Cucurbitacin B Inhibits the Proliferation of WPMY-1 Cells and HPRF Cells via the p53/MDM2 Axis
Abstract
1. Introduction
2. Results
2.1. Cu B Exhibited Antiproliferative Effects on WPMY-1 and HPRF Cells
2.2. Cu B Induced the Apoptosis of WPMY-1 and HPRF Cells
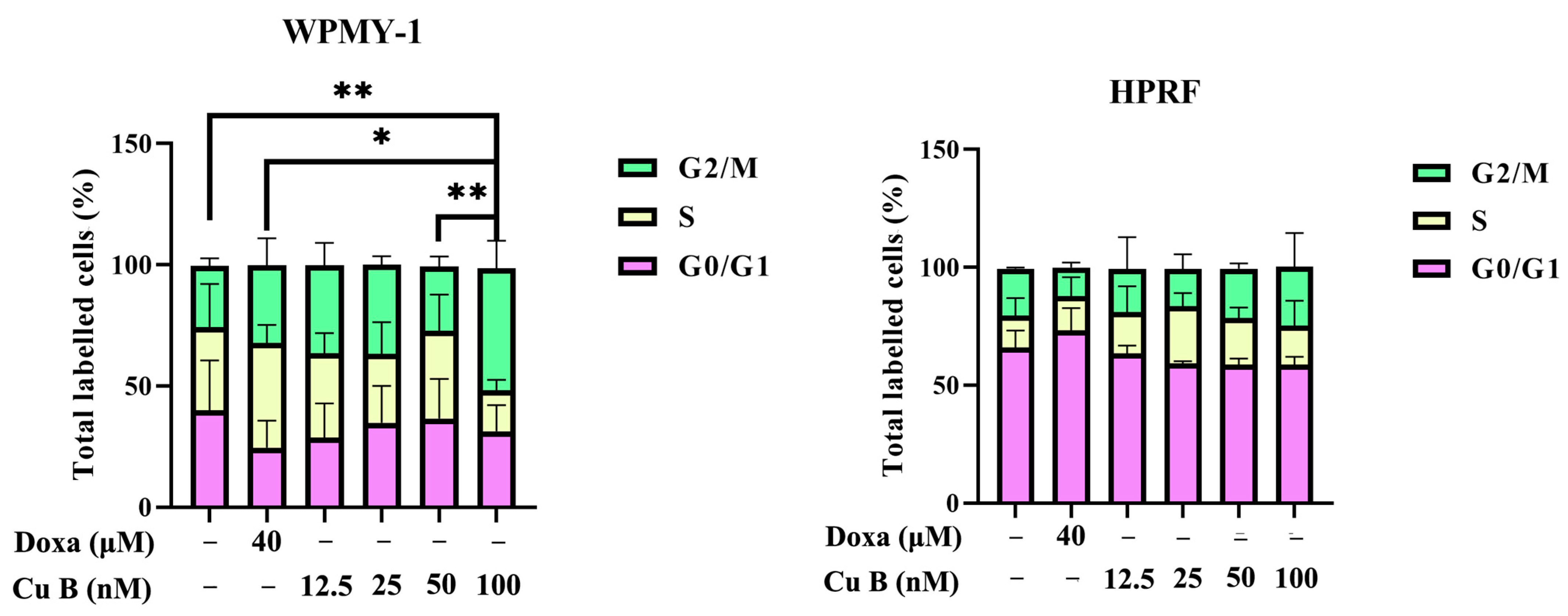
2.3. Cu B Caused G2/M Cell Cycle Arrest in WPMY-1 Cells
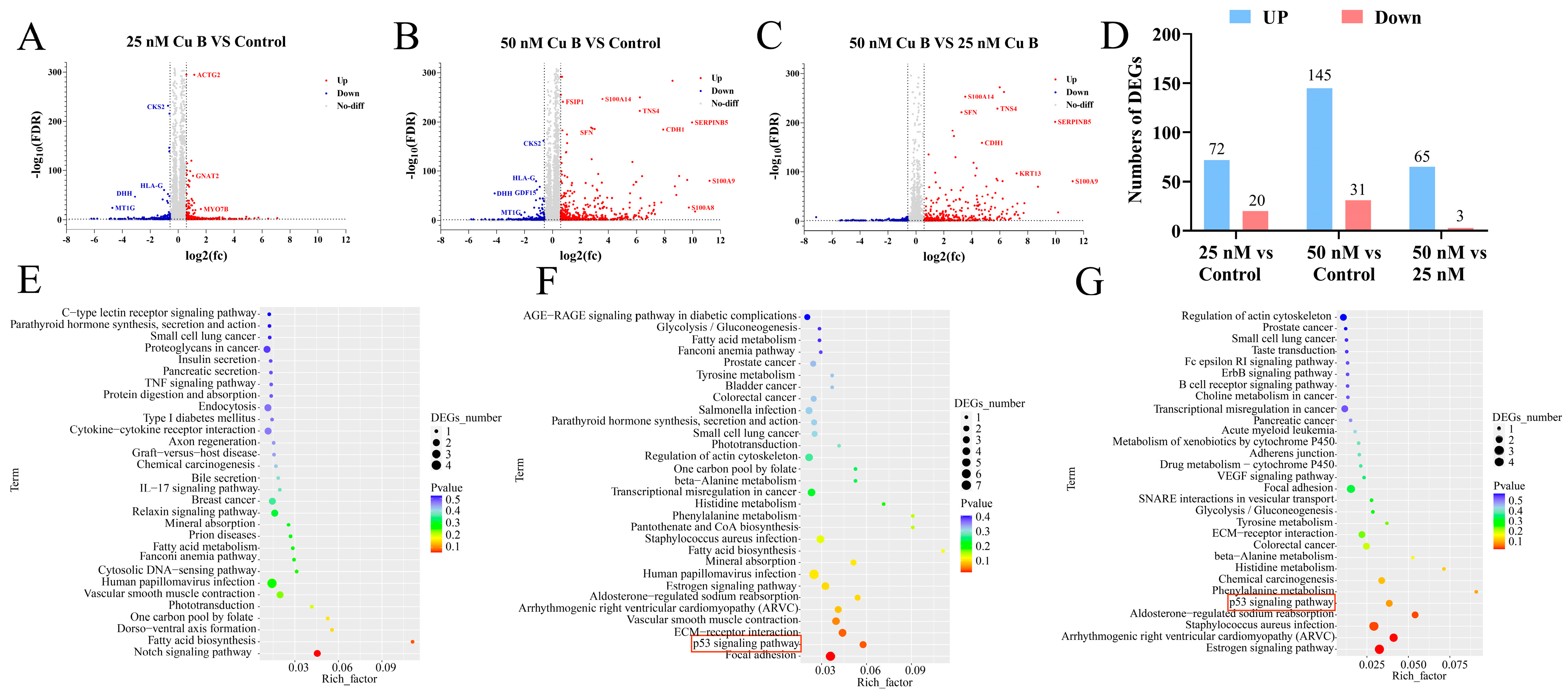
2.4. Cu B Affected p53 Signaling Pathway in WPMY-1 Cells
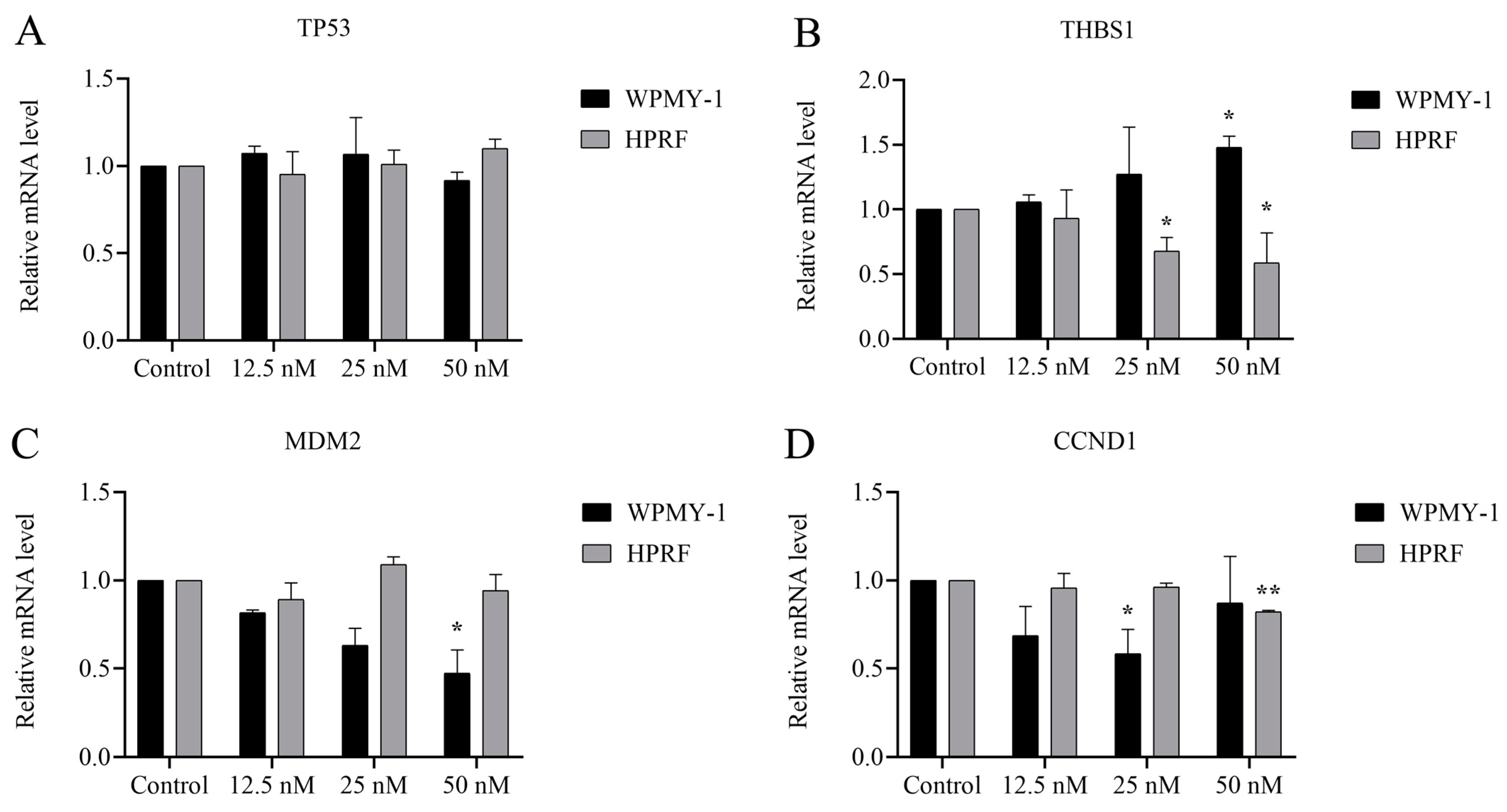
2.5. Cu B Regulated Gene Expression Levels of p53/MDM2 Signaling Axis
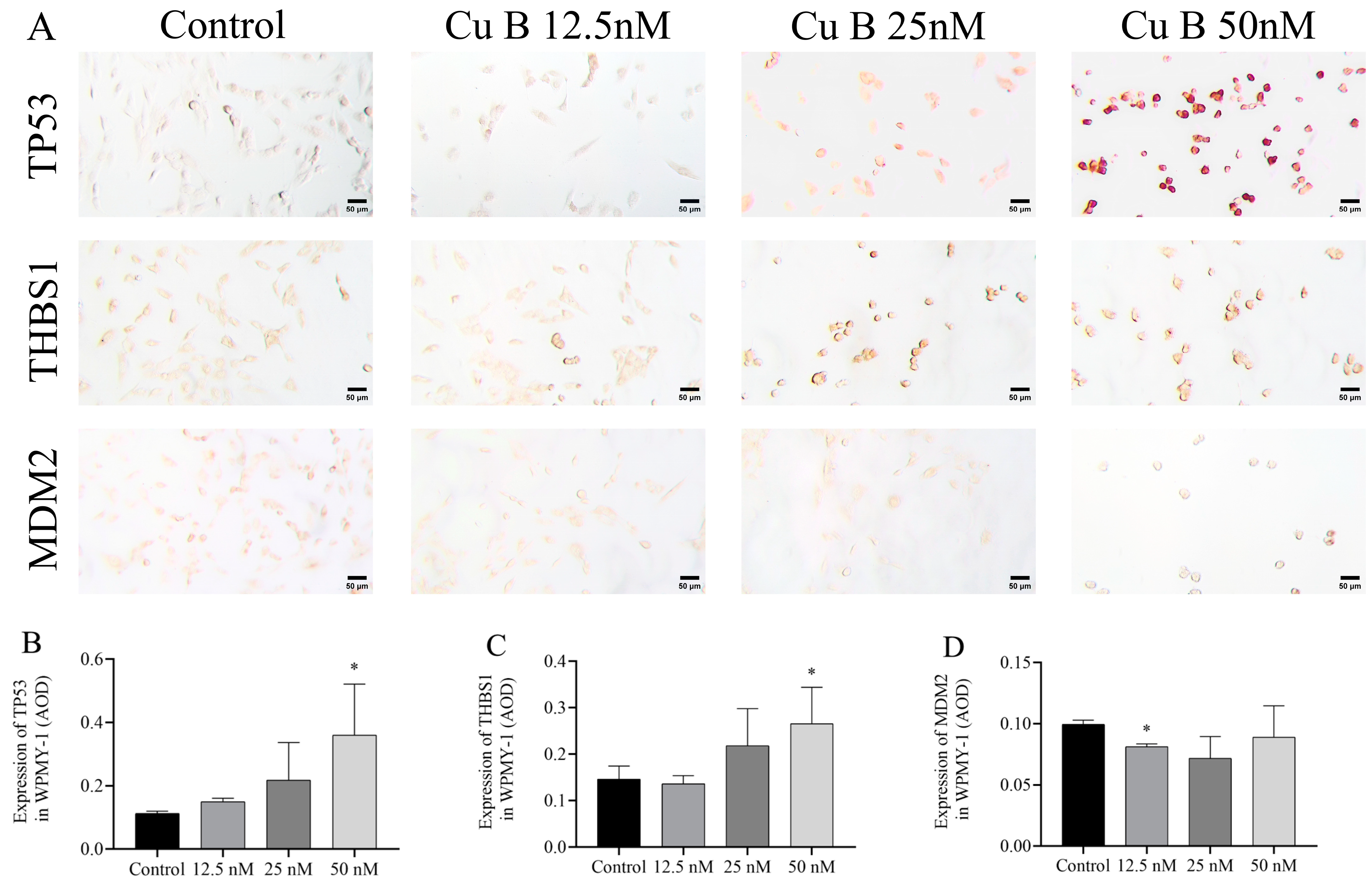
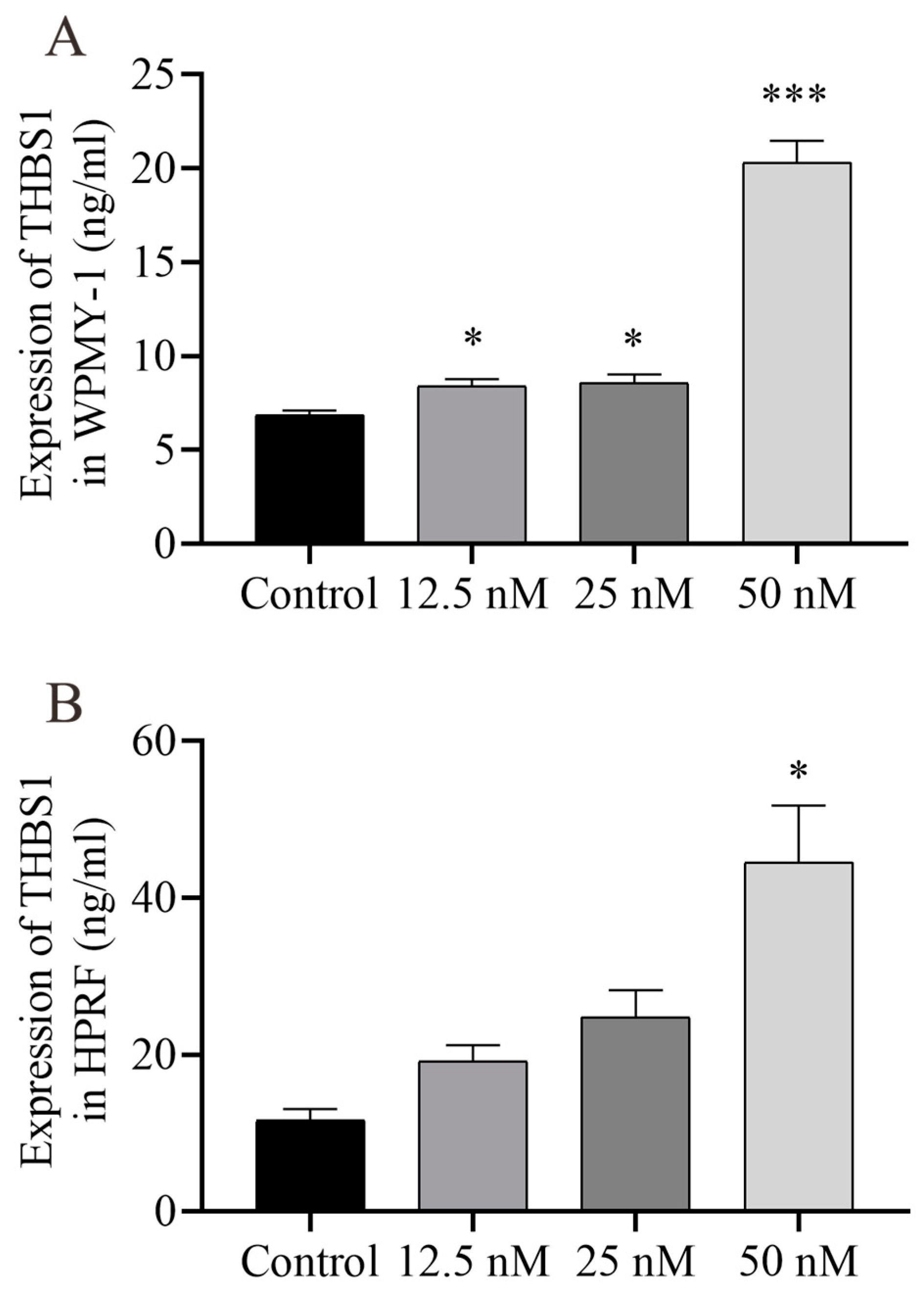
2.6. Cu B Up-Regulated TP53 and THBS1 Protein Expressions and Modulated MDM2 Protein Expression in Prostate Cell
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Cell Viability and Cell Morphology
4.4. Annexin V-FITC/PI Cell Apoptosis Detection
4.5. Cell Cycle Assay
4.6. RNA-Sequencing Analysis
4.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.8. Immunocytochemistry
4.9. Enzyme-Linked Immunosorbent Assay (ELISA)
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Flaxman, A.D.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; Aboyans, V.; et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.F.; Liu, G.X.; Guo, Y.S.; Zhu, H.Y.; He, D.L.; Qiao, X.M.; Li, X.H. Global, Regional, and National Incidence and Year Lived with Disability for Benign Prostatic Hyperplasia from 1990 to 2019. Am. J. Men’s Health 2021, 15, 15579883211036786. [Google Scholar] [CrossRef]
- Egan, K.B. The Epidemiology of Benign Prostatic Hyperplasia Associated with Lower Urinary Tract Symptoms: Prevalence and Incident Rates. Urol. Clin. N. Am. 2016, 43, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Steers, W.D. 5alpha-reductase activity in the prostate. Urology 2001, 58 (Suppl. 1), 17–24, discussion 24. [Google Scholar] [CrossRef] [PubMed]
