Pathological Mechanisms Involved in Epidermolysis Bullosa Simplex: Current Knowledge and Therapeutic Perspectives
Abstract
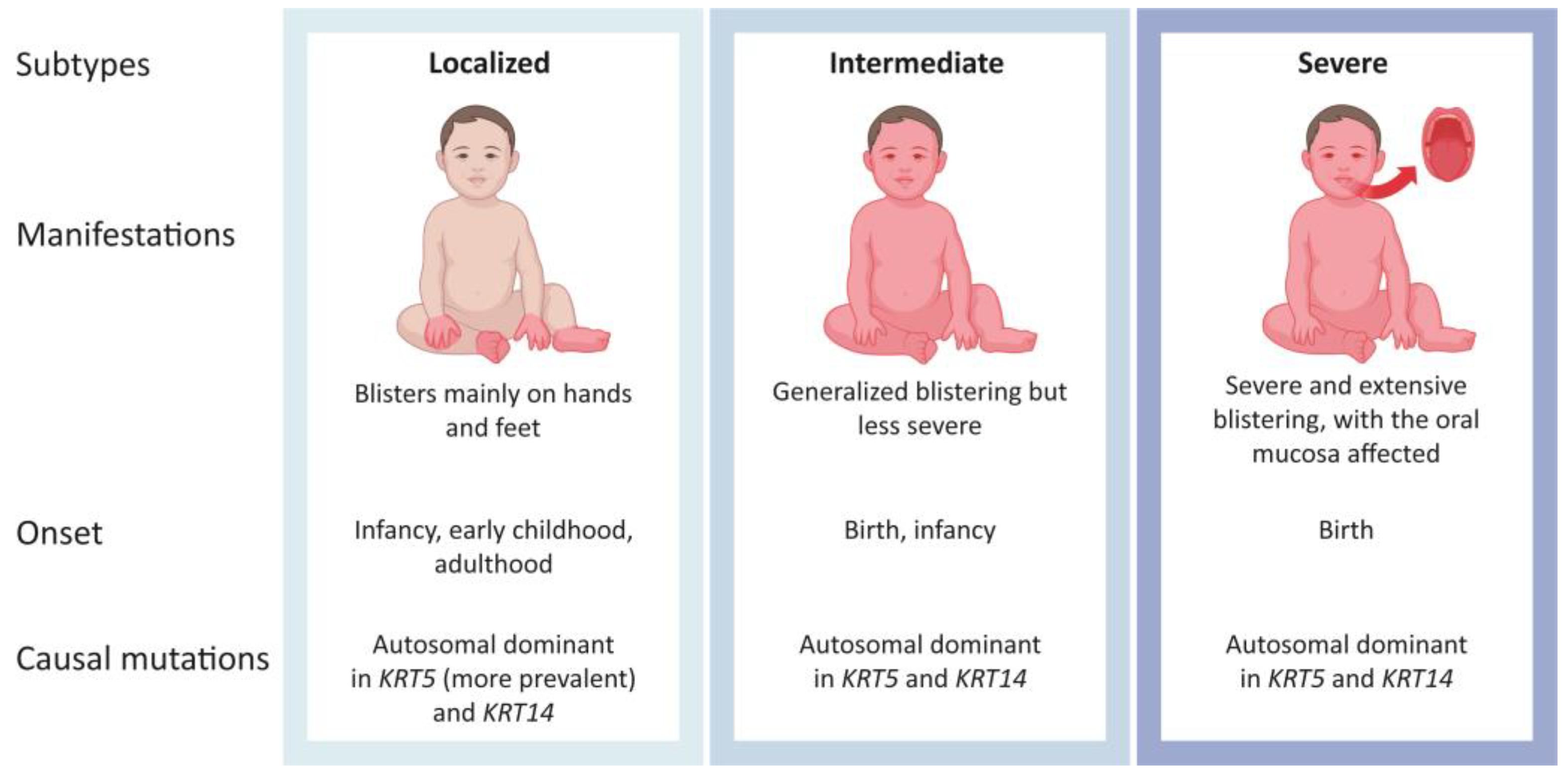
1. Introduction
2. EBS Gene Expression Profile Studies
2.1. Increase of Inflammation Components
2.1.1. c-Jun N-Terminal Kinase (JNK) Stress Pathway
2.1.2. IFN-γ Inflammatory Signaling Pathway
2.1.3. Phosphatidylinositol 3-Kinase (PI3K)-Protein Kinase B (Akt)-mTOR Pathway
2.1.4. Wnt-Receptor Signaling Pathway
2.1.5. Bone Morphogenetic Proteins Signaling (BMPs)
2.1.6. T Helper Type 17 (Th17) Immune Response
2.2. Keratins and Cell-Junction Components
3. Strategies to Improve and Alleviate EBS Manifestations
3.1. Therapeutic Molecules: Anti-Inflammatory
3.1.1. Tetracycline Antibiotics
3.1.2. Diacerein
3.1.3. Humanized IFN-γ Blocking Antibody
3.1.4. mTOR Inhibitors
3.1.5. 4-Phenyl Butyric Acid (4-PBA)
3.1.6. Apremilast (Anti-IL-17 Agent)
3.1.7. Afatinib (Epidermal Growth Factor Receptor (EGFR) Inhibitor)
3.2. Therapeutic Molecules: Beyond Anti-Inflammatory Mechanisms
3.2.1. Aluminium Chloride Hexahydrate
3.2.2. Botulinum Toxins (BoNT)
3.2.3. Parthenolide (PN)
3.2.4. PKC412 Kinase Inhibition
| Molecule | Reference | Type of Study | Patients | Vehicle | Target | Outcomes | Perspectives |
|---|---|---|---|---|---|---|---|
| 4-PBA | [62] | In vitro | EBS-loc, EBS-sev | 1 mM solution | Immortalized keratinocytes | Reduction of keratin aggregates upon heat shock | Reversing protein aggregates and reducing tissue fragility |
| [63] | In vitro | EBS-sev | 1 mM solution | Immortalized keratinocytes | Reduction of keratin aggregates and amelioration of inflammatory phenotype | Assay with low doses to avoid toxicity | |
| [23] | In vitro | EBS-sev | 1 μM solution | Keratinocytes | Reduction of around 40% of keratin aggregates | Reversing protein aggregates and reducing tissue fragility | |
| Afatinib | [69] | In vitro | EBS-sev | 1 μM solution | Immortalized keratinocytes | Reduction of keratin aggregates, induction of quiescent state to cells | Optimize afatinib for therapy (reduce adverse side effects when used in cancer therapy) |
| Aluminium chloride hexahydrate | [70] | Case report | EBS-loc | 20% in alcohol | Feet | Prevent new blisters | Study other cases to establish a pattern of efficiency |
| [71] | Case report | EBS-loc | 20% in alcohol | Hands and feet | Prevent new blisters | Study other cases to establish a pattern of efficiency | |
| [72] | Therapeutic assay | EBS-loc | 20% in alcohol | Feet | No significant reduction of blisters | Study the effect on more severe subtypes | |
| Apremilast | [26] | Therapeutic assay | EBS-sev | Oral treatment (10 mg/day to 30 mg twice/day) | Skin | Rapid and sustained (7–10 months) improvement in skin lesions | Trial with a higher number of patients (placebo–control) |
| Botulinum toxins | [74] | Case report | EBS-loc | 100 U BTX-A (Botox) in saline | Feet | 65% reduction in blister surface area, decrease in pain and perspiration | Trial with a higher number of patients (placebo–control), testing higher concentration of the molecule |
| [75] | Therapeutic assay | EBS-loc, EBS-sev | 170–700 U BTX-A (Dysport) in saline, 2500 U BTX-B (Neurobloc) in saline | Feet | Improvement in blistering and pedal pain | Trial with a higher number of patients (placebo–control) | |
| [76] | Case report | EBS | 100 U BTX-A (Botox) in saline | Feet | Reduction of blisters, smaller blisters, decreased pedal pain and odor | Treatment for pain management and improved quality of life | |
| Diacerein | [30] | In vitro | EBS-sev | 10 μg mL−1 solution | Keratinocytes | Stabilization of the IF network and reduction of inflammatory components | Trial with EBS-sev patients |
| [52] | Therapeutic assay | EBS-sev | 1% cream | Armpits | Reduction of blisters | Trial with a higher number of patients | |
| [53] | Phase 2/3 clinical trial | EBS-sev | 1% cream | Skin | More than 40% reduction in blister number | Trial with a higher number of patients and more invasive data acquisition | |
| [54] | Therapeutic assay | EBS-sev, EBS-intermed | 1% cream | Skin | No significant improvement compared with vehicule cream alone | Focus on EBS-sev individuals with a bigger sample size | |
| Doxycycline | [14] | In vivo | K5−/− mice | 50 μg/mL solution with 5% sucrose | Skin | Downregulation of MMPP13 and IL-1β | Screening similar compounds for additional targets |
