Moonlighting Crypto-Enzymes and Domains as Ancient and Versatile Signaling Devices
Abstract
1. What Are Crypto-Enzymes
2. Uncovering Crypto-Enzymes in Moonlighting Proteins—Plant Guanylate Cyclases as a Case in Point
3. Cryptic Moonlighting Enzymes in Animal Proteomes—Uncovering GCs in Humans
4. Structural and Functional Aspects of Cryptic Mononucleotide Cyclases
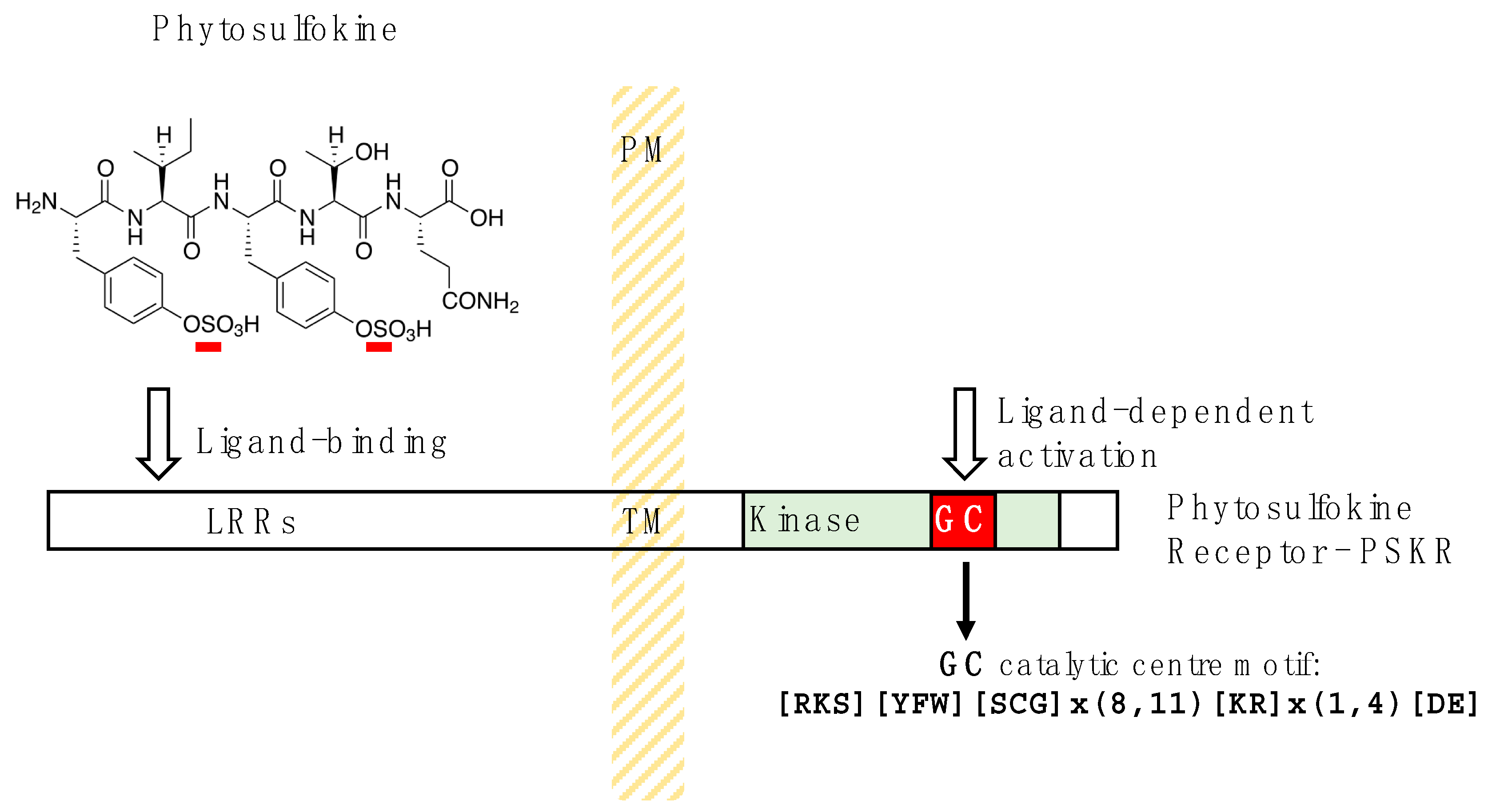
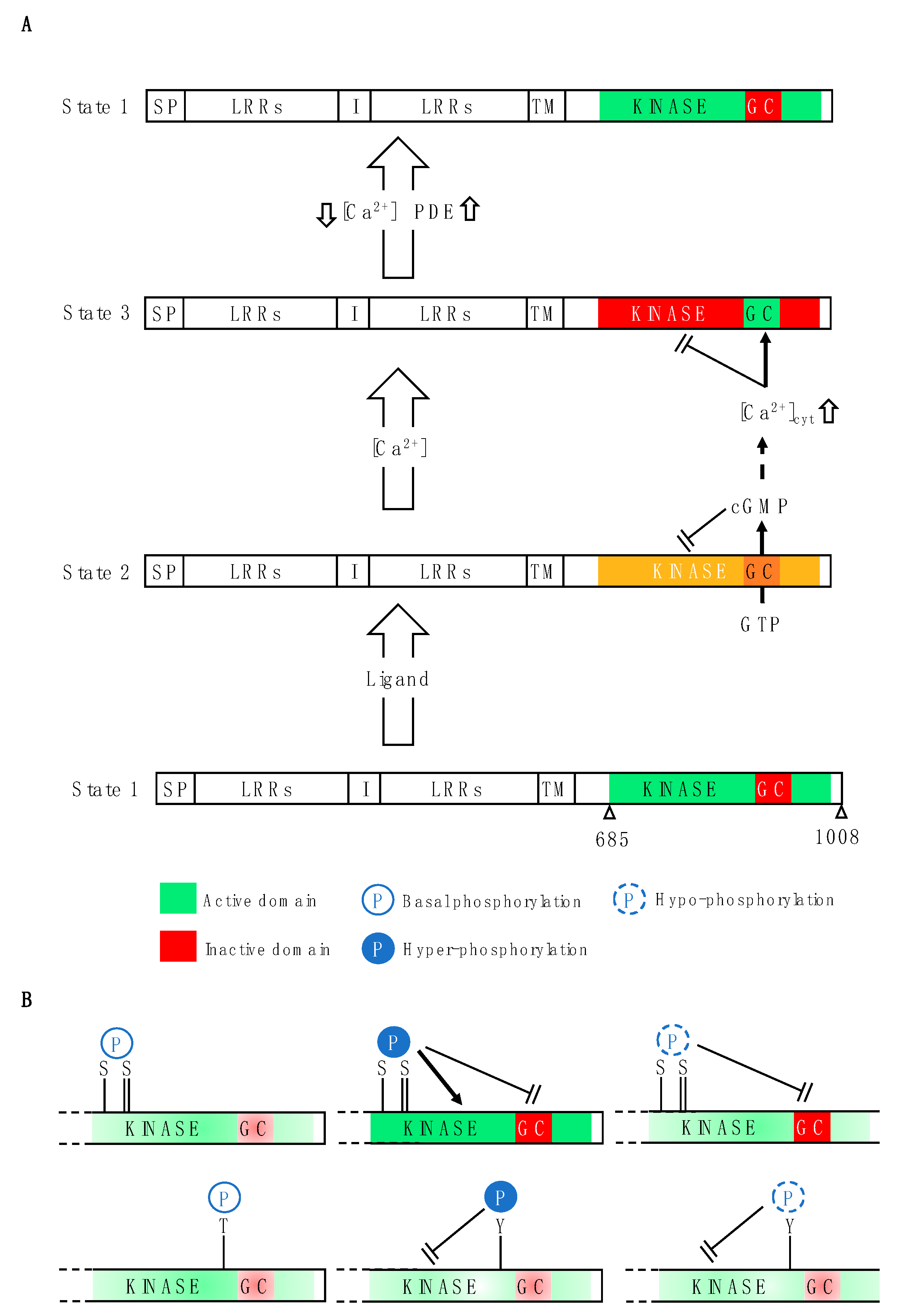
5. Phytosulfokine Receptors Harbor Hidden Nucleotide Cyclases
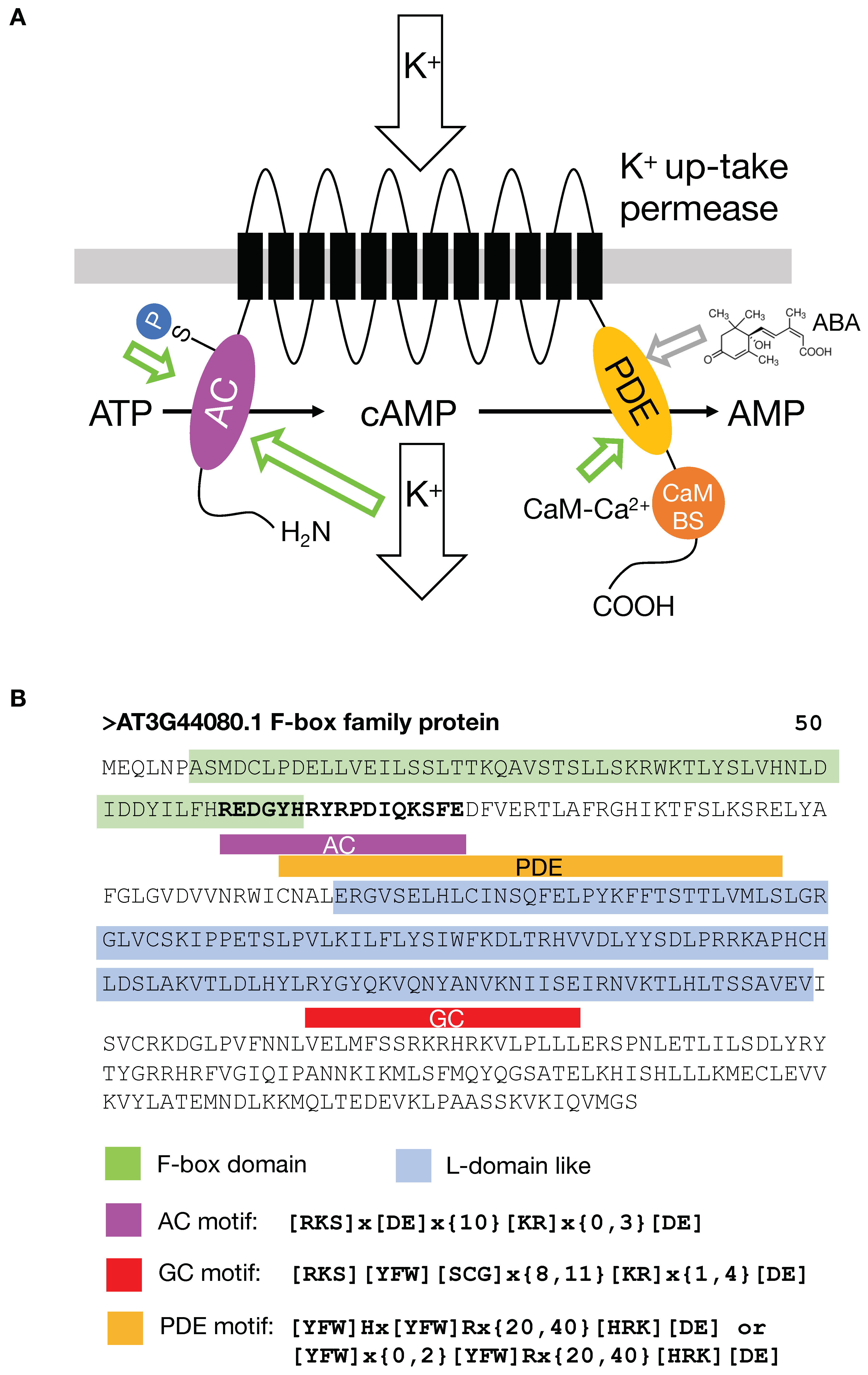
6. A K+ Channel with Multiple Crypto-Domains
7. Cryptic Mononucleotide Cyclases Can Tune Signal Networks
8. Ancient Mononucleotide Cyclases
9. Finding Conserved Hidden Functions beyond Crypto-Enzymes
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Previero, A.; Coletti-Previero, M.A.; Galzigna, L. Cryptic functions of enzymes in chemical catalysis. Monatshefte Für Chem./Chem. Mon. 1983, 114, 1059–1069. [Google Scholar] [CrossRef]
- Ludidi, N.N.; Gehring, C. Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 6490–6494. [Google Scholar] [CrossRef] [PubMed]
- Varfolomeev, S.D. Catalytic centres of enzymes: Structural paradoxes, the phenomenon of structural unity and new reactions. Mendeleev Commun. 2004, 14, 185–188. [Google Scholar] [CrossRef]
- Al-Younis, I.; Moosa, B.; Kwiatkowski, M.; Jaworski, K.; Wong, A.; Gehring, C. Functional crypto-adenylate cyclases operate in complex plant proteins. Front. Plant Sci. 2021, 12, 711749. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Seoighe, C.; Kwezi, L.; Irving, H.; Gehring, C. Plant nucleotide cyclases: An increasingly complex and growing family. Plant Signal. Behav. 2007, 2, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Gehring, C.; Irving, H.R. Conserved functional motifs and homology modeling to predict hidden moonlighting functional sites. Front. Bioengin. Biotech. 2015, 3, 82. [Google Scholar] [CrossRef]
