Loss of Lipooligosaccharide Synthesis in Acinetobacter baumannii Produces Changes in Outer Membrane Vesicle Protein Content
Abstract
:1. Introduction
2. Results
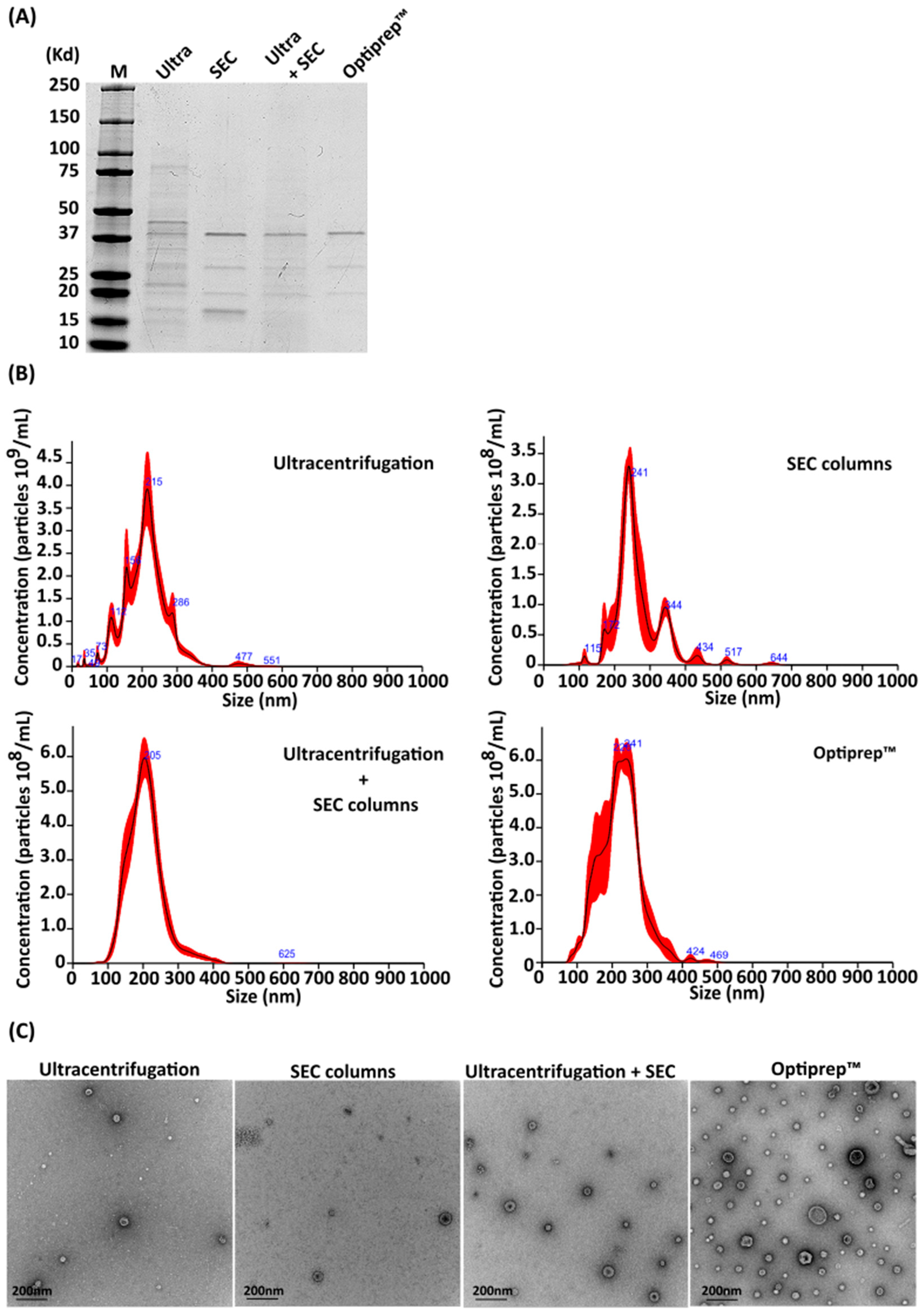
2.1. Comparison of Methods for Purifying A. baumannii OMVs
2.2. Comparison of ATCC 19606 and IB010 OMVs
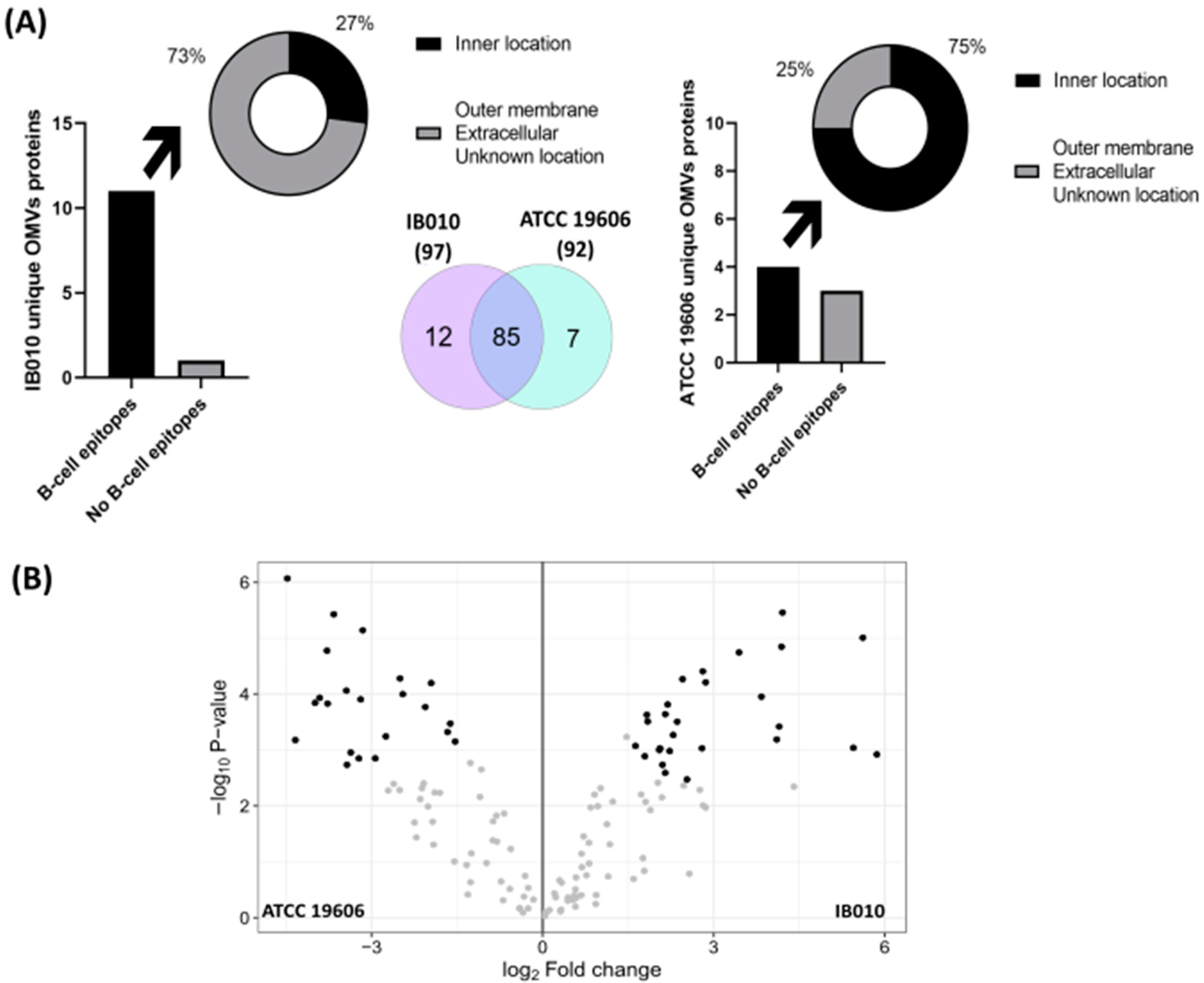
2.3. Proteomic Characterization of ATCC 19606 and IB010 OMVs
2.4. Computational Characterization of Differentially Expressed Proteins
3. Discussion
4. Materials and Methods
4.1. Purification of A. baumannii OMVs
4.2. Nanoparticle Tracking Analysis (NTA)
4.3. Electron Microscopy and Cryoelectron Microscopy
4.4. Mass Spectrometry Analysis (LC-MS/MS)
4.5. Subcellular Localization and Epitope Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal Experimental Requirements for Definition of Extracellular Vesicles and Their Functions: A Position Statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Delgado, L.; López-Iglesias, C.; Mercade, E. Outer-Inner Membrane Vesicles Naturally Secreted by Gram-Negative Pathogenic Bacteria. PLoS ONE 2015, 10, e0116896. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive Cell Lysis as a Mechanism for the Biogenesis of Bacterial Membrane Vesicles and Biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef]
- Orench-Rivera, N.; Kuehn, M.J. Differential Packaging into Outer Membrane Vesicles upon Oxidative Stress Reveals a General Mechanism for Cargo Selectivity. Front. Microbiol. 2021, 12, 561863. [Google Scholar] [CrossRef]