- Hennenberg, M.; Stief, C.G.; Gratzke, C. Prostatic alpha1-adrenoceptors: New concepts of function, regulation, and intracellular signaling. Neurourol. Urodyn. 2014, 33, 1074–1085. [Google Scholar] [CrossRef]
- Sandhu, J.S. Therapeutic options in the treatment of benign prostatic hyperplasia. Patient Prefer. Adherence 2009, 3, 213–223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Baker, D.D.; Chu, M.; Oza, U.; Rajgarhia, V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–1244. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, L.; Wang, H.; Xiong, Y. Recent Advances in Antiviral Activities of Triterpenoids. Pharmaceuticals 2022, 15, 1169. [Google Scholar] [CrossRef]
- Ren, Y.; Kinghorn, A.D. Natural Product Triterpenoids and Their Semi-Synthetic Derivatives with Potential Anticancer Activity. Planta Med. 2019, 85, 802–814. [Google Scholar] [CrossRef]
- Wu, N.; Wu, G.C.; Hu, R.; Li, M.; Feng, H. Ginsenoside Rh2 inhibits glioma cell proliferation by targeting microRNA-128. Acta Pharmacol. Sin. 2011, 32, 345–353. [Google Scholar] [CrossRef]
- Kim, S.H.; Ryu, H.G.; Lee, J.; Shin, J.; Harikishore, A.; Jung, H.Y.; Kim, Y.S.; Lyu, H.N.; Oh, E.; Baek, N.I.; et al. Ursolic acid exerts anti-cancer activity by suppressing vaccinia-related kinase 1-mediated damage repair in lung cancer cells. Sci. Rep. 2015, 5, 14570. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Guo, L.; Miao, L.; Bao, W.; Yang, J.; Li, X.; Xi, T.; Zhao, W. Ursolic acid inhibits epithelial-mesenchymal transition by suppressing the expression of astrocyte-elevated gene-1 in human nonsmall cell lung cancer A549 cells. Anticancer Drugs 2013, 24, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhao, S.; Tang, Q.; Zheng, F.; Chen, Y.; Yang, L.; Yang, X.; Li, L.; Wu, W.; Hann, S.S. Activation of SAPK/JNK mediated the inhibition and reciprocal interaction of DNA methyltransferase 1 and EZH2 by ursolic acid in human lung cancer cells. J. Exp. Clin. Cancer Res. 2015, 34, 99. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Wang, C.; Zhao, X.; Ma, C.; Fu, K.; Liu, Y.; Peng, C.; Li, Y. Cucurbitacin B: A review of its pharmacology, toxicity, and pharmacokinetics. Pharmacol. Res. 2023, 187, 106587. [Google Scholar] [CrossRef] [PubMed]