- Zhou, W.; Chi, W.; Shen, W.; Dou, W.; Wang, J.; Tian, X.; Gehring, C.; Wong, A. Computational identification of functional centers in complex proteins: A step-by-step guide with examples. Front. Bioinform. 2021, 1, 652286. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Axell, A.; Turek, I.; Wright, B.; Meehan-Andrews, T.; Irving, H.R. Modulation of inflammatory cytokine production in human monocytes by cGMP and IRAK3. Int. J. Mol. Sci. 2022, 23, 2552. [Google Scholar] [CrossRef]
- Ruzvidzo, O.; Gehring, C.; Wong, A. New Perspectives on Plant Adenylyl Cyclases. Front. Mol. Biosci. 2019, 6, 136. [Google Scholar] [CrossRef]
- Irving, H.R.; Kwezi, L.; Wheeler, J.I.; Gehring, C. Moonlighting kinases with guanylate cyclase activity can tune regulatory signal networks. Plant Signal. Behav. 2012, 7, 201–204. [Google Scholar] [CrossRef]
- Kwezi, L.; Wheeler, J.I.; Marondedze, C.; Gehring, C.; Irving, H.R. Intramolecular crosstalk between catalytic activities of receptor kinases. Plant Signal. Behav. 2018, 13, e1430544. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Kwiatkowski, M.; Chen, H.; Hoermayer, L.; Sinclair, S.; Zou, M.; del Genio, C.I.; Kubeš, M.F.; Napier, R.; Jaworski, K.; et al. Adenylate cyclase activity of TIR1/AFB auxin receptors in plants. Nature 2022, 611, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Moonlighting proteins. Trends Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. An introduction to protein moonlighting. Biochem. Soc. Trans. 2014, 42, 1679–1683. [Google Scholar] [CrossRef]
- Irving, H.R.; Cahill, D.M.; Gehring, C. Moonlighting proteins and their role in the control of signalling microenvironments, as exemplified by cGMP and phytosulfokine receptor 1 (PSKR1). Front. Plant Sci. 2018, 9, 415. [Google Scholar] [CrossRef]