| INF-γ blocking antibodies | [34] | In vitro | Cell model of EBS-sev | Humanized monoclonal antibody | Keratinocytes | Reversing effect of IFN-γ (less keratin aggregates, restored cell proliferation, increased cell–cell adhesion, accelerated wound closure) | Promising therapy for patients with EBS or other skin disease (in vivo assays) |
| mTOR inhibitors | [24] | Therapeutic assay | EBS | 2% sirolimus ointment | Feet | Reduction of blisters and keratoderma | Drug repositioning based on transcriptomic signature in EBS |
| Parthenolide | [78] | In vitro | Mice cell model of EBS | 5 µM solution | Keratinocytes | Increasing cell adhesion and resistance to mechanical stress | Trials with keratinocytes of patients to assess efficiency |
| PKC412 kinase inhibitor | [81] | In vitro | EBS-sev | 1 µM solution | Immortalized keratinocytes | Reduction of 40% of keratin aggregates and strengthening of cellular cohesion | Pretesting on animal models and skin explants before oral or topic administration in EBS patients |
| Tetracycline | [49] | Therapeutic assay | EBS-sev | Oral treatment (1500 mg/day) | Skin | Reduction of blisters and less fragile epidermis (dose-dependant response) | Trial with a higher number of patients |
3.3. Genome Editing Approaches
3.3.1. RNA Trans-Splicing
3.3.2. Transcription Activator-like Effector Nucleases (TALENS)
3.3.3. CRISPR-Cas9
4. Why Therapeutic Approaches Are Difficult to Find in Epidermolysis Bullosa?
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simpson, C.L.; Patel, D.M.; Green, K.J. Deconstructing the skin: Cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol. 2011, 12, 565–580. [Google Scholar] [CrossRef]
- Chambers, E.S.; Vukmanovic-Stejic, M. Skin barrier immunity and ageing. Immunology 2020, 160, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, P.A.; Kerns, M.L.; Fuchs, E. Epidermolysis bullosa simplex: A paradigm for disorders of tissue fragility. J. Clin. Investig. 2009, 119, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Evtushenko, N.A.; Beilin, A.K.; Kosykh, A.V.; Vorotelyak, E.A.; Gurskaya, N.G. Keratins as an Inflammation Trigger Point in Epidermolysis Bullosa Simplex. Int. J. Mol. Sci. 2021, 22, 12446. [Google Scholar] [CrossRef] [PubMed]
- Beriault, D.R.; Haddad, O.; McCuaig, J.V.; Robinson, Z.J.; Russell, D.; Lane, E.B.; Fudge, D.S. The mechanical behavior of mutant K14-R125P keratin bundles and networks in NEB-1 keratinocytes. PLoS ONE 2012, 7, e31320. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E.; Coulombe, P.; Cheng, J.; Chan, Y.M.; Hutton, E.; Syder, A.; Degenstein, L.; Yu, Q.C.; Letai, A.; Vassar, R. Genetic bases of epidermolysis bullosa simplex and epidermolytic hyperkeratosis. J. Investig. Dermatol. 1994, 103, 25S–30S. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, P.A.; Lee, C.H. Defining keratin protein function in skin epithelia: Epidermolysis bullosa simplex and its aftermath. J. Investig. Dermatol. 2012, 132, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Bar, J.; Kumar, V.; Roth, W.; Schwarz, N.; Richter, M.; Leube, R.E.; Magin, T.M. Skin fragility and impaired desmosomal adhesion in mice lacking all keratins. J. Investig. Dermatol. 2014, 134, 1012–1022. [Google Scholar] [CrossRef]
- Li, P.; Rietscher, K.; Jopp, H.; Magin, T.M.; Omary, M.B. Posttranslational modifications of keratins and their associated proteins as therapeutic targets in keratin diseases. Curr. Opin. Cell Biol. 2023, 85, 102264. [Google Scholar] [CrossRef]
- Snauwaert, J.J.; Yuen, W.Y.; Jonkman, M.F.; Moons, P.; Naulaers, G.; Morren, M.A. Burden of itch in epidermolysis bullosa. Br. J. Dermatol. 2014, 171, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Scheffschick, A.; Kiritsi, D.; Magin, T.M. Keratin defects trigger the itch-inducing cytokine thymic stromal lymphopoietin through amphiregulin-epidermal growth factor receptor signaling. J. Allergy Clin. Immunol. 2019, 144, 1719–1722. [Google Scholar] [CrossRef]
- Fine, J.D.; Bruckner-Tuderman, L.; Eady, R.A.; Bauer, E.A.; Bauer, J.W.; Has, C.; Heagerty, A.; Hintner, H.; Hovnanian, A.; Jonkman, M.F.; et al. Inherited epidermolysis bullosa: Updated recommendations on diagnosis and classification. J. Am. Acad. Dermatol. 2014, 70, 1103–1126. [Google Scholar] [CrossRef] [PubMed]
