 15, 6. [Google Scholar] [CrossRef]
- Li, K.W.; Wang, S.H.; Wei, X.; Hou, Y.Z.; Li, Z.H. Mechanism of miR-122-5p regulating the activation of PI3K-Akt-mTOR signaling pathway on the cell proliferation and apoptosis of osteosarcoma cells through targeting TP53 gene. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12655–12666. [Google Scholar] [CrossRef]
- Cao, F.; Yin, L.X. miR-122 enhances sensitivity of hepatocellular carcinoma to oxaliplatin via inhibiting MDR1 by targeting Wnt/β-catenin pathway. Exp. Mol. Pathol. 2019, 106, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Desi, N.; Teh, V.; Tong, Q.Y.; Lim, C.Y.; Tabatabaeian, H.; Chew, X.H.; Sanchez-Mejias, A.; Chan, J.J. MiR-138 is a potent regulator of the heterogenous MYC transcript population in cancers. Oncogene 2022, 41, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Xiao, T.; Fang, Z.; Sun, Y.; Li, F.; Gao, Y.; Feng, Y.; Li, L.; Wang, Y.; Liu, X.; et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 2012, 287, 23227–23235. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, Y.; Wang, S.; Sheng, X.; Zhang, S. MicroRNA-152 acts as a tumor suppressor MicroRNA by inhibiting kruppel-like factor 5 in human cervical cancer. Oncol. Res. 2019, 27, 335–340. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Li, X.; Sanda, T.; Look, A.T.; Novina, C.D.; von Boehmer, H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 2011, 208, 663–675. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Xing, C.; Wei, S.; Guo, N.; Wang, Y. miR-486 inhibited osteosarcoma cells invasion and epithelial-mesenchymal transition by targeting PIM1. Cancer Biomark. 2018, 23, 269–277. [Google Scholar] [CrossRef]
- Yin, A.; Chen, Q.; Zhong, M.; Jia, B. MicroRNA-138 improves LPS-induced trophoblast dysfunction through targeting RELA and NF-kappaB signaling. Cell Cycle 2021, 20, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Teng, G.G.; Wang, W.H.; Dai, Y.; Wang, S.J.; Chu, Y.X.; Li, J. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS ONE 2013, 8, e56709. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, H. miR-451 elevation relieves inflammatory pain by suppressing microglial activation-evoked inflammatory response via targeting TLR4. Cell Tissue Res. 2018, 374, 487–495. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, Y.Y.; Chen, P.P.; Xu, H. Genome-wide identification of microRNA targets reveals positive regulation of the Hippo pathway by miR-122 during liver development. Cell Death Dis. 2021, 12, 1161. [Google Scholar] [CrossRef] [PubMed]
- Brito, V.G.B.; Bell-Hensley, A.; McAlinden, A. MicroRNA-138: An emerging regulator of skeletal development, homeostasis, and disease. Am. J. Physiol. Cell Physiol. 2023, 325, C1387–C1400. [Google Scholar] [CrossRef] [PubMed]
- Nimmo, R.; Ciau-Uitz, A.; Ruiz-Herguido, C.; Soneji, S.; Bigas, A.; Patient, R.; Enver, T. MiR-142-3p controls the specification of definitive hemangioblasts during ontogeny. Dev. Cell 2013, 26, 237–249. [Google Scholar] [CrossRef]