- Turek, I.; Irving, H. Moonlighting proteins shine new light on molecular signaling niches. Int. J. Mol. Sci. 2021, 22, 1367. [Google Scholar] [CrossRef]
- Zeugolis, D.I. Bioinspired in vitro microenvironments to control cell fate: Focus on macromolecular crowding. Am. J. Physiol. Cell Physiol. 2021, 320, C842–C849. [Google Scholar] [CrossRef]
- Domingo, G.; Marsoni, M.; Chiodaroli, L.; Fortunato, S.; Bracale, M.; De Pinto, M.C.; Gehring, C.; Vannini, C. Quantitative phosphoproteomics reveals novel roles of cAMP in plants. Proteomics 2023, 23, 2300165. [Google Scholar] [CrossRef]
- Marondedze, C.; Groen, A.; Thomas, L.; Lilley, K.S.; Gehring, C. A quantitative phosphoproteome analysis of cGMP-dependent cellular responses in Arabidopsis thaliana. Mol. Plant 2016, 9, 621–623. [Google Scholar] [CrossRef]
- Newton, R.P.; Roef, L.; Witters, E.; van Onckelen, H. Tansley review no. 106—Cyclic nucleotides in higher plants: The enduring paradox. New Phytol. 1999, 143, 427–455. [Google Scholar] [CrossRef]
- Kwiatkowski, M.; Zhang, J.; Zhou, W.; Gehring, C.; Wong, A. Cyclic nucleotides—The rise of a family. Trends Plant Sci. 2024, 29, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Wong, A.; Thomas, L.; Irving, H.; Gehring, C. Cyclic nucleotide monophosphates in plants and plant signaling. Handb. Exp. Pharmacol. 2017, 238, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.P.; Kingston, E.E.; Evans, D.E.; Younis, L.M.; Brown, E.G. Occurrence of guanosine 3′,5′-cyclic monophosphate (cyclic GMP) and associated enzyme systems in Phaseolus vulgaris. Phytochemistry 1984, 23, 1367–1372. [Google Scholar] [CrossRef]
- Newton, R.P.; Smith, C.J. Cyclic nucleotides. Phytochemistry 2004, 65, 2423–2437. [Google Scholar] [CrossRef]
- Penson, S.P.; Schuurink, R.C.; Fath, A.; Gubler, F.; Jacobsen, J.V.; Jones, R.L. cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 1996, 8, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef]