- Has, C.; Bauer, J.W.; Bodemer, C.; Bolling, M.C.; Bruckner-Tuderman, L.; Diem, A.; Fine, J.D.; Heagerty, A.; Hovnanian, A.; Marinkovich, M.P.; et al. Consensus reclassification of inherited epidermolysis bullosa and other disorders with skin fragility. Br. J. Dermatol. 2020, 183, 614–627. [Google Scholar] [CrossRef]
- Lu, H.; Chen, J.; Planko, L.; Zigrino, P.; Klein-Hitpass, L.; Magin, T.M. Induction of inflammatory cytokines by a keratin mutation and their repression by a small molecule in a mouse model for EBS. J. Investig. Dermatol. 2007, 127, 2781–2789. [Google Scholar] [CrossRef] [PubMed]
- Liovic, M.; Lee, B.; Tomic-Canic, M.; D’Alessandro, M.; Bolshakov, V.N.; Lane, E.B. Dual-specificity phosphatases in the hypo-osmotic stress response of keratin-defective epithelial cell lines. Exp. Cell Res. 2008, 314, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Liovic, M.; D’Alessandro, M.; Tomic-Canic, M.; Bolshakov, V.N.; Coats, S.E.; Lane, E.B. Severe keratin 5 and 14 mutations induce down-regulation of junction proteins in keratinocytes. Exp. Cell Res. 2009, 315, 2995–3003. [Google Scholar] [CrossRef] [PubMed]
- Roth, W.; Reuter, U.; Wohlenberg, C.; Bruckner-Tuderman, L.; Magin, T.M. Cytokines as genetic modifiers in K5-/- mice and in human epidermolysis bullosa simplex. Hum. Mutat. 2009, 30, 832–841. [Google Scholar] [CrossRef]
- Wagner, M.; Hintner, H.; Bauer, J.W.; Onder, K. Gene expression analysis of an epidermolysis bullosa simplex Dowling-Meara cell line by subtractive hybridization: Recapitulation of cellular differentiation, migration and wound healing. Exp. Dermatol. 2012, 21, 111–117. [Google Scholar] [CrossRef]
- Wagner, M.; Trost, A.; Hintner, H.; Bauer, J.W.; Onder, K. Imbalance of intermediate filament component keratin 14 contributes to increased stress signalling in epidermolysis bullosa simplex. Exp. Dermatol. 2013, 22, 292–294. [Google Scholar] [CrossRef]
- Bchetnia, M.; Tremblay, M.L.; Leclerc, G.; Duperee, A.; Powell, J.; McCuaig, C.; Morin, C.; Legendre-Guillemin, V.; Laprise, C. Expression signature of epidermolysis bullosa simplex. Hum. Genet. 2012, 131, 393–406. [Google Scholar] [CrossRef]
- Lettner, T.; Lang, R.; Klausegger, A.; Hainzl, S.; Bauer, J.W.; Wally, V. MMP-9 and CXCL8/IL-8 are potential therapeutic targets in epidermolysis bullosa simplex. PLoS ONE 2013, 8, e70123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Herzog, J.; Rid, R.; Wagner, M.; Hundsberger, H.; Eger, A.; Bauer, J.; Onder, K. Whole-transcriptome gene expression profiling in an epidermolysis bullosa simplex Dowling-Meara model keratinocyte cell line uncovered novel, potential therapeutic targets and affected pathways. BMC Res. Notes 2015, 8, 785. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coutier, J.; Bonnette, M.; Martineau, S.; Mercadier, A.; Domingues, S.; Saidani, M.; Jarrige, M.; Polveche, H.; Darle, A.; Holic, N.; et al. Human-Induced Pluripotent Stem Cell–Derived Keratinocytes, a Useful Model to Identify and Explore the Pathological Phenotype of Epidermolysis Bullosa Simplex. J. Investig. Dermatol. 2022, 142, 2695–2705 e2611. [Google Scholar] [CrossRef]
- Lee, G.H.; Lekwuttikarn, R.; Tafoya, E.; Martin, M.; Sarin, K.Y.; Teng, J.M. Transcriptomic Repositioning Analysis Identifies mTOR Inhibitor as Potential Therapy for Epidermolysis Bullosa Simplex. J. Investig. Dermatol. 2022, 142, 382–389. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding Tissue-Specific Gene Regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Castela, E.; Tulic, M.K.; Rozieres, A.; Bourrat, E.; Nicolas, J.F.; Kanitakis, J.; Vabres, P.; Bessis, D.; Mazereeuw, J.; Morice-Picard, F.; et al. Epidermolysis bullosa simplex generalized severe induces a T helper 17 response and is improved by apremilast treatment. Br. J. Dermatol. 2019, 180, 357–364. [Google Scholar] [CrossRef]
- Mellerio, J.E. Potential therapeutic targeting of inflammation in epidermolysis bullosa simplex. Br. J. Dermatol. 2019, 180, 258–260. [Google Scholar] [CrossRef]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Richmond, J.M.; Harris, J.E. Immunology and skin in health and disease. Cold Spring Harb. Perspect. Med. 2014, 4, a015339. [Google Scholar] [CrossRef]
