Conjugation of CRAMP18–35 Peptide to Chitosan and Hydroxypropyl Chitosan via Copper-Catalyzed Azide–Alkyne Cycloaddition and Investigation of Antibacterial Activity
Abstract
:1. Introduction
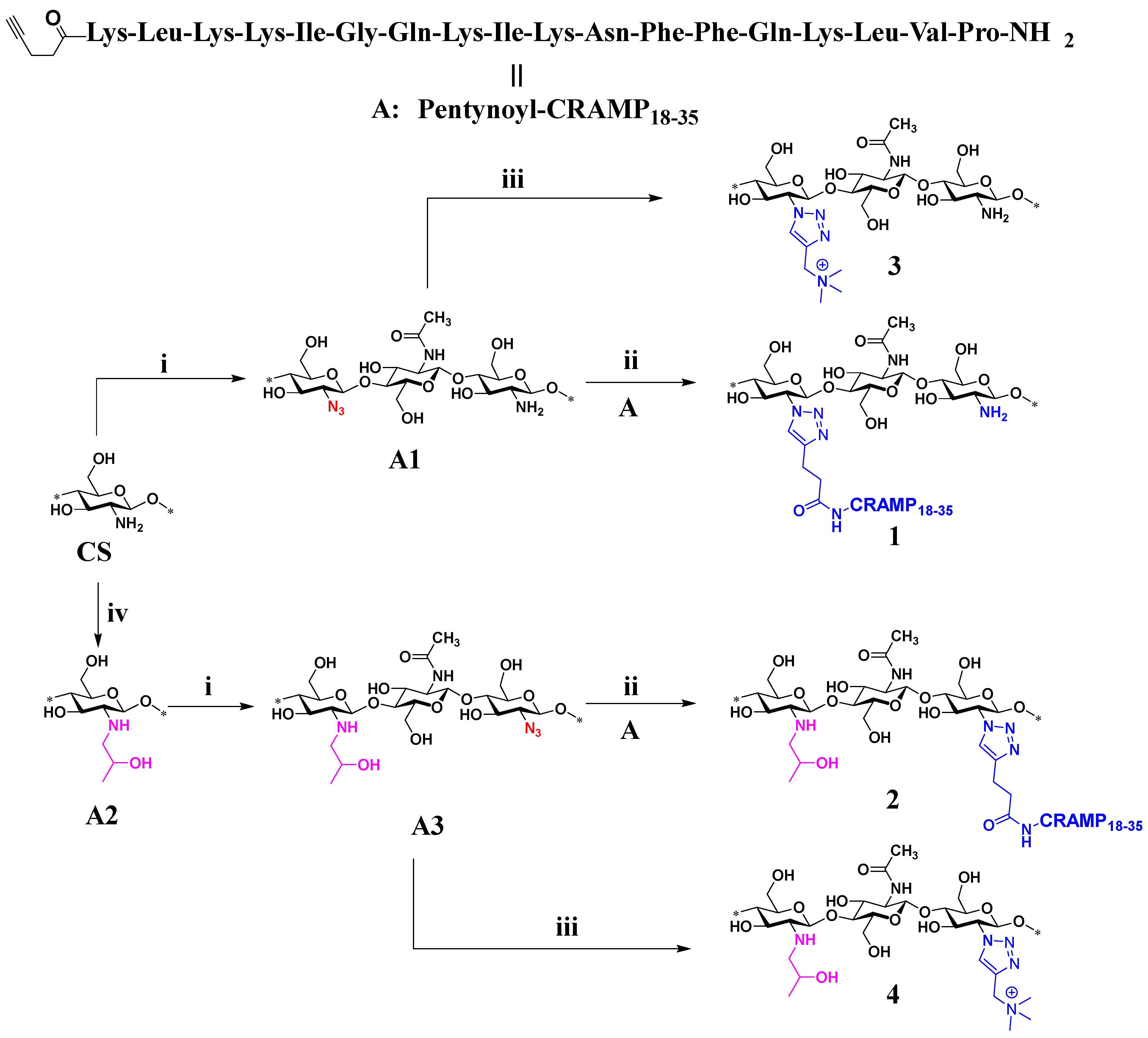
2. Results and Discussion
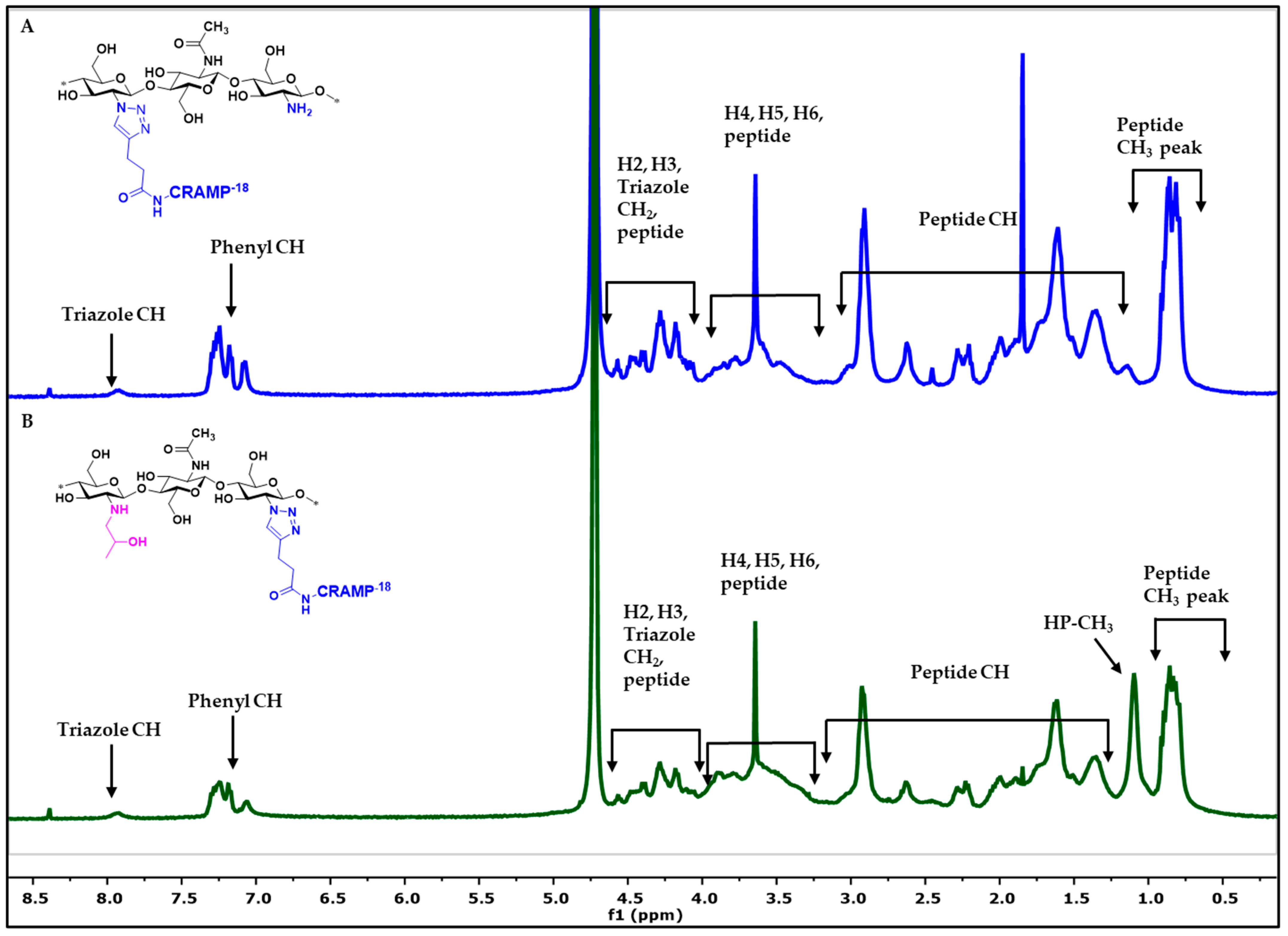
2.1. Characterization of Products by IR Spectroscopy
2.2. Characterization of Products by NMR Spectroscopy
2.3. Antibacterial Analysis of Antimicrobial Peptide Chitosan Derivatives
3. Materials and Methods
3.1. Material
3.1.1. Synthesis of Antimicrobial Peptide CRAMP18–35 (A)
3.1.2. Preparation of Chitosan Azide (A1)
3.1.3. Synthesis of Chitosan–CRAMP18–35 Peptide Conjugation via CuAAC Reaction (1)
3.1.4. Synthesis of N-Hydroxypropyl Chitosan (HPC) (A2)
3.1.5. Preparation of N-Hydroxypropyl Chitosan Azide (HPC-N3) (A3)
3.1.6. Synthesis of HPC-CRAMP18–35 Peptide Conjugation via CuAAC Reaction (2)
3.1.7. Synthesis of Chitotriazolan (3)
3.1.8. Synthesis of HP-Chitotriazolan (4)
3.2. Characterization
3.2.1. NMR and FTIR Spectroscopy
3.2.2. Gel Permeation Chromatography
3.3. Antibacterial Assay for the Antimicrobial Chitosan Peptide Derivatives
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E.W. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Song, Y. Mechanism of antimicrobial peptides: Antimicrobial, anti-inflammatory and antibiofilm activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, K.G.N.; Rodrigues, G.; Monges, B.E.D.; Cardoso, M.H.; Franco, O.L. Bioactive peptides against fungal biofilms. Front. Microbiol. 2019, 10, 2169. [Google Scholar] [CrossRef]
- Buda De Cesare, G.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial peptides: A new frontier in antifungal therapy. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef]
- Talapko, J.; Meštrović, T.; Juzbašić, M.; Tomas, M.; Erić, S.; Horvat Aleksijević, L.; Bekić, S.; Schwarz, D.; Matić, S.; Neuberg, M.; et al. Antimicrobial peptides-mechanisms of action, antimicrobial effects and clinical applications. Antibiotics 2022, 11, 1417. [Google Scholar] [CrossRef]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, B.; Davis, E.G.; Ross, C.R.; Blecha, F. Cathelicidins: Microbicidal activity, mechanisms of action, and roles in innate immunity. Microbes Infect. 2002, 4, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.-P.S.; Hancock, R.E.W. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef]
