Hypoxia Abrogates Tumor-Suppressive Activities of C/EBPδ in Pancreatic Cancer
Abstract
1. Introduction
2. Results
2.1. C/EBPδ Limits PDAC Cell Expansion without Exerting Paracrine Effects
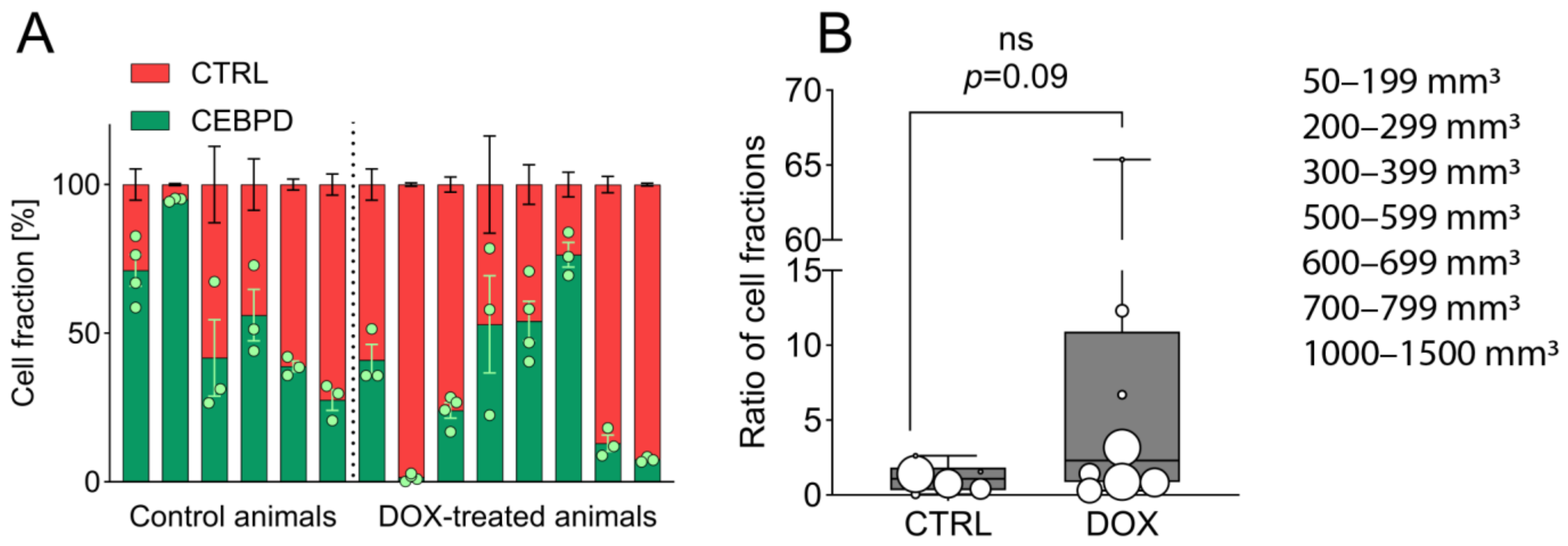
2.2. Growth-Limiting Effects of C/EBPδ Are Diminished In Vivo
2.3. C/EBPδ Is Expressed near Necrotic, HIF-1α-Positive Areas in Subcutaneous Tumors
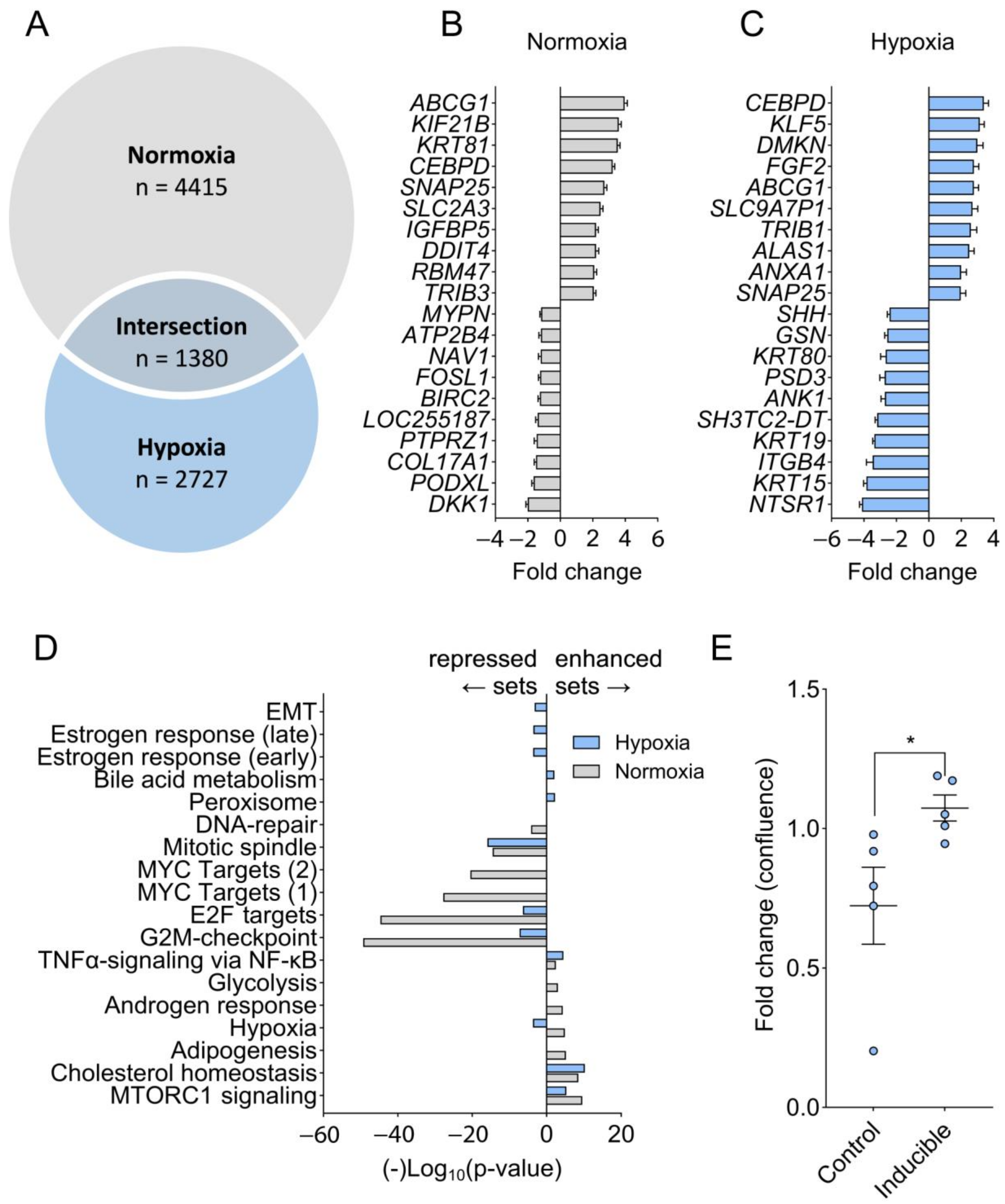
2.4. C/EBPδ Is Induced by Hypoxia and Boosts HIF-1a Protein Expression
2.5. Hypoxia Prevents C/EBPδ-Mediated Suppression of Proliferation-Associated Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Cloning Strategy and Lentiviral Transduction
4.3. Competition Assay
4.4. Animal Experiments and Declarations
4.5. Single Cell Dissociation and FACS Analysis
4.6. Fluorescence Analysis of Tumors
4.7. Immunohistochemistry
4.8. RNAscope
4.9. Protein Analysis Using Capillary Western Immunoassay (WES)
4.10. RNA Extraction and RNA Sequencing
4.11. Proliferation Assay
4.12. Bioinformatics Analyses
4.13. Quantitative Image Analysis
4.14. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2024; American Cancer Society: Atlanta, GA, USA, 2024. [Google Scholar]
- Tolomeo, M.; Grimaudo, S. The “Janus” Role of C/EBPs Family Members in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 4308. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K.; Sterneck, E. The many faces of C/EBPδ and their relevance for inflammation and cancer. Int. J. Biol. Sci. 2013, 9, 917–933. [Google Scholar] [CrossRef]
- Ko, C.-Y.; Chang, W.-C.; Wang, J.-M. Biological roles of CCAAT/Enhancer-binding protein delta during inflammation. J. Biomed. Sci. 2015, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Hardy, K.L.; Kitajewski, A.M.; Shawber, C.J.; Kitajewski, J.K.; Wu, J.K. Propranolol accelerates adipogenesis in hemangioma stem cells and causes apoptosis of hemangioma endothelial cells. Plast. Reconstr. Surg. 2012, 130, 1012–1021. [Google Scholar] [CrossRef]