- Wally, V.; Lettner, T.; Peking, P.; Peckl-Schmid, D.; Murauer, E.M.; Hainzl, S.; Hintner, H.; Bauer, J.W. The pathogenetic role of IL-1beta in severe epidermolysis bullosa simplex. J. Investig. Dermatol. 2013, 133, 1901–1903. [Google Scholar] [CrossRef] [PubMed]
- Gallucci, R.M.; Sloan, D.K.; Heck, J.M.; Murray, A.R.; O’Dell, S.J. Interleukin 6 indirectly induces keratinocyte migration. J. Investigig Dermatol. 2004, 122, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Cardoso, A.P.; Goncalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Gresser, I. Biologic effects of interferons. J. Investig. Dermatol. 1990, 95, 66S–71S. [Google Scholar] [CrossRef]
- Badowski, C.; Tan, T.S.; Aliev, T.; Trudil, D.; Larina, M.; Argentova, V.; Firdaus, M.J.; Benny, P.; Woo, V.S.T.; Lane, E.B. Detrimental Effects of IFN-gamma on an Epidermolysis Bullosa Simplex Cell Model and Protection by a Humanized Anti-IFN-gamma Monoclonal Antibody. JID Innov. 2022, 2, 100096. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Kerns, M.L.; DePianto, D.; Dinkova-Kostova, A.T.; Talalay, P.; Coulombe, P.A. Reprogramming of keratin biosynthesis by sulforaphane restores skin integrity in epidermolysis bullosa simplex. Proc. Natl. Acad. Sci. USA 2007, 104, 14460–14465. [Google Scholar] [CrossRef]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Spranger, S.; Fuchs, E.; López-Soto, A. WNT Signaling in Cancer Immunosurveillance. Trends Cell Biol. 2019, 29, 44–65. [Google Scholar] [CrossRef]
- Sneddon, J.B.; Zhen, H.H.; Montgomery, K.; van de Rijn, M.; Tward, A.D.; West, R.; Gladstone, H.; Chang, H.Y.; Morganroth, G.S.; Oro, A.E.; et al. Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc. Natl. Acad. Sci. USA 2006, 103, 14842–14847. [Google Scholar] [CrossRef]
- Sharov, A.A.; Mardaryev, A.N.; Sharova, T.Y.; Grachtchouk, M.; Atoyan, R.; Byers, H.R.; Seykora, J.T.; Overbeek, P.; Dlugosz, A.; Botchkarev, V.A. Bone morphogenetic protein antagonist noggin promotes skin tumorigenesis via stimulation of the Wnt and Shh signaling pathways. Am. J. Pathol. 2009, 175, 1303–1314. [Google Scholar] [CrossRef]
- Kim, C.H. Migration and function of Th17 cells. Inflamm. Allergy Drug Targets 2009, 8, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Harper, E.G.; Guo, C.; Rizzo, H.; Lillis, J.V.; Kurtz, S.E.; Skorcheva, I.; Purdy, D.; Fitch, E.; Iordanov, M.; Blauvelt, A. Th17 cytokines stimulate CCL20 expression in keratinocytes in vitro and in vivo: Implications for psoriasis pathogenesis. J. Investig. Dermatol. 2009, 129, 2175–2183. [Google Scholar] [CrossRef] [PubMed]
- Szeverenyi, I.; Cassidy, A.J.; Chung, C.W.; Lee, B.T.; Common, J.E.; Ogg, S.C.; Chen, H.; Sim, S.Y.; Goh, W.L.; Ng, K.W.; et al. The Human Intermediate Filament Database: Comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 2008, 29, 351–360. [Google Scholar] [CrossRef]
- Autio, P.; Keski-Oja, J. Tetracyclines as anti-inflammatory treatment in skin diseases. Nord. Med. 1996, 111, 348–351. [Google Scholar]
- Khanna, N. Follow up of patients of acne vulgaris treated with tetracyclines. Indian. J. Dermatol. Venereol. Leprol. 1996, 62, 301–303. [Google Scholar]
- Yang, Y.L.; Wang, L.X.; Fei, X.M.; Lei, F.; Lu, W.P.; Yu, X.Q.; Zhang, S. The Effect of Doxycycline on the Expression of MMP-2 and MMP-9 in Multiple Myeloma. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2022, 30, 487–492. [Google Scholar] [CrossRef]
- Forloni, G.; Iussich, S.; Awan, T.; Colombo, L.; Angeretti, N.; Girola, L.; Bertani, I.; Poli, G.; Caramelli, M.; Grazia Bruzzone, M.; et al. Tetracyclines affect prion infectivity. Proc. Natl. Acad. Sci. USA 2002, 99, 10849–10854. [Google Scholar] [CrossRef] [PubMed]
- Bendeck, M.P.; Conte, M.; Zhang, M.; Nili, N.; Strauss, B.H.; Farwell, S.M. Doxycycline modulates smooth muscle cell growth, migration, and matrix remodeling after arterial injury. Am. J. Pathol. 2002, 160, 1089–1095. [Google Scholar] [CrossRef]
- Retief, C.R.; Malkinson, F.D.; Pearson, R.W. Two familial cases of epidermolysis bullosa simplex successfully treated with tetracycline. Arch. Dermatol. 1999, 135, 997–998. [Google Scholar] [CrossRef]
- Sapadin, A.N.; Fleischmajer, R. Tetracyclines: Nonantibiotic properties and their clinical implications. J. Am. Acad. Dermatol. 2006, 54, 258–265. [Google Scholar] [CrossRef]