- Lieberman, L.A. Outer Membrane Vesicles: A Bacterial-Derived Vaccination System. Front. Microbiol. 2022, 13, 1029146. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Schild, S.; Kaparakis-Liaskos, M.; Eberl, L. Composition and Functions of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2023, 21, 415–430. [Google Scholar] [CrossRef]
- Micoli, F.; MacLennan, C.A. Outer Membrane Vesicle Vaccines. Semin. Immunol. 2020, 50, 101433. [Google Scholar] [CrossRef]
- van der Pol, L.; Stork, M.; van der Ley, P. Outer Membrane Vesicles as Platform Vaccine Technology. Biotechnol. J. 2015, 10, 1689–1706. [Google Scholar] [CrossRef]
- Sartorio, M.G.; Pardue, E.J.; Feldman, M.F.; Haurat, M.F. Bacterial Outer Membrane Vesicles: From Discovery to Applications. Annu. Rev. Microbiol. 2021, 75, 609–630. [Google Scholar] [CrossRef]
- Fingermann, M.; Avila, L.; De Marco, M.B.; Vázquez, L.; Di Biase, D.N.; Müller, A.V.; Lescano, M.; Dokmetjian, J.C.; Fernández Castillo, S.; Pérez Quiñoy, J.L. OMV-Based Vaccine Formulations against Shiga Toxin Producing Escherichia coli Strains Are Both Protective in Mice and Immunogenic in Calves. Hum. Vaccines Immunother. 2018, 14, 2208–2213. [Google Scholar] [CrossRef] [PubMed]