- England, R.W.B.; Hardy, K.L.B.; Kitajewski, A.M.B.; Wong, A.; Kitajewski, J.K.; Shawber, C.J.; Wu, J.K. Propranolol promotes accelerated and dysregulated adipogenesis in hemangioma stem cells. Ann. Plat. Surg. 2014, 73 (Suppl. 1), S119–S124. [Google Scholar] [CrossRef]
- Balamurugan, K.; Mendoza-Villanueva, D.; Sharan, S.; Summers, G.H.; Dobrolecki, L.E.; Lewis, M.T.; Sterneck, E. C/EBPδ links IL-6 and HIF-1 signaling to promote breast cancer stem cell-associated phenotypes. Oncogene 2019, 38, 3765–3780. [Google Scholar] [CrossRef]
- Wang, S.-M.; Lin, H.-Y.; Chen, Y.-L.; Hsu, T.-I.; Chuang, J.-Y.; Kao, T.-J.; Ko, C.-Y. CCAAT/enhancer-binding protein delta regulates the stemness of glioma stem-like cells through activating PDGFA expression upon inflammatory stimulation. J. Neuroinflammation 2019, 16, 146. [Google Scholar] [CrossRef]
- Balamurugan, K.; Wang, J.M.; Tsai, H.H.; Sharan, S.; Anver, M.; Leighty, R.; Sterneck, E. The tumour suppressor C/EBPδ inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 2010, 29, 4106–4117. [Google Scholar] [CrossRef]
- Cooper, L.A.; Gutman, D.A.; Chisolm, C.; Appin, C.; Kong, J.; Rong, Y.; Kurc, T.; Van Meir, E.G.; Saltz, J.H.; Moreno, C.S.; et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am. J. Pathol. 2012, 180, 2108–2119. [Google Scholar] [CrossRef]
- Min, Y.; Li, J.; Qu, P.; Lin, P.C. C/EBP-δ positively regulates MDSC expansion and endothelial VEGFR2 expression in tumor development. Oncotarget 2017, 8, 50582–50593. [Google Scholar] [CrossRef] [PubMed]
- Pathak, R.R.; Grover, A.; Malaney, P.; Quarni, W.; Pandit, A.; Allen-Gipson, D.; Davé, V. Loss of phosphatase and tensin homolog (PTEN) induces leptin-mediated leptin gene expression: Feed-forward loop operating in the lung. J. Biol. Chem. 2013, 288, 29821–29835. [Google Scholar] [CrossRef] [PubMed]
- Hartl, L.; Duitman, J.; Aberson, H.L.; Chen, K.; Dijk, F.; Roelofs, J.; Dings, M.P.G.; Hooijer, G.K.J.; Hernanda, P.Y.; Pan, Q.; et al. CCAAT/Enhancer-Binding Protein Delta (C/EBPδ): A Previously Unrecognized Tumor Suppressor that Limits the Oncogenic Potential of Pancreatic Ductal Adenocarcinoma Cells. Cancers 2020, 12, 2546. [Google Scholar] [CrossRef] [PubMed]