- Moldovan, F.; Pelletier, J.P.; Jolicoeur, F.C.; Cloutier, J.M.; Martel-Pelletier, J. Diacerhein and rhein reduce the ICE-induced IL-1beta and IL-18 activation in human osteoarthritic cartilage. Osteoarthr. Cartil. 2000, 8, 186–196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wally, V.; Kitzmueller, S.; Lagler, F.; Moder, A.; Hitzl, W.; Wolkersdorfer, M.; Hofbauer, P.; Felder, T.K.; Dornauer, M.; Diem, A.; et al. Topical diacerein for epidermolysis bullosa: A randomized controlled pilot study. Orphanet J. Rare Dis. 2013, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Wally, V.; Hovnanian, A.; Ly, J.; Buckova, H.; Brunner, V.; Lettner, T.; Ablinger, M.; Felder, T.K.; Hofbauer, P.; Wolkersdorfer, M.; et al. Diacerein orphan drug development for epidermolysis bullosa simplex: A phase 2/3 randomized, placebo-controlled, double-blind clinical trial. J. Am. Acad. Dermatol. 2018, 78, 892–901 e897. [Google Scholar] [CrossRef]
- Teng, J.; Paller, A.S.; Bruckner, A.L. Diacerein 1% Ointment for the Treatment of Epidermolysis Bullosa Simplex: A Randomized, Controlled Trial. J. Drugs Dermatol. 2023, 22, 599–604. [Google Scholar] [CrossRef]
- Skurkovich, B.; Skurkovich, S. Autoimmune diseases are connected with disturbances in cytokine synthesis, and therapy with IFN-gamma blockers is their main pathogenetic treatment. Ann. N. Y. Acad. Sci. 2007, 1109, 167–177. [Google Scholar] [CrossRef]
- Larina, M.V.; Aliev, T.K.; Solopova, O.N.; Pozdnyakova, L.P.; Korobova, S.V.; Yakimov, S.A.; Sveshnikov, P.G.; Dolgikh, D.A.; Kirpichnikov, M.P. Neutralizing Monoclonal and Chimeric Antibodies to Human IFN-gamma. Bioorg. Khim. 2015, 41, 316–326. [Google Scholar] [CrossRef]
- Kuo, K.Y.; Cho, H.G.; Sarin, K.Y. Identification of Atorvastatin for Moderate to Severe Hidradenitis through Drug Repositioning Using Public Gene Expression Datasets. J. Investig. Dermatol. 2018, 138, 1209–1212. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.N.; Fry, M.A.; Urman, N.M.; Atwood, S.X.; Roffey, J.; Ott, G.R.; Chen, B.; Lee, A.; Brown, A.S.; Aasi, S.Z.; et al. Combined inhibition of atypical PKC and histone deacetylase 1 is cooperative in basal cell carcinoma treatment. JCI Insight 2017, 2, e97071. [Google Scholar] [CrossRef]
- Cho, H.G.; Fiorentino, D.; Lewis, M.; Sirota, M.; Sarin, K.Y. Identification of Alpha-Adrenergic Agonists as Potential Therapeutic Agents for Dermatomyositis through Drug-Repurposing Using Public Expression Datasets. J. Investig. Dermatol. 2016, 136, 1517–1520. [Google Scholar] [CrossRef][Green Version]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef]
- Bchetnia, M.; Lacroix, J.; Farez, T.; Larouche, M.; Powell, J.; McCuaig, C.; Dupere, A.; Morin, C.; Legendre-Guillemin, V.; Laprise, C. Reduction in keratin aggregates in epidermolysis bullosa simplex keratinocytes after pretreatment with trimethylamine N-oxide. Exp. Dermatol. 2016, 25, 229–230. [Google Scholar] [CrossRef]
- Chamcheu, J.C.; Navsaria, H.; Pihl-Lundin, I.; Liovic, M.; Vahlquist, A.; Torma, H. Chemical chaperones protect epidermolysis bullosa simplex keratinocytes from heat stress-induced keratin aggregation: Involvement of heat shock proteins and MAP kinases. J. Investig. Dermatol. 2011, 131, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Sporrer, M.; Prochnicki, A.; Tolle, R.C.; Nystrom, A.; Esser, P.R.; Homberg, M.; Athanasiou, I.; Zingkou, E.; Schilling, A.; Gerum, R.; et al. Treatment of keratinocytes with 4-phenylbutyrate in epidermolysis bullosa: Lessons for therapies in keratin disorders. EBioMedicine 2019, 44, 502–515. [Google Scholar] [CrossRef]
- Papp, K.; Reich, K.; Leonardi, C.L.; Kircik, L.; Chimenti, S.; Langley, R.G.; Hu, C.; Stevens, R.M.; Day, R.M.; Gordon, K.B.; et al. Apremilast, an oral phosphodiesterase 4 (PDE4) inhibitor, in patients with moderate to severe plaque psoriasis: Results of a phase III, randomized, controlled trial (Efficacy and Safety Trial Evaluating the Effects of Apremilast in Psoriasis [ESTEEM] 1). J. Am. Acad. Dermatol. 2015, 73, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, L.I.; Bogdanos, D.P. Are psoriasis and psoriatic arthritis the same disease? The IL-23/IL-17 axis data. Autoimmun. Rev. 2017, 16, 10–15. [Google Scholar] [CrossRef]
- Aushev, M.; Koller, U.; Mussolino, C.; Cathomen, T.; Reichelt, J. Traceless Targeting and Isolation of Gene-Edited Immortalized Keratinocytes from Epidermolysis Bullosa Simplex Patients. Mol. Ther. Methods Clin. Dev. 2017, 6, 112–123. [Google Scholar] [CrossRef]