- Gallo, R.L.; Kim, K.J.; Bernfield, M.; Kozak, C.A.; Zanetti, M.; Merluzzi, L.; Gennaro, R. Identification of cramp, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J. Biol. Chem. 1997, 272, 13088–13093. [Google Scholar] [CrossRef] [PubMed]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef]
- Shin, S.Y.; Kang, S.W.; Lee, D.G.; Eom, S.H.; Song, W.K.; Kim, J.I. Cramp analogues having potent antibiotic activity against bacterial, fungal, and tumor cells without hemolytic activity. Biochem. Biophys. Res. Commun. 2000, 275, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wubbolts, R.W.; Haagsman, H.P.; Veldhuizen, E.J.A. Inhibition and eradication of pseudomonas aeruginosa biofilms by host defence peptides. Sci. Rep. 2018, 8, 10446. [Google Scholar] [CrossRef]
- Ha, J.M.; Shin, S.Y.; Kang, S.W. Synthesis and antibiotic activities of cramp, a cathelin-related antimicrobial peptide and its fragments. Bull. Korean Chem. Soc. 1999, 20, 1073–1077. [Google Scholar] [CrossRef]
- Ray, S.; Dhaked, H.P.; Panda, D. Antimicrobial peptide cramp (16–33) stalls bacterial cytokinesis by inhibiting ftsz assembly. Biochemistry 2014, 53, 6426–6429. [Google Scholar] [CrossRef]
- De Brucker, K.; Delattin, N.; Robijns, S.; Steenackers, H.; Verstraeten, N.; Landuyt, B.; Luyten, W.; Schoofs, L.; Dovgan, B.; Fröhlich, M.; et al. Derivatives of the mouse cathelicidin-related antimicrobial peptide (cramp) inhibit fungal and bacterial biofilm formation. Antimicrob. Agents Chemother. 2014, 58, 5395–5404. [Google Scholar] [CrossRef] [PubMed]
- Winderickx, S.; De Brucker, K.; Bird, M.J.; Windmolders, P.; Meert, E.; Cammue, B.P.A.; Thevissen, K. Structure-activity relationship study of the antimicrobial cramp-derived peptide cramp20-33. Peptides 2018, 109, 33–38. [Google Scholar] [CrossRef]