- Zurita, M.E.; Wilk, M.M.; Carriquiriborde, F.; Bartel, E.; Moreno, G.; Misiak, A.; Mills, K.H.G.; Hozbor, D. A Pertussis Outer Membrane Vesicle-Based Vaccine Induces Lung-Resident Memory CD4 T Cells and Protection Against Bordetella Pertussis, Including Pertactin Deficient Strains. Front. Cell. Infect. Microbiol. 2019, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- König, E.; Gagliardi, A.; Riedmiller, I.; Andretta, C.; Tomasi, M.; Irene, C.; Frattini, L.; Zanella, I.; Berti, F.; Grandi, A.; et al. Multi-Antigen Outer Membrane Vesicle Engineering to Develop Polyvalent Vaccines: The Staphylococcus Aureus Case. Front. Immunol. 2021, 12, 752168. [Google Scholar] [CrossRef]
- Ministry of Health. The Meningococcal B Immunisation Programme—A Response to an Epidemic, National Implementation Strategy; Ministry of Health: Wellington, New Zealand, 2004.
- Pérez, A.E.; Dickinson, F.O.; Rodríguez, M. Community Acquired Bacterial Meningitis in Cuba: A Follow up of a Decade. BMC Infect. Dis. 2010, 10, 130. [Google Scholar] [CrossRef]
- McConnell, M.J.; Actis, L.; Pachón, J. Acinetobacter Baumannii: Human Infections, Factors Contributing to Pathogenesis and Animal Models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef]
- Valencia, R.; Arroyo, L.A.; Conde, M.; Aldana, J.M.; Torres, M.-J.; Fernández-Cuenca, F.; Garnacho-Montero, J.; Cisneros, J.M.; Ortiz, C.; Pachón, J.; et al. Nosocomial Outbreak of Infection With Pan–Drug-Resistant Acinetobacter Baumannii in a Tertiary Care University Hospital. Infect. Control Hosp. Epidemiol. 2009, 30, 257–263. [Google Scholar] [CrossRef]
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 14 June 2024).