- Alafnan, A.; Alamri, A.; Hussain, T.; Rizvi, S.M.D. Cucurbitacin-B Exerts Anticancer Effects through Instigation of Apoptosis and Cell Cycle Arrest within Human Prostate Cancer PC3 Cells via Downregulating JAK/STAT Signaling Cascade. Pharmaceuticals 2022, 15, 1229. [Google Scholar] [CrossRef]
- Gao, Y.; Islam, M.S.; Tian, J.; Lui, V.W.; Xiao, D. Inactivation of ATP citrate lyase by Cucurbitacin B: A bioactive compound from cucumber, inhibits prostate cancer growth. Cancer Lett. 2014, 349, 15–25. [Google Scholar] [CrossRef]
- Zhou, P.; Huang, S.; Shao, C.; Huang, D.; Hu, Y.; Su, X.; Yang, R.; Jiang, J.; Wu, J. The Antiproliferative and Proapoptotic Effects of Cucurbitacin B on BPH-1 Cells via the p53/MDM2 Axis. Int. J. Mol. Sci. 2023, 25, 442. [Google Scholar] [CrossRef]
- Shapiro, E.; Becich, M.J.; Hartanto, V.; Lepor, H. The Relative Proportion of Stromal and Epithelial Hyperplasia Is Related To the Development of Symptomatic Benign Prostate Hyperplasia. J. Urology 1992, 147, 1293–1297. [Google Scholar] [CrossRef]
- Siejka, A.; Schally, A.V.; Barabutis, N. The effect of LHRH antagonist cetrorelix in crossover conditioned media from epithelial (BPH-1) and stromal (WPMY-1) prostate cells. Horm. Metab. Res. 2014, 46, 21–26. [Google Scholar] [CrossRef]
- Webber, M.M.; Trakul, N.; Thraves, P.S.; Bello-DeOcampo, D.; Chu, W.W.; Storto, P.D.; Huard, T.K.; Rhim, J.S.; Williams, D.E. A human prostatic stromal myofibroblast cell line WPMY-1: A model for stromal-epithelial interactions in prostatic neoplasia. Carcinogenesis 1999, 20, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Zemskova, M.Y.; Song, J.H.; Cen, B.; Cerda-Infante, J.; Montecinos, V.P.; Kraft, A.S. Regulation of prostate stromal fibroblasts by the PIM1 protein kinase. Cell. Signal. 