- Raheem, N.; Straus, S.K. Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Reassessing the host defense peptide landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef]
- Riool, M.; de Breij, A.; Drijfhout, J.W.; Nibbering, P.H.; Zaat, S.A.J. Antimicrobial peptides in biomedical device manufacturing. Front. Chem. 2017, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, C.; Kumar, A.; Lang, A.; Ohlenschläger, O. Cysteines and disulfide bonds as structure-forming units: Insights from different domains of life and the potential for characterization by nmr. Front. Chem. 2020, 8, 280. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Devaraj, N.K.; Finn, M.G. Introduction: Click chemistry. Chem. Rev. 2021, 121, 6697–6698. [Google Scholar] [CrossRef]
- Geng, Z.; Shin, J.J.; Xi, Y.; Hawker, C.J. Click chemistry strategies for the accelerated synthesis of functional macromolecules. J. Polym. Sci. 2021, 59, 963–1042. [Google Scholar] [CrossRef]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef]
- Rabea, E.I.; Badawy, M.E.T.; Stevens, C.V.; Smagghe, G.; Steurbaut, W. Chitosan as antimicrobial agent: Applications and mode of action. Biomacromolecules 2003, 4, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Dash, M.; Chiellini, F.; Ottenbrite, R.M.; Chiellini, E. Chitosan—A versatile semi-synthetic polymer in biomedical applications. Prog. Polym. Sci. 2011, 36, 981–1014. [Google Scholar] [CrossRef]
- Kong, M.; Chen, X.G.; Xing, K.; Park, H.J. Antimicrobial properties of chitosan and mode of action: A state of the art review. Int. J. Food Microbiol. 2010, 144, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, S.; Ólafsdóttir, S.; Jónsdóttir, S.; Hjálmarsdóttir, M.; Másson, M. Selective synthesis of n,n,n-trimethylated chitosan derivatives at different degree of substitution and investigation of structure-activity relationship for activity against p. Aeruginosa and mrsa. Int. J. Biol. Macromol. 2020, 160, 548–557. [Google Scholar] [CrossRef]
- Rathinam, S.; Solodova, S.; Kristjánsdóttir, I.; Hjálmarsdóttir, M.Á.; Másson, M. The antibacterial structure-activity relationship for common chitosan derivatives. Int. J. Biol. Macromol. 2020, 165, 1686–1693. [Google Scholar] [CrossRef]