- Hartl, L.; Maarschalkerweerd, P.A.F.; Butler, J.M.; Manz, X.D.; Thijssen, V.L.J.L.; Bijlsma, M.F.; Duitman, J.; Spek, C.A. C/EBPδ Suppresses Motility-Associated Gene Signatures and Reduces PDAC Cell Migration. Cells 2022, 11, 3334. [Google Scholar] [CrossRef] [PubMed]
- Sheshadri, N.; Poria, D.K.; Sharan, S.; Hu, Y.; Yan, C.; Koparde, V.N.; Balamurugan, K.; Sterneck, E. PERK signaling through C/EBPδ contributes to ER stress-induced expression of immunomodulatory and tumor promoting chemokines by cancer cells. Cell Death Dis. 2021, 12, 1038. [Google Scholar] [CrossRef]
- Chi, J.-Y.; Hsiao, Y.-W.; Li, C.-F.; Lo, Y.-C.; Lin, Z.-Y.; Hong, J.-Y.; Liu, Y.-M.; Han, X.; Wang, S.-M.; Chen, B.-K.; et al. Targeting chemotherapy-induced PTX3 in tumor stroma to prevent the progression of drug-resistant cancers. Oncotarget 2015, 6, 23987–24001. [Google Scholar] [CrossRef]
- Urbanova, M.; Buocikova, V.; Trnkova, L.; Strapcova, S.; Kajabova, V.H.; Melian, E.B.; Novisedlakova, M.; Tomas, M.; Dubovan, P.; Earl, J.; et al. DNA Methylation Mediates EMT Gene Expression in Human Pancreatic Ductal Adenocarcinoma Cell Lines. Int. J. Mol. Sci. 2022, 23, 2117. [Google Scholar] [CrossRef]
- Pandol, S.; Edderkaoui, M.; Gukovsky, I.; Lugea, A.; Gukovskaya, A. Desmoplasia of pancreatic ductal adenocarcinoma. Clin. Gastroenterol. Hepatol. 2009, 7 (Suppl. 11), S44–S47. [Google Scholar] [CrossRef]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef]
- Uchida, T.; Rossignol, F.; Matthay, M.A.; Mounier, R.; Couette, S.; Clottes, E.; Clerici, C. Prolonged Hypoxia Differentially Regulates Hypoxia-inducible Factor (HIF)-1α and HIF-2α Expression in Lung Epithelial Cells. JBC 2004, 15, 14871–14878. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- R2: Genomics Analysis and Visualization Platform. Available online: http://r2.amc.nl (accessed on 15 June 2022).
- Min, Y.; Ghose, S.; Boelte, K.; Li, J.; Yang, L.; Lin, P.C. C/EBP-δ regulates VEGF-C autocrine signaling in lymphangiogenesis and metastasis of lung cancer through HIF-1α. Oncogene 2011, 30, 4901–4909. [Google Scholar] [CrossRef] [PubMed]