- López-Siles, M.; Corral-Lugo, A.; McConnell, M.J. Vaccines for Multidrug Resistant Gram Negative Bacteria: Lessons from the Past for Guiding Future Success. FEMS Microbiol. Rev. 2021, 45, fuaa054. Available online: https://academic.oup.com/femsre/article/45/3/fuaa054/6027553?login=true (accessed on 20 September 2023). [CrossRef] [PubMed]
- Tan, Y.C.; Lahiri, C. Promising Acinetobacter Baumannii Vaccine Candidates and Drug Targets in Recent Years. Front. Immunol. 2022, 13, 900509. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, D.-Q.; Ji, L.; Sun, S.; Jin, Z.; Jin, Z.-L.; Sun, H.-W.; Zeng, H.; Zhang, W.-J.; Lu, D.-S.; et al. Development of Different Methods for Preparing Acinetobacter Baumannii Outer Membrane Vesicles Vaccine: Impact of Preparation Method on Protective Efficacy. Front. Immunol. 2020, 11, 1069. [Google Scholar] [CrossRef]
- Pulido, M.R.; García-Quintanilla, M.; Pachón, J.; McConnell, M.J. A Lipopolysaccharide-Free Outer Membrane Vesicle Vaccine Protects against Acinetobacter Baumannii Infection. Vaccine 2020, 38, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Yao, Y.; Long, Q.; Yang, X.; Sun, W.; Liu, C.; Jin, X.; Li, Y.; Chu, X.; Chen, B.; et al. Immunization against Multidrug-Resistant Acinetobacter Baumannii Effectively Protects Mice in Both Pneumonia and Sepsis Models. PLoS ONE 2014, 9, e100727. [Google Scholar] [CrossRef]
- Jun, S.H.; Lee, J.H.; Kim, B.R.; Kim, S.I.; Park, T.I.; Lee, J.C.; Lee, Y.C. Acinetobacter Baumannii Outer Membrane Vesicles Elicit a Potent Innate Immune Response via Membrane Proteins. PLoS ONE 2013, 8, e71751. [Google Scholar] [CrossRef] [PubMed]
- Higham, S.L.; Baker, S.; Flight, K.E.; Krishna, A.; Kellam, P.; Reece, S.T.; Tregoning, J.S. Intranasal Immunization with Outer Membrane Vesicles (OMV) Protects against Airway Colonization and Systemic Infection with Acinetobacter Baumannii. J. Infect. 2023, 86, 563–573. [Google Scholar] [CrossRef]
- Huang, W.; Wang, S.; Yao, Y.; Xia, Y.; Yang, X.; Li, K.; Sun, P.; Liu, C.; Sun, W.; Bai, H.; et al. Employing Escherichia Coli-Derived Outer Membrane Vesicles as an Antigen Delivery Platform Elicits Protective Immunity against Acinetobacter Baumannii Infection. Sci. Rep. 2016, 6, 37242. [Google Scholar] [CrossRef]
- Gnopo, Y.M.D.; Watkins, H.C.; Stevenson, T.C.; DeLisa, M.P.; Putnam, D. Designer Outer Membrane Vesicles as Immunomodulatory Systems—Reprogramming Bacteria for Vaccine Delivery. Adv. Drug Deliv. Rev. 2017, 114, 132–142. [Google Scholar] [CrossRef]
- Daneshian, M.; Guenther, A.; Wendel, A.; Hartung, T.; von Aulock, S. In Vitro Pyrogen Test for Toxic or Immunomodulatory Drugs. J. Immunol. Methods 2006, 313, 169–175. [Google Scholar] [CrossRef]
- Zhang, G.; Meredith, T.C.; Kahne, D. On the Essentiality of Lipopolysaccharide to Gram-Negative Bacteria. Curr. Opin. Microbiol. 2013, 16, 779–785. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin Resistance in Acinetobacter Baumannii Is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.J.; Hall, R.M. Variation in the Complex Carbohydrate Biosynthesis Loci of Acinetobacter Baumannii Genomes. PLoS ONE 2013, 8, e62160. [Google Scholar] [CrossRef]