- Kean, T.; Roth, S.; Thanou, M. Trimethylated chitosans as non-viral gene delivery vectors: Cytotoxicity and transfection efficiency. J. Control. Release 2005, 103, 643–653. [Google Scholar] [CrossRef]
- Benediktsdóttir, B.E.; Baldursson, Ó.; Másson, M. Challenges in evaluation of chitosan and trimethylated chitosan (tmc) as mucosal permeation enhancers: From synthesis to in vitro application. J. Control. Release 2014, 173, 18–31. [Google Scholar] [CrossRef]
- Croisier, F.; Jérôme, C. Chitosan-based biomaterials for tissue engineering. Eur. Polym. J. 2013, 49, 780–792. [Google Scholar] [CrossRef]
- Rathinam, S.; Hjálmarsdóttir, M.Á.; Thygesen, M.B.; Másson, M. Chitotriazolan (poly(β(1-4)-2-(1h-1,2,3-triazol-1-yl)-2-deoxy-d-glucose)) derivatives: Synthesis, characterization, and evaluation of antibacterial activity. Carbohydr. Polym. 2021, 267, 118162. [Google Scholar] [CrossRef]
- Rathinam, S.; Magdadaro, R.; Hjálmarsdóttir, M.Á.; Másson, M. Water-soluble quaternary and protonable basic chitotriazolans: Synthesis by click chemistry conversion of chitosan azides and investigation of antibacterial activity. J. Funct. Biomater. 2024, 15, 63. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Z.; Chen, L.; Gu, W.; Li, Y. Synthesis of 6-n,n,n-trimethyltriazole chitosan via “click chemistry” and evaluation for gene delivery. Biomacromolecules 2009, 10, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Kulbokaite, R.; Ciuta, G.; Netopilik, M.; Makuska, R. N-peg’ylation of chitosan via “click chemistry” reactions. React. Funct. Polym. 2009, 69, 771–778. [Google Scholar] [CrossRef]
- Sahariah, P.; Sørensen, K.K.; Hjálmarsdóttir, M.Á.; Sigurjónsson, Ó.E.; Jensen, K.J.; Másson, M.; Thygesen, M.B. Antimicrobial peptide shows enhanced activity and reduced toxicity upon grafting to chitosan polymers. Chem. Commun. 2015, 51, 11611–11614. [Google Scholar] [CrossRef]
- Barbosa, M.; Vale, N.; Costa, F.M.T.A.; Martins, M.C.L.; Gomes, P. Tethering antimicrobial peptides onto chitosan: Optimization of azide-alkyne “click” reaction conditions. Carbohydr. Polym. 2017, 165, 384–393. [Google Scholar] [CrossRef]