- Kirfel, G.; Herzog, V. Migration of epidermal keratinocytes: Mechanisms, regulation, and biological significance. Protoplasma 2004, 223, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.G.; McCawley, L.J. Contributions of the epidermal growth factor receptor to keratinocyte motility. Microsc. Res. Tech. 1998, 43, 444–455. [Google Scholar] [CrossRef]
- Tan, T.S.; Common, J.E.A.; Lim, J.S.Y.; Badowski, C.; Firdaus, M.J.; Leonardi, S.S.; Lane, E.B. A cell-based drug discovery assay identifies inhibition of cell stress responses as a new approach to treatment of epidermolysis bullosa simplex. J. Cell Sci. 2021, 134, jcs258409. [Google Scholar] [CrossRef]
- Tkach, J.R. Treatment of recurrent bullous eruption of the hand and feet (Weber-Cockayne disease) with topical aluminum chloride. J. Am. Acad. Dermatol. 1982, 6, 1095–1096. [Google Scholar] [CrossRef]
- Jennings, J.L. Aluminum chloride hexahydrate treatment of localized epidermolysis bullosa. Arch. Dermatol. 1984, 120, 1382. [Google Scholar] [CrossRef] [PubMed]
- Younger, I.R.; Priestley, G.C.; Tidman, M.J. Aluminum chloride hexahydrate and blistering in epidermolysis bullosa simplex. J. Am. Acad. Dermatol. 1990, 23, 930–931. [Google Scholar] [CrossRef]
- Choudhury, S.; Baker, M.R.; Chatterjee, S.; Kumar, H. Botulinum Toxin: An Update on Pharmacology and Newer Products in Development. Toxins 2021, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Abitbol, R.J.; Zhou, L.H. Treatment of Epidermolysis Bullosa Simplex, Weber-Cockayne Type, With Botulinum Toxin Type A. Arch. Dermatol. 2009, 145, 13–15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Swartling, C.; Karlqvist, M.; Hymnelius, K.; Weis, J.; Vahlquist, A. Botulinum toxin in the treatment of sweat-worsened foot problems in patients with epidermolysis bullosa simplex and pachyonychia congenita. Br. J. Dermatol. 2010, 163, 1072–1076. [Google Scholar] [CrossRef] [PubMed]
- Holahan, H.M.; Farah, R.S.; Ferguson, N.N.; Paller, A.S.; Legler, A.A. Treatment of symptomatic epidermolysis bullosa simplex with botulinum toxin in a pediatric patient. JAAD Case Rep. 2016, 2, 259–260. [Google Scholar] [CrossRef][Green Version]
- Freund, R.R.A.; Gobrecht, P.; Fischer, D.; Arndt, H.D. Advances in chemistry and bioactivity of parthenolide. Nat. Prod. Rep. 2020, 37, 541–565. [Google Scholar] [CrossRef]
- Sun, J.; Li, P.; Gui, H.; Rittié, L.; Lombard, D.B.; Rietscher, K.; Magin, T.M.; Xie, Q.; Liu, L.; Omary, M.B. Deacetylation via SIRT2 prevents keratin-mutation-associated injury and keratin aggregation. JCI Insight 2023, 8, e166314. [Google Scholar] [CrossRef]
- Luskin, M.R.; DeAngelo, D.J. Midostaurin/PKC412 for the treatment of newly diagnosed FLT3 mutation-positive acute myeloid leukemia. Expert. Rev. Hematol. 2017, 10, 1033–1045. [Google Scholar] [CrossRef]
- Kwan, R.; Chen, L.; Looi, K.; Tao, G.Z.; Weerasinghe, S.V.; Snider, N.T.; Conti, M.A.; Adelstein, R.S.; Xie, Q.; Omary, M.B. PKC412 normalizes mutation-related keratin filament disruption and hepatic injury in mice by promoting keratin-myosin binding. Hepatology 2015, 62, 1858–1869. [Google Scholar] [CrossRef]
- Rietscher, K.; Jahnke, H.G.; Rübsam, M.; Lin, E.W.; Has, C.; Omary, M.B.; Niessen, C.M.; Magin, T.M. Kinase Inhibition by PKC412 Prevents Epithelial Sheet Damage in Autosomal Dominant Epidermolysis Bullosa Simplex through Keratin and Cell Contact Stabilization. J. Investig. Dermatol. 2022, 142, 3282–3293. [Google Scholar] [CrossRef]
- Omary, M.B.; Ku, N.O.; Tao, G.Z.; Toivola, D.M.; Liao, J. “Heads and tails” of intermediate filament phosphorylation: Multiple sites and functional insights. Trends Biochem. Sci. 2006, 31, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Tockner, B.; Kocher, T.; Hainzl, S.; Reichelt, J.; Bauer, J.W.; Koller, U.; Murauer, E.M. Construction and validation of an RNA trans-splicing molecule suitable to repair a large number of COL7A1 mutations. Gene Ther. 2016, 23, 775–784. [Google Scholar] [CrossRef]
- Wally, V.; Brunner, M.; Lettner, T.; Wagner, M.; Koller, U.; Trost, A.; Murauer, E.M.; Hainzl, S.; Hintner, H.; Bauer, J.W. K14 mRNA reprogramming for dominant epidermolysis bullosa simplex. Hum. Mol. Genet. 2010, 19, 4715–4725. [Google Scholar] [CrossRef] [PubMed]