2015, 27, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Song, J.; Lee, J.; Moon, S.K.; Moon, B. Resveratrol Attenuates the Proliferation of Prostatic Stromal Cells in Benign Prostatic Hyperplasia by Regulating Cell Cycle Progression, Apoptosis, Signaling Pathways, BPH Markers, and NF-κB Activity. Int. J. Mol. Sci. 2021, 22, 5969. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.Y.; Jiang, C.Y.; Dai, C.Y.; Zhao, R.Z.; Wang, X.J.; Zhu, Y.P.; Qian, Y.X.; Yin, F.L.; Fu, X.Y.; Jing, Y.F.; et al. 5-ARI induces autophagy of prostate epithelial cells through suppressing IGF-1 expression in prostate fibroblasts. Cell Prolif. 2019, 52, e12590. [Google Scholar] [CrossRef]
- Wang, Z.L.; Strasser, A.; Kelly, G.L. Should mutant TP53 be targeted for cancer therapy? Cell Death Differ. 2022, 29, 911–920. [Google Scholar] [CrossRef]
- Mircetic, J.; Dietrich, A.; Paszkowski-Rogacz, M.; Krause, M.; Buchholz, F. Development of a genetic sensor that eliminates p53 deficient cells. Nat. Commun. 2017, 8, 1463. [Google Scholar] [CrossRef]
- Jain, A.K.; Barton, M.C. p53: Emerging roles in stem cells, development and beyond. Development 2018, 145, dev158360. [Google Scholar] [CrossRef]
- Mendoza, M.; Mandani, G.; Momand, J. The MDM2 gene family. Biomol. Concepts 2014, 5, 9–19. [Google Scholar] [CrossRef]
- Yu, Q.; Li, Y.; Mu, K.; Li, Z.; Meng, Q.; Wu, X.; Wang, Y.; Li, L. Amplification of Mdmx and overexpression of MDM2 contribute to mammary carcinogenesis by substituting for p53 mutations. Diagn. Pathol. 2014, 9, 71. [Google Scholar] [CrossRef]
- Dong, M.; Ma, G.; Tu, W.; Guo, K.J.; Tian, Y.L.; Dong, Y.T. Clinicopathological significance of p53 and mdm2 protein expression in human pancreatic cancer. World J. Gastroenterol. 2005, 11, 2162–2165. [Google Scholar] [CrossRef]
- Schmitz-Drager, B.J.; Kushima, M.; Goebell, P.; Jax, T.W.; Gerharz, C.D.; Bultel, H.; Schulz, W.A.; Ebert, T.; Ackermann, R. p53 and MDM2 in the development and progression of bladder cancer. Eur. Urol. 1997, 32, 487–493. [Google Scholar]