- Hao, J. HIF-1 is a critical target of pancreatic cancer. Oncoimmunology 2015, 4, e1026535. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, J.; Tanaka, T.; Eto, N.; Nangaku, M. Inflammation and hypoxia linked to renal injury by CCAAT/enhancer-binding protein δ. Kidney Int. 2015, 88, 262–275. [Google Scholar] [CrossRef]
- Mao, X.-G.; Xue, X.-Y.; Lv, R.; Ji, A.; Shi, T.-Y.; Chen, X.-Y.; Jiang, X.-F.; Zhang, X. CEBPD is a master transcriptional factor for hypoxia regulated proteins in glioblastoma and augments hypoxia induced invasion through extracellular matrix-integrin mediated EGFR/PI3K pathway. Cell Death Dis. 2023, 14, 269. [Google Scholar] [CrossRef]
- Balamurugan, K.; Sharan, S.; Klarmann, K.D.; Zhang, Y.; Coppola, V.; Summers, G.H.; Roger, T.; Morrison, D.K.; Keller, J.R.; Sterneck, E. FBXW7α attenuates inflammatory signalling by downregulating C/EBPδ and its target gene Tlr4. Nat. Commun. 2013, 4, 1662. [Google Scholar] [CrossRef]
- Morales-Oyarvide, V.; Rubinson, D.A.; Dunne, R.F.; Kozak, M.M.; Bui, J.L.; Yuan, C.; Qian, Z.R.; Babic, A.; Da Silva, A.; Nowak, J.A.; et al. Lymph node metastases in resected pancreatic ductal adenocarcinoma: Predictors of disease recurrence and survival. Br. J. Cancer 2017, 117, 1874–1882. [Google Scholar] [CrossRef]
- Tiwari, A.; Tashiro, K.; Dixit, A.; Soni, A.; Vogel, K.; Hall, B.; Shafqat, I.; Slaughter, J.; Param, N.; Le, A.; et al. Loss of HIF1A From Pancreatic Cancer Cells Increases Expression of PPP1R1B and Degradation of p53 to Promote Invasion and Metastasis. Gastroenterology 2020, 159, 1882–1897.e5. [Google Scholar] [CrossRef]
- Zoa, A.; Yang, Y.; Huang, W.; Yang, J.; Wang, J.; Wang, H.; Dong, M.; Tian, Y. High expression of hypoxia-inducible factor 1-alpha predicts poor prognosis in pancreatic ductal adenocarcinoma: A meta-analysis and database validation protocol. Transl. Cancer Res. 2022, 11, 3080–3091. [Google Scholar] [CrossRef]