- Fernández-Reyes, M.; Rodríguez-Falcón, M.; Chiva, C.; Pachón, J.; Andreu, D.; Rivas, L. The Cost of Resistance to Colistin in Acinetobacter Baumannii: A Proteomic Perspective. Proteomics 2009, 9, 1632–1645. [Google Scholar] [CrossRef] [PubMed]
- Webber, J.; Clayton, A. How Pure Are Your Vesicles? J. Extracell. Vesicles 2013, 2, 19861. [Google Scholar] [CrossRef]
- Jha, C.; Ghosh, S.; Gautam, V.; Malhotra, P.; Ray, P. In Vitro Study of Virulence Potential of Acinetobacter Baumannii Outer Membrane Vesicles. Microb. Pathog. 2017, 111, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Koning, R.I.; de Breij, A.; Oostergetel, G.T.; Nibbering, P.H.; Koster, A.J.; Dijkshoorn, L. Cryo-Electron Tomography Analysis of Membrane Vesicles from Acinetobacter Baumannii ATCC19606T. Res. Microbiol. 2013, 164, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Dell’Annunziata, F.; Dell’Aversana, C.; Doti, N.; Donadio, G.; Dal Piaz, F.; Izzo, V.; De Filippis, A.; Galdiero, M.; Altucci, L.; Boccia, G.; et al. Outer Membrane Vesicles Derived from Klebsiella Pneumoniae Are a Driving Force for Horizontal Gene Transfer. Int. J. Mol. Sci. 2021, 22, 8732. [Google Scholar] [CrossRef]
- Jasim, R.; Han, M.-L.; Zhu, Y.; Hu, X.; Hussein, M.H.; Lin, Y.-W.; Zhou, Q.; Dong, C.Y.D.; Li, J.; Velkov, T. Lipidomic Analysis of the Outer Membrane Vesicles from Paired Polymyxin-Susceptible and -Resistant Klebsiella Pneumoniae Clinical Isolates. Int. J. Mol. Sci. 2018, 19, 2356. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.; Vizio, D.D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques Used for the Isolation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. Available online: https://onlinelibrary.wiley.com/doi/10.1080/20013078.2018.1535750 (accessed on 21 September 2023). [CrossRef]
- Steć, A.; Jońca, J.; Waleron, K.; Waleron, M.; Płoska, A.; Kalinowski, L.; Wielgomas, B.; Dziomba, S. Quality Control of Bacterial Extracellular Vesicles with Total Protein Content Assay, Nanoparticles Tracking Analysis, and Capillary Electrophoresis. Int. J. Mol. Sci. 2022, 23, 4347. [Google Scholar] [CrossRef]
- Lau, Y.T.; Tan, H.S. Acinetobacter baumannii Subunit Vaccines: Recent Progress and Challenges. Crit. Rev. Microbiol. 2024, 50, 434–449. Available online: https://www.tandfonline.com/doi/abs/10.1080/1040841X.2023.2215303 (accessed on 26 September 2023). [CrossRef]
- Moon, D.C.; Choi, C.H.; Lee, J.H.; Choi, C.-W.; Kim, H.-Y.; Park, J.S.; Kim, S.I.; Lee, J.C. Acinetobacter Baumannii Outer Membrane Protein a Modulates the Biogenesis of Outer Membrane Vesicles. J. Microbiol. 2012, 50, 155–160. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, M.H.; Kim, S.I.; Son, J.H.; Kim, S.; Lee, Y.C.; Shin, M.; Oh, M.H.; Lee, J.C. The Sensor Kinase BfmS Controls Production of Outer Membrane Vesicles in Acinetobacter Baumannii. BMC Microbiol. 2019, 19, 301. [Google Scholar] [CrossRef]