- Wade, M.; Wang, Y.V.; Wahl, G.M. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010, 20, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Mairinger, F.D.; Walter, R.F.H.; Ting, S.; Vollbrecht, C.; Kollmeier, J.; Griff, S.; Hager, T.; Mairinger, T.; Christoph, D.C.; Theegarten, D.; et al. Mdm2 protein expression is strongly associated with survival in malignant pleural mesothelioma. Future Oncol. 2014, 10, 995–1005. [Google Scholar] [CrossRef]
- Hou, H.; Sun, D.; Zhang, X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 2019, 19, 216. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Zong, C.S.; Xia, W.; Wei, Y.; Ali-Seyed, M.; Li, Z.; Broglio, K.; Berry, D.A.; Hung, M.C. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell Biol. 2006, 26, 7269–7282. [Google Scholar] [CrossRef]
- Ding, H.; Zhao, J.; Zhang, Y.; Yu, J.; Liu, M.; Li, X.; Xu, L.; Lin, M.; Liu, C.; He, Z.; et al. Systematic Analysis of Drug Vulnerabilities Conferred by Tumor Suppressor Loss. Cell Rep. 2019, 27, 3331–3344.e6. [Google Scholar] [CrossRef]
- Slabakova, E.; Kharaishvili, G.; Smejova, M.; Pernicova, Z.; Suchankova, T.; Remsik, J.; Lerch, S.; Strakova, N.; Bouchal, J.; Kral, M.; et al. Opposite regulation of MDM2 and MDMX expression in acquisition of mesenchymal phenotype in benign and cancer cells. Oncotarget 2015, 6, 36156–36171. [Google Scholar] [CrossRef]
- Dang, J.; Kuo, M.L.; Eischen, C.M.; Stepanova, L.; Sherr, C.J.; Roussel, M.F. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002, 62, 1222–1230. [Google Scholar]
- Brown, D.R.; Thomas, C.A.; Deb, S.P. The human oncoprotein MDM2 arrests the cell cycle: Elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998, 17, 2513–2525. [Google Scholar] [PubMed]
- Cohen, A.A.; Geva-Zatorsky, N.; Eden, E.; Frenkel-Morgenstern, M.; Issaeva, I.; Sigal, A.; Milo, R.; Cohen-Saidon, C.; Liron, Y.; Kam, Z.; et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 2008, 322, 1511–1516. [Google Scholar] [CrossRef]