- Moutinho, A.; Hussey, P.J.; Trewavas, A.J.; Malhó, R. cAMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. USA 2001, 98, 10481–10486. [Google Scholar] [CrossRef]
- Liang, S.; Sun, J.; Luo, Y.; Lv, S.; Chen, J.; Liu, Y.; Hu, X. cAMP is a promising regulatory molecule for plant adaptation to heat stress. Life 2022, 12, 885. [Google Scholar] [CrossRef]
- Xu, R.; Guo, Y.; Peng, S.; Liu, J.; Li, P.; Jia, W.; Zhao, J. Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis. Biomolecules 2021, 11, 688. [Google Scholar] [CrossRef]
- Qi, L.; Friml, J. Tale of cAMP as a second messenger in auxin signaling and beyond. New Phytol. 2023, 240, 489–495. [Google Scholar] [CrossRef]
- Lomovatskaya, L.A.; Romanenko, A.S.; Filinova, N.V. Plant adenylate cyclases. J. Recept. Signal Transduct. 2008, 28, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, N.E.; Franz, P.; Luzarowski, M.; Martinez-Seidel, F.; Moreno, J.C.; Childs, D.; Ziemblicka, A.; Sampathkumar, A.; Andersen, T.G.; Tsiavaliaris, G.; et al. Protein interactome of 3′,5′-cAMP reveals its role in regulating the actin cytoskeleton. Plant J. 2023, 115, 1214–1230. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, T. Regulation of voltage dependence of the KAT1 channel by intracellular factors. J. Gen. Physiol. 1995, 105, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.; Sanders, D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001, 127, 1617–1625. [Google Scholar] [CrossRef]
- Li, W.; Luan, S.; Schreiber, S.L.; Assmann, S.M. Cyclic AMP stimulates K+ channel activity in mesophyll cells of Vicia faba L. Plant Physiol. 1994, 106, 957–961. [Google Scholar] [CrossRef]
- Gehring, C.A.; Irving, H.R. Natriuretic peptides—A class of heterologous molecules in plants. Int. J. Biochem. Cell Biol. 2003, 35, 1318–1322. [Google Scholar] [CrossRef]