- Yun, S.H.; Park, E.C.; Lee, S.-Y.; Lee, H.; Choi, C.-W.; Yi, Y.-S.; Ro, H.-J.; Lee, J.C.; Jun, S.; Kim, H.-Y.; et al. Antibiotic Treatment Modulates Protein Components of Cytotoxic Outer Membrane Vesicles of Multidrug-Resistant Clinical Strain, Acinetobacter Baumannii DU202. Clin. Proteom. 2018, 15, 28. [Google Scholar] [CrossRef]
- Song, X.; Zhang, H.; Zhang, D.; Xie, W.; Zhao, G. Bioinformatics Analysis and Epitope Screening of a Potential Vaccine Antigen TolB from Acinetobacter Baumannii Outer Membrane Protein. Infect. Genet. Evol. 2018, 62, 73–79. [Google Scholar] [CrossRef]
- Kesavan, D.; Vasudevan, A.; Wu, L.; Chen, J.; Su, Z.; Wang, S.; Xu, H. Integrative Analysis of Outer Membrane Vesicles Proteomics and Whole-Cell Transcriptome Analysis of Eravacycline Induced Acinetobacter Baumannii Strains. BMC Microbiol. 2020, 20, 31. [Google Scholar] [CrossRef]
- Hassan, A.; Naz, A.; Obaid, A.; Paracha, R.Z.; Naz, K.; Awan, F.M.; Muhmmad, S.A.; Janjua, H.A.; Ahmad, J.; Ali, A. Pangenome and Immuno-Proteomics Analysis of Acinetobacter Baumannii Strains Revealed the Core Peptide Vaccine Targets. BMC Genom. 2016, 17, 732. [Google Scholar] [CrossRef]
- Hua, X.; Liu, L.; Fang, Y.; Shi, Q.; Li, X.; Chen, Q.; Shi, K.; Jiang, Y.; Zhou, H.; Yu, Y. Colistin Resistance in Acinetobacter Baumannii MDR-ZJ06 Revealed by a Multiomics Approach. Front. Cell. Infect. Microbiol. 2017, 7, 45. [Google Scholar] [CrossRef]
- Henry, R.; Vithanage, N.; Harrison, P.; Seemann, T.; Coutts, S.; Moffatt, J.H.; Nation, R.L.; Li, J.; Harper, M.; Adler, B.; et al. Colistin-Resistant, Lipopolysaccharide-Deficient Acinetobacter Baumannii Responds to Lipopolysaccharide Loss through Increased Expression of Genes Involved in the Synthesis and Transport of Lipoproteins, Phospholipids, and Poly-β-1,6-N-Acetylglucosamine. Antimicrob. Agents Chemother. 2012, 56, 59–69. [Google Scholar] [CrossRef]
- Labrador-Herrera, G.; Pérez-Pulido, A.J.; Álvarez-Marín, R.; Casimiro-Soriguer, C.S.; Cebrero-Cangueiro, T.; Morán-Barrio, J.; Pachón, J.; Viale, A.M.; Pachón-Ibáñez, M.E. Virulence Role of the Outer Membrane Protein CarO in Carbapenem-Resistant Acinetobacter Baumannii. Virulence 2020, 11, 1727–1737. [Google Scholar] [CrossRef]
- García-Quintanilla, M.; Pulido, M.R.; Pachón, J.; McConnell, M.J. Immunization with Lipopolysaccharide-Deficient Whole Cells Provides Protective Immunity in an Experimental Mouse Model of Acinetobacter baumannii Infection. PLoS ONE 2014, 9, e114410. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0114410#pone.0114410-Moffatt1 (accessed on 19 September 2023). [CrossRef] [PubMed]
- Tulkens, J.; De Wever, O.; Hendrix, A. Analyzing Bacterial Extracellular Vesicles in Human Body Fluids by Orthogonal Biophysical Separation and Biochemical Characterization. Nat. Protoc. 2020, 15, 40–67. [Google Scholar] [CrossRef]
- Brunelle, J.L.; Green, R. Chapter Thirteen—Coomassie Blue Staining. In Methods in Enzymology; Lorsch, J., Ed.; Laboratory Methods in Enzymology: Protein Part C; Academic Press: Cambridge, MA, USA, 2014; Volume 541, pp. 161–167. [Google Scholar]
- FragPipe. Available online: https://fragpipe.nesvilab.org/ (accessed on 4 October 2023).