- Kaur, S.; Bronson, S.M.; Pal-Nath, D.; Miller, T.W.; Soto-Pantoja, D.R.; Roberts, D.D. Functions of Thrombospondin-1 in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 4570. [Google Scholar] [CrossRef]
- Li, Q.; Ahuja, N.; Burger, P.C.; Issa, J.P. Methylation and silencing of the Thrombospondin-1 promoter in human cancer. Oncogene 1999, 18, 3284–3289. [Google Scholar] [CrossRef] [PubMed]
- Omatsu, M.; Nakanishi, Y.; Iwane, K.; Aoyama, N.; Duran, A.; Muta, Y.; Martinez-Ordonez, A.; Han, Q.; Agatsuma, N.; Mizukoshi, K.; et al. THBS1-producing tumor-infiltrating monocyte-like cells contribute to immunosuppression and metastasis in colorectal cancer. Nat. Commun. 2023, 14, 5534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, D.; Zhou, T.; Song, H.; Hulsurkar, M.; Su, N.; Liu, Y.; Wang, Z.; Shao, L.; Ittmann, M.; et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 2018, 9, 4080. [Google Scholar] [CrossRef] [PubMed]
- Hulsurkar, M.; Li, Z.; Zhang, Y.; Li, X.; Zheng, D.; Li, W. Beta-adrenergic signaling promotes tumor angiogenesis and prostate cancer progression through HDAC2-mediated suppression of thrombospondin-1. Oncogene 2017, 36, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Maitisha, G.; Aimaiti, M.; An, Z.; Li, X. Allicin induces cell cycle arrest and apoptosis of breast cancer cells in vitro via modulating the p53 pathway. Mol. Biol. Rep. 2021, 48, 7261–7272. [Google Scholar] [CrossRef]
- Tchakarska, G.; Sola, B. The double dealing of cyclin D1. Cell Cycle 2020, 19, 163–178. [Google Scholar] [CrossRef]
- Tanyi, J.; Tory, K.; Bankfalvi, A.; Shroder, W.; Rath, W.; Fuzesi, L. Analysis of p53 mutation and cyclin D1 expression in breast tumors. Pathol. Oncol. Res. 1999, 5, 90–94. [Google Scholar] [CrossRef]
- Nakashima, T.; Clayman, G.L. Antisense inhibition of cyclin D1 in human head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2000, 126, 957–961. [Google Scholar] [CrossRef][Green Version]