- Pharmawati, M.; Billington, T.; Gehring, C.A. Stomatal guard cell responses to kinetin and natriuretic peptides are cGMP dependent. Cell. Mol. Life Sci. 1998, 54, 272–276. [Google Scholar] [CrossRef]
- Pharmawati, M.; Maryani, M.M.; Nikolakopoulos, T.; Gehring, C.A.; Irving, H.R. Cyclic GMP modulates stomatal opening induced by natriuretic peptides and immunoreactive analogues. Plant Physiol. Biochem. 2001, 39, 385–394. [Google Scholar] [CrossRef]
- Bowler, C.; Neuhaus, G.; Yamagata, H.; Chua, N.H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 1994, 77, 73–81. [Google Scholar] [CrossRef]
- Bowler, C.; Yamagata, H.; Neuhaus, G.; Chua, N.H. Phytochrome signal transduction pathways are regulated by reciprocal control mechanisms. Genes Dev. 1994, 8, 2188–2202. [Google Scholar] [CrossRef]
- Quail, P.H. Photosensory perception and signal transduction in plants. Curr. Opin. Genet. Dev. 1994, 4, 652–661. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ruoho, A.E.; Rao, V.D.; Hurley, J.H. Catalytic mechanism of the adenylyl and guanylyl cyclases: Modeling and mutational analysis. Proc. Nat. Acad. Sci. USA 1997, 94, 13414–13419. [Google Scholar] [CrossRef] [PubMed]
- McCue, L.A.; McDonough, K.A.; Lawrence, C.E. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 2000, 10, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.J.; Hurley, J.H. Catalytic mechanism and regulation of mammalian adenylyl cyclases. Mol. Pharmacol. 1998, 54, 231–240. [Google Scholar] [CrossRef]
- Thompson, D.K.; Garbers, D.L. Dominant negative mutations of the guanylyl cyclase-A receptor. Extracellular domain deletion and catalytic domain point mutations J. Biol. Chem. 1995, 270, 425–430. [Google Scholar] [CrossRef]
- Thorpe, D.S.; Morkin, E. The carboxyl region contains the catalytic domain of the membrane form of guanylate cyclase. J. Biol. Chem. 1990, 265, 14717–14720. [Google Scholar] [CrossRef]