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved Protein Subcellular Localization Prediction with Refined Localization Subcategories and Predictive Capabilities for All Prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Clifford, J.N.; Høie, M.H.; Deleuran, S.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-3.0: Improved B-Cell Epitope Prediction Using Protein Language Models. Protein Sci. 2022, 31, e4497. [Google Scholar] [CrossRef]
- Shashkova, T.I.; Umerenkov, D.; Salnikov, M.; Strashnov, P.V.; Konstantinova, A.V.; Lebed, I.; Shcherbinin, D.N.; Asatryan, M.N.; Kardymon, O.L.; Ivanisenko, N.V. SEMA: Antigen B-Cell Conformational Epitope Prediction Using Deep Transfer Learning. Front. Immunol. 2022, 13, 960985. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Bagos, P.G.; Liakopoulos, T.D.; Spyropoulos, I.C.; Hamodrakas, S.J. PRED-TMBB: A Web Server for Predicting the Topology of β-Barrel Outer Membrane Proteins. Nucleic Acids Res. 2004, 32, W400–W404. [Google Scholar] [CrossRef]

| Protein ID | Gene Name | Protein Description | Combined Total Peptides |
|---|---|---|---|
| D0CG85 | tuf | Elongation factor Tu (Fragment) | 5 |
| D0CDQ1 | bpeB | Efflux pump membrane transporter | 4 |
| D0C629 | HMPREF0010_00209 | Translocation and assembly module subunit TamA | 3 |
| D0CEK8 | atpF | ATP synthase subunit b | 3 |
| D0C807 | gltI | Glutamate-aspartate periplasmic-binding protein | 2 |
| D0CCD3 | HMPREF0010_02413 | Uncharacterized protein | 6 |
| D0C7Q4 | HMPREF0010_00784 | Uncharacterized protein | 2 |
| Protein ID | Gene Name | Protein Description | Combined Total Peptides |
|---|---|---|---|
| D0C7A8 | HMPREF0010_00638 | Lipoprotein | 3 |
| D0CDQ8 | lolA | Outer-membrane lipoprotein carrier protein | 3 |
| D0C9M0 | lolB | Outer-membrane lipoprotein LolB | 2 |
| D0CC74 | rpsB | 30S ribosomal protein S2 | 2 |
| D0CDB4 | HMPREF0010_02744 | DUF306 domain-containing protein | 2 |
| D0CDY8 | HMPREF0010_02968 | TonB-dependent siderophore receptor | 2 |
| D0CF71 | HMPREF0010_03537 | Carbapenem susceptibility porin CarO | 2 |
| D0CFX6 | HMPREF0010_03656 | Metallo-beta-lactamase domain protein | 2 |
| D0CBN6 | HMPREF0010_02166 | Uncharacterized protein | 3 |
| D0CF26 | HMPREF0010_03356 | Uncharacterized protein | 3 |
| D0C7G2 | HMPREF0010_00692 | Uncharacterized protein | 2 |
| D0CB14 | HMPREF0010_01944 | Uncharacterized protein | 2 |
| Protein ID | Gene Name | Protein Description | IB010 vs. ATCC 19606 log2 Fold Change | p Value |
|---|---|---|---|---|
| ||||
| D0C9R5 | HMPREF0010_01378 | OmpA family protein | 4.19 | <0.01 |
| D0C907 | HMPREF0010_01565 | META domain protein (Fragment) | 2.81 | <0.01 |
| D0CEN2 | HMPREF0010_03212 | Lipoprotein | 2.8 | <0.01 |
| D0CCX7 | ttg2D | Toluene tolerance protein Ttg2D | 2.46 | <0.01 |
| D0C6H4 | HMPREF0010_00354 | Outer membrane protein | 2.19 | <0.01 |
| D0C5N9 | HMPREF0010_00069 | Signal peptide protein | 2.15 | <0.01 |
| D0CDW1 | HMPREF0010_02941 | Lipoprotein | 2.15 | <0.01 |