- Xie, M.; Zhao, F.; Zou, X.; Jin, S.; Xiong, S. The association between CCND1 G870A polymorphism and colorectal cancer risk: A meta-analysis. Medicine 2017, 96, e8269. [Google Scholar] [CrossRef]
- Gumbiner, L.M.; Gumerlock, P.H.; Mack, P.C.; Chi, S.G.; White, R.W.D.; Mohler, J.L.; Pretlow, T.G.; Tricoli, J.V. Overexpression of cyclin D1 is rare in human prostate carcinoma. Prostate 1999, 38, 40–45. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, Q.; Cui, Y.; Liu, Z.Y.; Zhao, W.; Wang, C.L.; Dong, Y.; Hou, L.J.; Hu, G.H.; Luo, C.; et al. Knockdown of cyclin D1 inhibits proliferation, induces apoptosis, and attenuates the invasive capacity of human glioblastoma cells. J. Neuro-Oncol. 2012, 106, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Guardavaccaro, D.; Corrente, G.; Covone, F.; Micheli, L.; D’Agnano, I.; Starace, G.; Caruso, M.; Tirone, F. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol. Cell Biol. 2000, 20, 1797–1815. [Google Scholar] [CrossRef]
- Cheon, S.Y.; Jin, B.R.; Kim, H.J.; An, H.J. Oleanolic Acid Ameliorates Benign Prostatic Hyperplasia by Regulating PCNA-Dependent Cell Cycle Progression In Vivo and In Vitro. J. Nat. Prod. 2020, 83, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.R.; Cheon, S.Y.; Kim, H.J.; Kim, M.S.; Lee, K.H.; An, H.J. Anti-Proliferative Effects of Standardized Cornus officinalis on Benign Prostatic Epithelial Cells via the PCNA/E2F1-Dependent Cell Cycle Pathway. Int. J. Mol. Sci. 2020, 21, 9567. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, M.; Wei, H.; Chen, Y. Targeting p53 pathways: Mechanisms, structures, and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 92. [Google Scholar] [CrossRef]
- Wei, J.; Chen, X.; Li, Y.; Li, R.; Bao, K.; Liao, L.; Xie, Y.; Yang, T.; Zhu, J.; Mao, F.; et al. Cucurbitacin B-induced G2/M cell cycle arrest of conjunctival melanoma cells mediated by GRP78-FOXM1-KIF20A pathway. Acta Pharm. Sin. B 2022, 12, 3861–3876. [Google Scholar] [CrossRef]