- Peking, P.; Breitenbach, J.S.; Ablinger, M.; Muss, W.H.; Poetschke, F.J.; Kocher, T.; Koller, U.; Hainzl, S.; Kitzmueller, S.; Bauer, J.W.; et al. An ex vivo RNA trans-splicing strategy to correct human generalized severe epidermolysis bullosa simplex. Br. J. Dermatol. 2019, 180, 141–148. [Google Scholar] [CrossRef]
- Hainzl, S.; Peking, P.; Kocher, T.; Murauer, E.M.; Larcher, F.; Del Rio, M.; Duarte, B.; Steiner, M.; Klausegger, A.; Bauer, J.W.; et al. COL7A1 Editing via CRISPR/Cas9 in Recessive Dystrophic Epidermolysis Bullosa. Mol. Ther. 2017, 25, 2573–2584. [Google Scholar] [CrossRef]
- Li, P.; Kleinstiver, B.P.; Leon, M.Y.; Prew, M.S.; Navarro-Gomez, D.; Greenwald, S.H.; Pierce, E.A.; Joung, J.K.; Liu, Q. Allele-Specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin-Associated Dominant Retinitis Pigmentosa. CRISPR J. 2018, 1, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, S.G.; Luoni, M.; Castoldi, V.; Massimino, L.; Cabassi, T.; Angeloni, D.; Demontis, G.C.; Leocani, L.; Andreazzoli, M.; Broccoli, V. Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum. Mol. Genet. 2018, 27, 761–779. [Google Scholar] [CrossRef]
- Kocher, T.; Peking, P.; Klausegger, A.; Murauer, E.M.; Hofbauer, J.P.; Wally, V.; Lettner, T.; Hainzl, S.; Ablinger, M.; Bauer, J.W.; et al. Cut and Paste: Efficient Homology-Directed Repair of a Dominant Negative KRT14 Mutation via CRISPR/Cas9 Nickases. Mol. Ther. 2017, 25, 2585–2598. [Google Scholar] [CrossRef]
- Bchetnia, M.; Dionne Gagné, R.; Powell, J.; Morin, C.; McCuaig, C.; Dupérée, A.; Germain, L.; Tremblay, J.P.; Laprise, C. Allele-Specific Inactivation of an Autosomal Dominant Epidermolysis Bullosa Simplex Mutation Using CRISPR-Cas9. CRISPR J. 2022, 5, 586–597. [Google Scholar] [CrossRef]
- Pfendner, E.; Uitto, J.; Fine, J.D. Epidermolysis bullosa carrier frequencies in the US population. J. Investig. Dermatol. 2001, 116, 483–484. [Google Scholar] [CrossRef] [PubMed]

| Reference | Model/Cell Lines | Upregulated Genes | Downregulated Genes | ||
|---|---|---|---|---|---|
| [14] | WT mouse skin vs. K5-−/− mouse skin | Gene symbol | Method | Gene symbol | Method |
| CXCL1, CXCL2, HPRT, KRT6a | MA | ||||
| IL6, IL1-β | MA, RTq-PCR | ||||
| [16] | WT vs. EBS-sev KEB-7 | ACTN1, AJAP1, CDH2, CDKN2A, CST6, DBN1, FLNB, FN1, GNA11, IGSF4, KIAA0992, KRT18, KRT8, KRTHB1, LMNB2, MYL9, PODXL, PRSS2, RHOA, TGM2, TNC, TPM2, TUBB2, VIM | MA | ABLIM1, AMOTL2, ANXA1, ARHGDIB, CLDN4, COL17A1, CSTA, CTGF, CXADR, DDR1, DST, EGFR, ENC1, FRMD4B, GABARAPL1, GPNMB, HSPB1, IL18, IL8, KRT13, KRT15, KRT16, KRT19, KRT5, LPXN, LY6D, MTSS1, OLR1, PCDH7, PDLIM1, PDZK3, PFN2, RND3, SAA1, SCEL, SLC9A3R1, SNCG, PRR1B, STEAP1, TSPAN1 | MA |
| CDH1, GJB5, ITGA6, LAMA5, LAMB3, PPL | MA, RTq-PCR | ||||
| CLDN3, CLDN7, DSC2, PKP1 | RTq-PCR | ||||
| DSG3, GJA1, JUP | MA, RTq-PCR, WB, IF | ||||
| DSP | MA, WB, IF | ||||
| WT vs. EBS-sev KEB-1 | GJA1, DSG3, DSP, JUP | WB | |||
| WT vs. EBS-loc KEB-4 | ACTA2, AJAP1, ANK3, CLDN3, CLDN7, GSN, KRT14, KRT15, KRT16, KRT17, KRT4, LGALS7, HLA-B, HLA-C, NEFH | MA | PCDH7, BASP1, CTGF, CYR61, DST, ENC1, FRMD4B, IL18, KRT19, LPXN, OLR1, RDX, RND3, STK6 | MA | |
| [17] | WT mouse skin vs. K5−/− mouse skin | MCP-1, CCL2, MIP-3a/CCL20, MIP-3b/CCL19, CXCL16 | |||
| [18] | WT vs. EBS-sev KEB-7 | ACSL3, ANXA2, ARPC3, CAV1, CCT5, EEF1G, EIF1, ENO1, GLUD1, H2AFZ, HNRPDL, SP90AB1, MAN2A1, PDCD4, RAB27B, RPL13, RPL19, RPL23A, RPL28, RPL31, RPL4, RPL5, RPL7A, RPLP0, RPS18, RPS2, PS25, RPS3, RPS4X, RPS6, RPS8, SFN, TAX1BP1, TMBIM6, TPI1, TPT1, EEF1A1, GSN, HNRNPA1, KRT5, KRT6A | SSH | ||
| ANXA8, DDIT4, DSG3, DSP, F3, KRT14, KRT16, KRT17, KRT6B, MALAT1, PERP, TXNIP, UBE2K, YWHAZ | SSH, RTq-PCR | ||||
| [20] | WT human epidermis tissue vs. EBS-sev and EBS-loc human epidermis tissue | SPRR2B, AREG, BDP1, NR4A2, GAL, LYVE1, PSG4, H19, PDE6A, C5orf27, THRSP, FAR2, AADACL3, CRAT, AGR2, IGFL4, KLK6, PM20D1, ATP12A, FSIP2 | MA | ||
| AWAT2, DGAT2L6, FADS2, ACSBG1, SPRR4, KRT79 | MA, RTq-PCR | ||||
| ELOVL3 | MA, RTq-PCR, WB | ||||
| WT human epidermis vs. EBS-sev human epidermis | FADS1, CYP4F8, AWAT1, ALOX15B, ACSM3, SOAT1, SLC27A2, HAO2, INSIG1, KRTAP5-8, KRT25, KRT71, KRT74, KRT27, TCHH, CHI3L1, MUC1, SLCO4C1, TGIF2LX, IGLJ3, IGJ, IGHA1, TMEM56, LRCC37A2, THRSP, FAR2, AADACL3, CRAT, AGR2, IGFL4, KLK6, PM20D1, ATP12A, FSIP2 | ||||
| AWAT2, DGAT2L6, FADS2, ACSBG1, SPRR4, KRT79 | MA, RTq-PCR | ||||
| ELOVL3 | MA, RTq-PCR, WB | ||||