| D0CDU8 | HMPREF0010_02928 | Lipoprotein | 2.1 | 0.02 |
| D0C6J4 | gdhB | Quinoprotein glucose dehydrogenase B | 2.06 | <0.01 |
| D0CBP8 | mucD | Periplasmic serine endoprotease DegP-like | 1.84 | <0.01 |
| D0C608 | HMPREF0010_00188 | DJ-1/PfpI family protein | 1.83 | <0.01 |
| D0C8L9 | HMPREF0010_01099 | Hemolysin | 1.79 | 0.01 |
| D0CDA9 | HMPREF0010_02739 | Uncharacterized protein | 5.62 | <0.01 |
| D0CBW9 | HMPREF0010_02249 | Uncharacterized protein | 3.44 | <0.01 |
| D0CFX4 | HMPREF0010_03654 | Uncharacterized protein | 2.86 | <0.01 |
| D0C5H7 | HMPREF0010_00007 | Uncharacterized protein | 2.05 | 0.01 |
| ||||
| D0CEW0 | ptk | Tyrosine-protein kinase Ptk (Fragment) | −4.48 | <0.01 |
| D0CBK4 | HMPREF0010_02134 | Rhombotarget A | −3.99 | <0.01 |
| D0C5L7 | mrcB | Penicillin-binding protein 1B | −3.78 | <0.01 |
| D0CBK5 | HMPREF0010_02135 | Gammaproteobacterial enzyme transmembrane domain protein | −3.36 | 0.01 |
| D0CD03 | rpsC | 30S ribosomal protein S3 | −3.22 | <0.01 |
| D0C9Z6 | mqo | Probable malate:quinone oxidoreductase | −3.15 | <0.01 |
| D0C8P3 | HMPREF0010_01123 | Type VI secretion system effector, Hcp1 family | −2.93 | 0.01 |
| D0CDE5 | gcd | Quinoprotein glucose dehydrogenase | −2.45 | <0.01 |
| D0CEV8 | HMPREF0010_03288 | Polysaccharide biosynthesis/export protein | −2.06 | <0.01 |
| D0CC83 | mltB | Lytic murein transglycosylase B | −1.67 | <0.01 |
| D0CBZ2 | HMPREF0010_02272 | Peptidase, M23 family | −1.62 | <0.01 |
| D0CDN5 | HMPREF0010_02865 | Tat pathway signal sequence domain protein | −1.53 | <0.01 |
| D0C6H8 | HMPREF0010_00358 | Uncharacterized protein | −3.66 | <0.01 |
| D0C6N7 | HMPREF0010_00417 | Uncharacterized protein | −3.44 | <0.01 |
| D0C985 | HMPREF0010_01198 | Uncharacterized protein | −2.75 | <0.01 |
| D0CD52 | HMPREF0010_02682 | Uncharacterized protein | −2.5 | <0.01 |
| D0C5L6 | HMPREF0010_00046 | Uncharacterized protein | −1.95 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cano-Castaño, B.; Corral-Lugo, A.; Gato, E.; Terrón, M.C.; Martín-Galiano, A.J.; Sotillo, J.; Pérez, A.; McConnell, M.J. Loss of Lipooligosaccharide Synthesis in Acinetobacter baumannii Produces Changes in Outer Membrane Vesicle Protein Content. Int. J. Mol. Sci. 2024, 25, 9272. https://doi.org/10.3390/ijms25179272
Cano-Castaño B, Corral-Lugo A, Gato E, Terrón MC, Martín-Galiano AJ, Sotillo J, Pérez A, McConnell MJ. Loss of Lipooligosaccharide Synthesis in Acinetobacter baumannii Produces Changes in Outer Membrane Vesicle Protein Content. International Journal of Molecular Sciences. 2024; 25(17):9272. https://doi.org/10.3390/ijms25179272
Chicago/Turabian StyleCano-Castaño, Beatriz, Andrés Corral-Lugo, Eva Gato, María C. Terrón, Antonio J. Martín-Galiano, Javier Sotillo, Astrid Pérez, and Michael J. McConnell. 2024. "Loss of Lipooligosaccharide Synthesis in Acinetobacter baumannii Produces Changes in Outer Membrane Vesicle Protein Content" International Journal of Molecular Sciences 25, no. 17: 9272. https://doi.org/10.3390/ijms25179272