- Li, Q.Z.; Chen, Y.Y.; Liu, Q.P.; Feng, Z.H.; Zhang, L.; Zhang, H. Cucurbitacin B suppresses hepatocellular carcinoma progression through inducing DNA damage-dependent cell cycle arrest. Phytomedicine 2024, 126, 155177. [Google Scholar] [CrossRef]
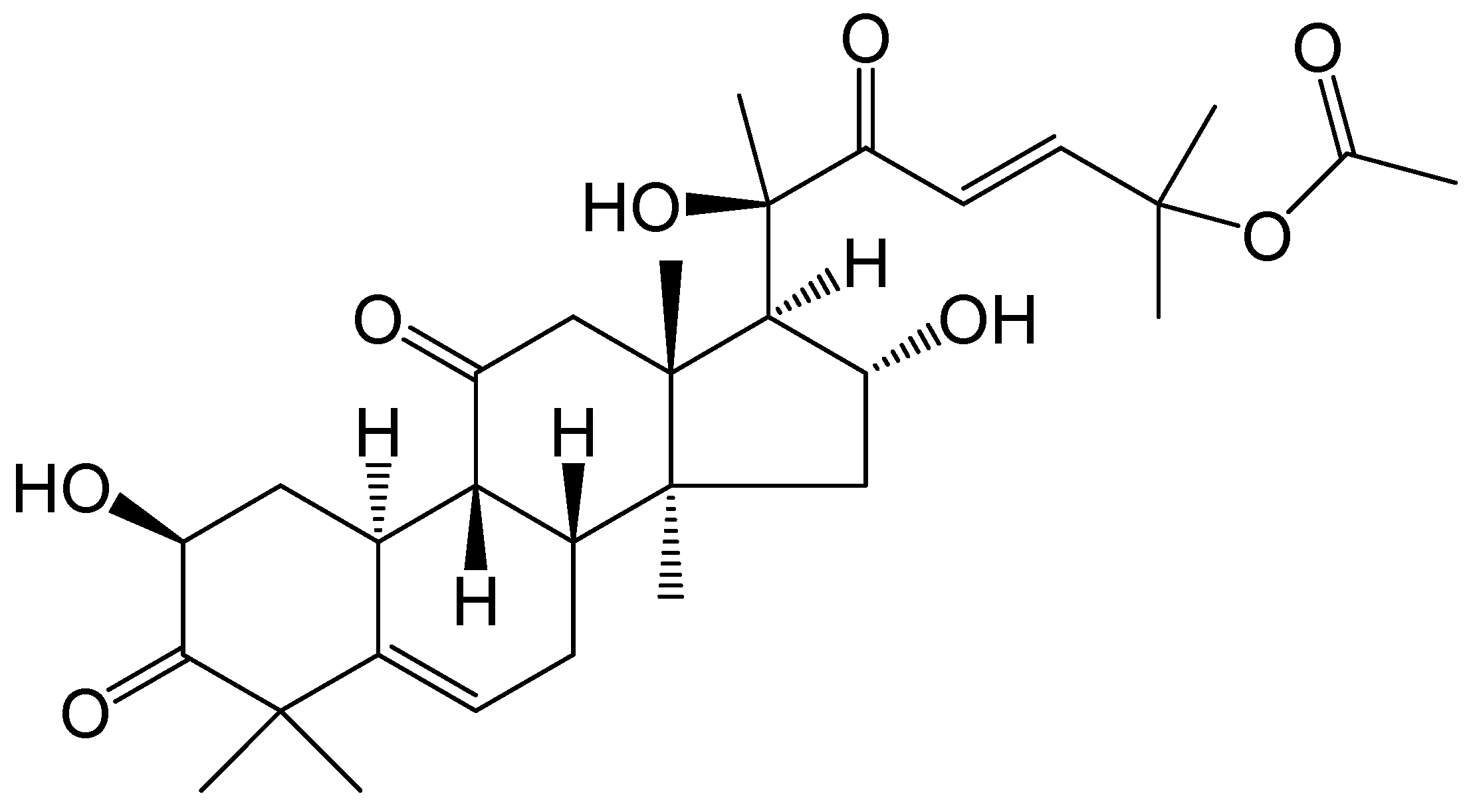
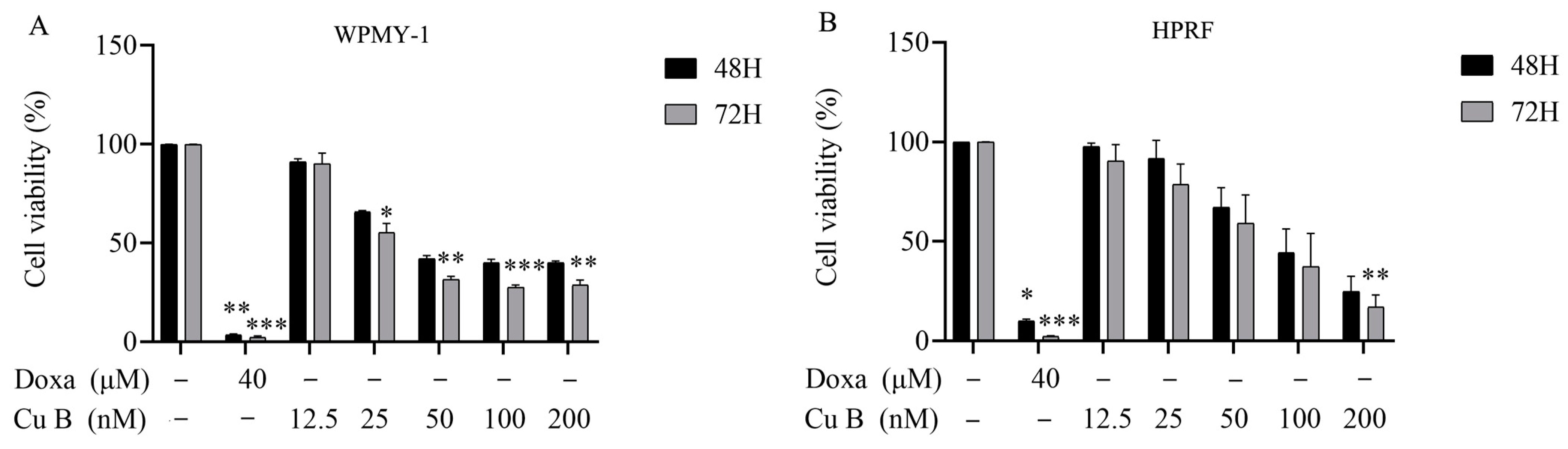

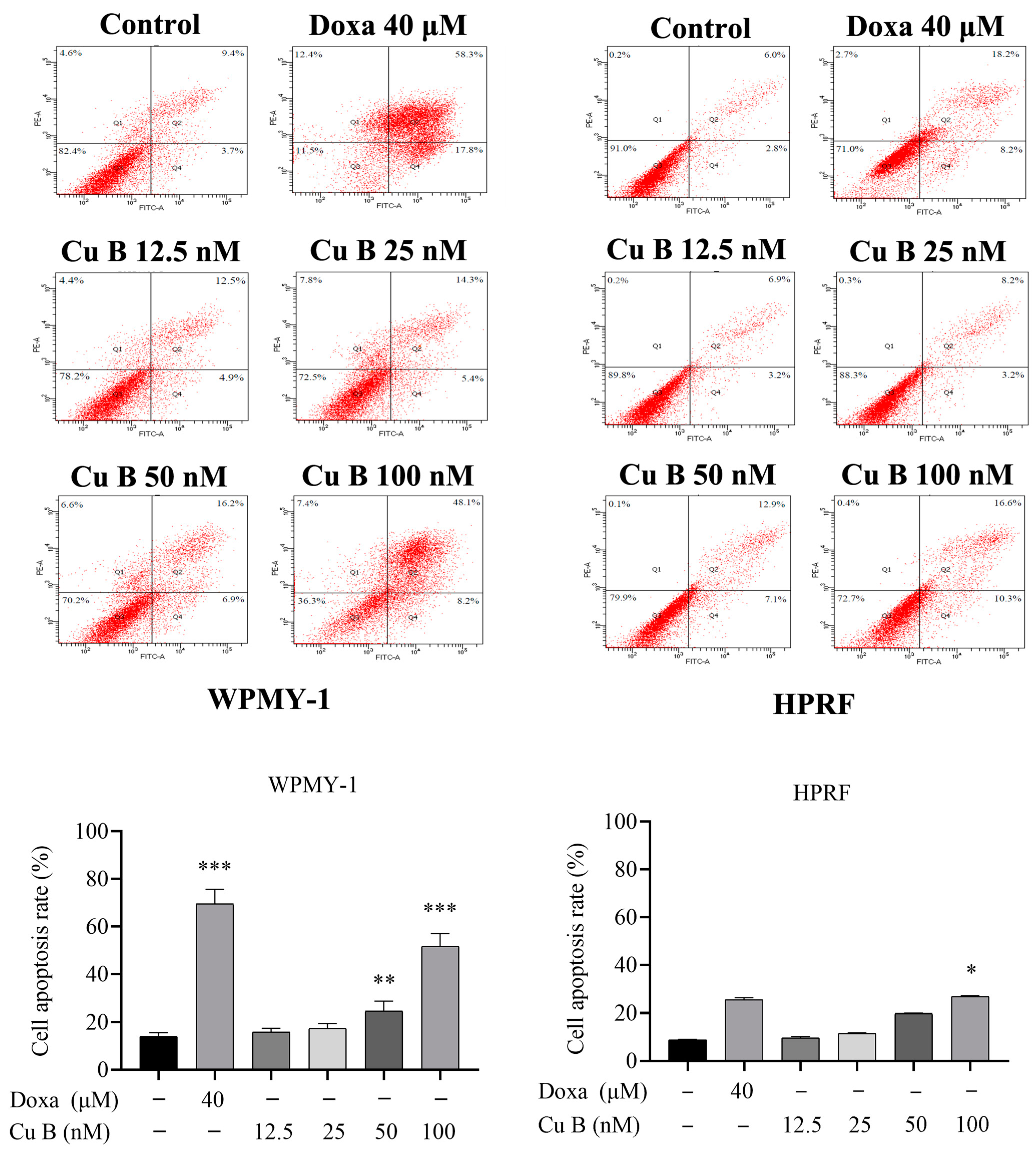

| Cell Types | IC50 (48 h) | IC50 (72 h) |
|---|---|---|
| WPMY-1 | 66.42 nM | 38.84 nM |
| HPRF | 88.94 nM | 65.98 nM |
| Genes | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| TP53 | CACTAAGCGAGCACTGCCCAACA | GCCTCATTCAGCTCTCGGAACATCT |
| MDM2 | TTGGCGTGCCAAGCTTCTCTGTG | ACCTGAGTCCGATGATTCCTGCTGA |
| THBS1 | ATGGAGAATGCTGTCCTCGCTGTTG | CGGTTGTTGAGGCTATCGCAGGAG |
| CCND1 | CGCCCTCGGTGTCCTACTTCAAATG | AGACCTCCTCCTCGCACTTCTGTTC |
| GAPDH | CAGGAGGCATTGCTGATGAT | GAAGGCTGGGGCTCATTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Zhou, P.; Huang, S.; Shao, C.; Huang, D.; Su, X.; Yang, R.; Jiang, J.; Wu, J. Cucurbitacin B Inhibits the Proliferation of WPMY-1 Cells and HPRF Cells via the p53/MDM2 Axis. Int. J. Mol. Sci. 2024, 25, 9333. https://doi.org/10.3390/ijms25179333
Jin Y, Zhou P, Huang S, Shao C, Huang D, Su X, Yang R, Jiang J, Wu J. Cucurbitacin B Inhibits the Proliferation of WPMY-1 Cells and HPRF Cells via the p53/MDM2 Axis. International Journal of Molecular Sciences. 2024; 25(17):9333. https://doi.org/10.3390/ijms25179333
Chicago/Turabian StyleJin, Yangtao, Ping Zhou, Sisi Huang, Congcong Shao, Dongyan Huang, Xin Su, Rongfu Yang, Juan Jiang, and Jianhui Wu. 2024. "Cucurbitacin B Inhibits the Proliferation of WPMY-1 Cells and HPRF Cells via the p53/MDM2 Axis" International Journal of Molecular Sciences 25, no. 17: 9333. https://doi.org/10.3390/ijms25179333
APA StyleJin, Y., Zhou, P., Huang, S., Shao, C., Huang, D., Su, X., Yang, R., Jiang, J., & Wu, J. (2024). Cucurbitacin B Inhibits the Proliferation of WPMY-1 Cells and HPRF Cells via the p53/MDM2 Axis. International Journal of Molecular Sciences, 25(17), 9333. https://doi.org/10.3390/ijms25179333