- Barbosa, M.; Costa, F.; Monteiro, C.; Duarte, F.; Martins, M.C.L.; Gomes, P. Antimicrobial coatings prepared from dhvar-5-click-grafted chitosan powders. Acta Biomater. 2019, 84, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, S.; Hjálmarsdóttir, M.Á.; Thygesen, M.B.; Másson, M. Water-soluble chitotriazolans derived from cationic, neutral, and anionic common chitosan derivatives: Synthesis, characterization, and antibacterial activity. Eur. Polym. J. 2023, 196, 112311. [Google Scholar] [CrossRef]
- Hong, V.; Presolski, S.I.; Ma, C.; Finn, M.G. Analysis and optimization of copper-catalyzed azide–alkyne cycloaddition for bioconjugation. Angew. Chem. Int. 2009, 48, 9879–9883. [Google Scholar] [CrossRef]
- Crownover, E.; Duvall, C.L.; Convertine, A.; Hoffman, A.S.; Stayton, P.S. Raft-synthesized graft copolymers that enhance ph-dependent membrane destabilization and protein circulation times. J. Control. Release 2011, 155, 167–174. [Google Scholar] [CrossRef]
- Másson, M. The quantitative molecular weight-antimicrobial activity relationship for chitosan polymers, oligomers, and derivatives. Carbohydr. Polym. 2024, 337, 122159. [Google Scholar] [CrossRef]
- Wikler, M.A.; Cockerill, F.R.; Bush, K.; Dudley, M.N.; Eliopoulos, G.M.; Hardy, D.J.; Hecht, D.W.; Ferraro, M.J.; Swenson, J.M.; Hindler, J.F.; et al. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Eighth Edition Clsi Document m07-a8; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2009; Volume 29. [Google Scholar]

| Derivatives | DS for Triazole CH | DS for Pep-phenyl | DS for Pep-Methyl | DS for HP-Methyl | DS for Triazole TM | DA | MW (Da) | Poly Dispersity Index (D) |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.20 | 0.23 | 0.22 | NA | NA | ND | 1.331 × 105 | 1.08 |
| 2 | 0.13 | 0.12 | 0.12 | 0.65 | NA | ND | 2.931 × 105 | 2.06 |
| 3 | 0.26 | NA | NA | NA | 0.34 | 0.11 | 4.138 × 104 | 1.01 |
| 4 | 0.22 | NA | NA | 0.61 | 0.25 | 0.05 | 1.916 × 105 | 3.59 |
| A2 | NA * | NA | NA | 0.80 | NA | 0.06 | 1.959 × 105 | 1.90 |