| [30] | WT immortalized keratinocytes vs. immortalized EBS-sev cell lines (KEB-7, EBDM-1) | KRT14,IL-1b, KRT6A | SqRT-PCR | ||
| [21] | WT immortalized keratinocytes vs. immortalized EBS-sev cell lines (KEB-7, EBDM-1) | KLK6, KLK8, KLK10, KLK11, KLK13, KLK14, MMP1, MMP13, WIPF1, ARHGEF4, ARHGEF37, CDC42BPG, KRT6B | MA | ||
| KLK5, KLK7, KRT14, KRT15, KRT16 KRT17, KRT5 | MA, sqRT-PCR, WB | ||||
| MMP7, MMP9, MMP19, ARHGEF9, DSC1, DSC2, DSC3, DSG1, DSG3, DSG4, GJA1, GJB2, GJB6, CXCL1, CXCL8/IL-8, CXCL14 | MA, sqRT-PCR | ||||
| [22] | WT vs. EBS-sev KEB-7 | KGFLP1, ANKRD2, PIK3R3, PTPN20A, FAM21A, APBB2, DUOX1, ZNF627, MCOLN2, APOB, AJAP1, BNC2, LOC100652860, FKTN, TMEM204, BMS1P1, MAN1A1, STOX1, RPL10, CDC144C, NOX5EVC2, PTPN20C, PTPN20B, NID1, ASAH2, FLJ20444, TP53INP1, FRG1B, MSLN, DENND1B, IFFO2, STRBP, DSEL, AOX1, LRP12, ADHFE1, FAM21D, SNORD64, FAM21A, EFEMP1, TSPYL5, RNF212, DDX43, ZNF136, CCDC144A, CCDC144A, ZNF700, BGN, H2AFY2, SNORD116-21, HOXA9, RPS23 | MA | MIR492, H19, EYA4, TMPRSS15, ITGBL1, EDIL3, CDR1, NEFL, SMOC2, GHR, TFPI2, ARHGAP28, NNMT, SOX2, HIST1H, PCCA, ZNF570, CDK14, MEST, CYP7B1, GALNTL4, CRIP2, IPO7, SAAL1, KRTCAP3, FAM159A, EYA1, CDC25B, NKX2-6, HTATIP2, ILK, ACSF2, PDZD2, CENPH, TOX, VSTM2L, SYT17, SLC7A2, IKZF3 | MA |
| TDRD12, NEFH, NLRP2, KLK5, ENPP1, ZFP42, DKK1, CYYR1, C10orf99, SYCP2, PRICKLE1, SLC44A5, PLA2G7, MOXD1, WNT5A, WISP3, ARHGEF9, HSD17B11, ADAMTSL3, FAM102B SGMS1, ARHGAP29, SLC15A2, ROBO1, ERCC6, NREP, KIAA1324L, ROR1, ZFAND4, SELENBP1, NF334 PTPN20C, IRX4, PTPN20A, PNMAL1, NID1, ZNF32, AHI1, ELAVL2, SLC16A9, FAM25A, MAPK8, CDC14B, GABPB2 TCHH, LMF1, CSTF2T, SGK1, UAP1, POPDC2, ZNF502 | MA, RTq-PCR | KRT19, KYNU, PDZK1, OLFM4, SLC38A4, BST2, PPARGC1A, GALNT5, FKBP10, GIPC2, AMOT, ZNF114, CLEC2B, FAM198B, SLC2A3, CAPNS2, TBX18, LRCH2, NEFM, CPT1C, ZNF43, LY75, GLDC, TMTC1, SLCO1B3, SLC6A14, SLC24A3, EPSTI1, SATB2, HSD17B2, AKR1B10, GPC3, IFITM3, HOXD10, MSX2, IL17RB, BLMH, SLC9A2, CPNE1, WDR17, RB1, DPYD, PRTFDC1, GLRX, PPP1R16B, GTF2H2D, REPS2, GPR143, GTF2H2, CYP7B1, BCL11A, MERTK, PRDM5, ACOXL, AHCY, ARMCX2, PAX6, HOXD11, SMARCA1, IFI44L, PITRM1, NAP1L5, PIGU | MA, RTq-PCR | ||
| [23] | WT human keratinocytes vs. EBS-loc and EBS-intermed keratinocytes | ACOT1, ALDH1A3, ALOX15B, ALPL 6, ANGPTL4, ASPN 34, ATP6V0A4, BEX1, C6orf223, CARD17, CCDC9B, CCL2, CCL20, CD99L2, CNIH3, CNTN3, COL1A1, COL6A6, CRYBB2P1, CXCL10, CXCL11, CXCL5, CXCL6, CYP1A1, DAAM2, DLK1, ECHDC1, ELN, FIBIN, FMOD, FNDC1, G0S2, GALNT14, GALNT16, HLA-DRB1, HMGB3, HS3ST2, HSD17B2, IER3, IFI27, IFI44, IL1A, IL1B, IL7R, INHBA, INSYN2B, ITGA10, ITGBL1, KRT6B, LAMB3, LIF, LINC00520, LINC-PINT, LOC100996732, LOX14, LSP1P4, LSP1P5, MANCR, MFAP4, MT1L, MX2, NFIX, NOV, NPAS2, NPIPB12, NPIPB13, NPIPB3, NPIPB5, NUPR1, LFML2B, PCDHA6, PDGFRB, PLD5, POSTN, PTGS1, RTEL1-TNFRSF6B, SAA1, SAA2, SAA2-SAA4, SERPINB2, SFRP2, SLC9A7, SMOC1, SMOC2, ST6GAL2, SYNDIG1, TAC1, TAGLN, TFCP2L1, TMEM255B, TNFRSF6B, TNMD, TPST1, TRNP1, VNN1, WIPI1, XDH | RNA seq | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bchetnia, M.; Powell, J.; McCuaig, C.; Boucher-Lafleur, A.-M.; Morin, C.; Dupéré, A.; Laprise, C. Pathological Mechanisms Involved in Epidermolysis Bullosa Simplex: Current Knowledge and Therapeutic Perspectives. Int. J. Mol. Sci. 2024, 25, 9495. https://doi.org/10.3390/ijms25179495
Bchetnia M, Powell J, McCuaig C, Boucher-Lafleur A-M, Morin C, Dupéré A, Laprise C. Pathological Mechanisms Involved in Epidermolysis Bullosa Simplex: Current Knowledge and Therapeutic Perspectives. International Journal of Molecular Sciences. 2024; 25(17):9495. https://doi.org/10.3390/ijms25179495
Chicago/Turabian StyleBchetnia, Mbarka, Julie Powell, Catherine McCuaig, Anne-Marie Boucher-Lafleur, Charles Morin, Audrey Dupéré, and Catherine Laprise. 2024. "Pathological Mechanisms Involved in Epidermolysis Bullosa Simplex: Current Knowledge and Therapeutic Perspectives" International Journal of Molecular Sciences 25, no. 17: 9495. https://doi.org/10.3390/ijms25179495
APA StyleBchetnia, M., Powell, J., McCuaig, C., Boucher-Lafleur, A.-M., Morin, C., Dupéré, A., & Laprise, C. (2024). Pathological Mechanisms Involved in Epidermolysis Bullosa Simplex: Current Knowledge and Therapeutic Perspectives. International Journal of Molecular Sciences, 25(17), 9495. https://doi.org/10.3390/ijms25179495