- Vacante, F.; Denby, L.; Sluimer, J.C.; Baker, A.H. The function of miR-143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vasc. Pharmacol. 2019, 112, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, G.; Li, S.; Lang, M.F.; Yang, S.; Li, W.; Shi, Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Inyawilert, W.; Fu, T.Y.; Lin, C.T.; Tang, P.C. Let-7-mediated suppression of mucin 1 expression in the mouse uterus during embryo implantation. J. Reprod. Dev. 2015, 61, 138–144. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- Su, Q.; Lv, X.; Ye, Z.; Sun, Y.; Kong, B.; Qin, Z.; Li, L. The mechanism of miR-142-3p in coronary microembolization-induced myocardiac injury via regulating target gene IRAK-1. Cell Death Dis. 2019, 10, 61. [Google Scholar] [CrossRef]
- Bei, Y.; Lu, D.; Bar, C.; Chatterjee, S.; Costa, A.; Riedel, I.; Mooren, F.C.; Zhu, Y.; Huang, Z.; Wei, M.; et al. miR-486 attenuates cardiac ischemia/reperfusion injury and mediates the beneficial effect of exercise for myocardial protection. Mol. Ther. 2022, 30, 1675–1691. [Google Scholar] [CrossRef]
- Del Vecchio, V.; Rehman, A.; Panda, S.K.; Torsiello, M.; Marigliano, M.; Nicoletti, M.M.; Ferraro, G.A.; De Falco, V.; Lappano, R.; Lieto, E.; et al. Mitochondrial transfer from Adipose stem cells to breast cancer cells drives multi-drug resistance. J. Exp. Clin. Cancer Res. 2024, 43, 166. [Google Scholar] [CrossRef] [PubMed]
- Papaccio, F.; Garcia-Mico, B.; Gimeno-Valiente, F.; Cabeza-Segura, M.; Gambardella, V.; Gutierrez-Bravo, M.F.; Alfaro-Cervello, C.; Martinez-Ciarpaglini, C.; Rentero-Garrido, P.; Zuniga-Trejos, S.; et al. Proteotranscriptomic analysis of advanced colorectal cancer patient derived organoids for drug sensitivity prediction. J. Exp. Clin. Cancer Res. 2023, 42, 8. [Google Scholar] [CrossRef] [PubMed]
- Pizzicannella, J.; Marconi, G.D.; Guarnieri, S.; Fonticoli, L.; Della Rocca, Y.; Konstantinidou, F.; Rajan, T.S.; Gatta, V.; Trubiani, O.; Diomede, F. Role of ascorbic acid in the regulation of epigenetic processes induced by Porphyromonas gingivalis in endothelial-committed oral stem cells. Histochem. Cell Biol. 2021, 156, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Lanuti, P.; Serafini, F.; Pierdomenico, L.; Simeone, P.; Bologna, G.; Ercolino, E.; Di Silvestre, S.; Guarnieri, S.; Canosa, C.; Impicciatore, G.G.; et al. Human mesenchymal stem cells reendothelialize porcine heart valve scaffolds: Novel perspectives in heart valve tissue engineering. BioResearch Open Access 2015, 4, 288–297. [Google Scholar] [CrossRef]
- Cossarizza, A.; Chang, H.D.; Radbruch, A.; Acs, A.; Adam, D.; Adam-Klages, S.; Agace, W.W.; Aghaeepour, N.; Akdis, M.; Allez, M.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019, 49, 1457–1973. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Della Rocca, Y.; Diomede, F.; Konstantinidou, F.; Gatta, V.; Stuppia, L.; Benedetto, U.; Zimarino, M.; Lanuti, P.; Trubiani, O.; Pizzicannella, J. Autologous hGMSC-Derived iPS: A New Proposal for Tissue Regeneration. Int. J. Mol. Sci. 2024, 25, 9169. https://doi.org/10.3390/ijms25179169
Della Rocca Y, Diomede F, Konstantinidou F, Gatta V, Stuppia L, Benedetto U, Zimarino M, Lanuti P, Trubiani O, Pizzicannella J. Autologous hGMSC-Derived iPS: A New Proposal for Tissue Regeneration. International Journal of Molecular Sciences. 2024; 25(17):9169. https://doi.org/10.3390/ijms25179169
Chicago/Turabian StyleDella Rocca, Ylenia, Francesca Diomede, Fanì Konstantinidou, Valentina Gatta, Liborio Stuppia, Umberto Benedetto, Marco Zimarino, Paola Lanuti, Oriana Trubiani, and Jacopo Pizzicannella. 2024. "Autologous hGMSC-Derived iPS: A New Proposal for Tissue Regeneration" International Journal of Molecular Sciences 25, no. 17: 9169. https://doi.org/10.3390/ijms25179169