| MIC Values in µg/mL | |||||
|---|---|---|---|---|---|
| Chitotriazolan Peptide Derivatives | Structures | S. aureus (ATCC 29213) | E. faecalis (ATCC 29212) | E. coli (ATCC 25922) | P. aeruginosa (ATCC 27853) |
| 1 | 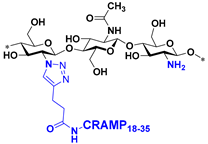 | ≥4096 * | ≥4096 * | 256 | 1024 |
| 2 | 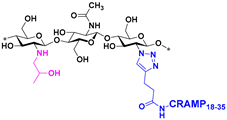 | ≥4096 * | ≥4096 * | 256 | 512 |
| 3 | 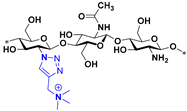 | 1024 | 4096 | 4096 | 4096 |
| 4 | 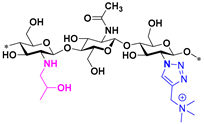 | 4096 | 4096 | 2048 | 4096 |
| A2 | 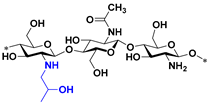 | 4096 | 4096 | 4096 | 4096 |
| A | Pentynoyl–CRAMP18–35 | 128 | 128 | 8 | 64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathinam, S.; Sørensen, K.K.; Hjálmarsdóttir, M.Á.; Thygesen, M.B.; Másson, M. Conjugation of CRAMP18–35 Peptide to Chitosan and Hydroxypropyl Chitosan via Copper-Catalyzed Azide–Alkyne Cycloaddition and Investigation of Antibacterial Activity. Int. J. Mol. Sci. 2024, 25, 9440. https://doi.org/10.3390/ijms25179440
Rathinam S, Sørensen KK, Hjálmarsdóttir MÁ, Thygesen MB, Másson M. Conjugation of CRAMP18–35 Peptide to Chitosan and Hydroxypropyl Chitosan via Copper-Catalyzed Azide–Alkyne Cycloaddition and Investigation of Antibacterial Activity. International Journal of Molecular Sciences. 2024; 25(17):9440. https://doi.org/10.3390/ijms25179440
Chicago/Turabian StyleRathinam, Sankar, Kasper K. Sørensen, Martha Á. Hjálmarsdóttir, Mikkel B. Thygesen, and Már Másson. 2024. "Conjugation of CRAMP18–35 Peptide to Chitosan and Hydroxypropyl Chitosan via Copper-Catalyzed Azide–Alkyne Cycloaddition and Investigation of Antibacterial Activity" International Journal of Molecular Sciences 25, no. 17: 9440. https://doi.org/10.3390/ijms25179440
APA StyleRathinam, S., Sørensen, K. K., Hjálmarsdóttir, M. Á., Thygesen, M. B., & Másson, M. (2024). Conjugation of CRAMP18–35 Peptide to Chitosan and Hydroxypropyl Chitosan via Copper-Catalyzed Azide–Alkyne Cycloaddition and Investigation of Antibacterial Activity. International Journal of Molecular Sciences, 25(17), 9440. https://doi.org/10.3390/ijms25179440