- Chan, T.C.; Chen, Y.T.; Tan, K.T.; Wu, C.L.; Wu, W.J.; Li, W.M.; Wang, J.M.; Shiue, Y.L.; Li, C.F. Biological significance of MYC and CEBPD coamplification in urothelial carcinoma: Multilayered genomic, transcriptional and posttranscriptional positive feedback loops enhance oncogenic glycolysis. Clin. Transl. Med. 2021, 11, e674. [Google Scholar] [CrossRef]
- Li, Y.; Sun, X.-X.; Qian, D.Z.; Dai, M.-S. Molecular Crosstalk between MYC and HIF in Cancer. Front. Cell Dev. Biol. 2020, 8, 590576. [Google Scholar] [CrossRef]
- Huang, L.E. Carrot and stick: HIF-α engages c-Myc in hypoxic adaptation. Cell Death Differ. 2008, 15, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.; Schreyer, D.; Gaul, L.-M.; Pfeffer, S.; Pilarsky, C.; Queiroz, K. A microfluidic-based PDAC organoid system reveals the impact of hypoxia in response to treatment. Cell Death Discov. 2023, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartl, L.; ten Brink, M.S.; Aberson, H.L.; Koster, J.; Zwijnenburg, D.A.; Duitman, J.; Bijlsma, M.F.; Spek, C.A. Hypoxia Abrogates Tumor-Suppressive Activities of C/EBPδ in Pancreatic Cancer. Int. J. Mol. Sci. 2024, 25, 9449. https://doi.org/10.3390/ijms25179449
Hartl L, ten Brink MS, Aberson HL, Koster J, Zwijnenburg DA, Duitman J, Bijlsma MF, Spek CA. Hypoxia Abrogates Tumor-Suppressive Activities of C/EBPδ in Pancreatic Cancer. International Journal of Molecular Sciences. 2024; 25(17):9449. https://doi.org/10.3390/ijms25179449
Chicago/Turabian StyleHartl, Leonie, Marieke S. ten Brink, Hella L. Aberson, Jan Koster, Danny A. Zwijnenburg, JanWillem Duitman, Maarten F. Bijlsma, and C. Arnold Spek. 2024. "Hypoxia Abrogates Tumor-Suppressive Activities of C/EBPδ in Pancreatic Cancer" International Journal of Molecular Sciences 25, no. 17: 9449. https://doi.org/10.3390/ijms25179449
APA StyleHartl, L., ten Brink, M. S., Aberson, H. L., Koster, J., Zwijnenburg, D. A., Duitman, J., Bijlsma, M. F., & Spek, C. A. (2024). Hypoxia Abrogates Tumor-Suppressive Activities of C/EBPδ in Pancreatic Cancer. International Journal of Molecular Sciences, 25(17), 9449. https://doi.org/10.3390/ijms25179449