- Wedel, B.J.; Garbers, D.L. New insights on the functions of the guanylyl cyclase receptors. FEBS Lett. 1997, 410, 29–33. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Kwezi, L.; Meier, S.; Mungur, L.; Ruzvidzo, O.; Irving, H.; Gehring, C. The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS ONE 2007, 2, e449. [Google Scholar] [CrossRef]
- Kwezi, L.; Ruzvidzo, O.; Wheeler, J.I.; Govender, K.; Iacuone, S.; Thompson, P.E.; Gehring, C.; Irving, H.R. The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependant signaling in plants. J. Biol. Chem. 2011, 286, 22580–22588. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Ruzvidzo, O.; Morse, M.; Donaldson, L.; Kwezi, L.; Gehring, C. The Arabidopsis Wall Associated Kinase-Like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE 2010, 5, e8904. [Google Scholar] [CrossRef]
- Wong, A.; Gehring, C. The Arabidopsis thaliana proteome harbors undiscovered multi-domain molecules with functional guanylyl cyclase catalytic centers. Cell Commun. Signal. 2013, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Tian, X.; Gehring, C.; Marondedze, C. Discovery of novel functional centers with rationally designed amino acid motifs. Comput. Struct. Biotechnol. J. 2018, 16, 70–76. [Google Scholar] [CrossRef]
- Freihat, L.; Muleya, V.; Manallack, D.T.; Wheeler, J.I.; Irving, H.R. Comparison of moonlighting guanylate cyclases—Roles in signal direction? Biochem. Soc. Trans. 2014, 42, 1773–1779. [Google Scholar] [CrossRef]
- Turek, I.; Freihat, L.; Vyas, J.; Wheeler, J.; Muleya, V.; Manallack, D.T.; Gehring, C.; Irving, H. The discovery of hidden guanylate cyclases (GCs) in the Homo sapiens proteome. Comput. Struct. Biotech. J. 2023, 21, 5523–5529. [Google Scholar] [CrossRef] [PubMed]
- Goetze, J.P.; Bruneau, B.G.; Ramos, H.R.; Ogawa, T.; Kuroski de Bold, M.; de Bold, A.J. Cardiac natriuretic peptides. Nat. Rev. Cardiol. 2020, 17, 698–717. [Google Scholar] [CrossRef]
- Freihat, L.A.; Wheeler, J.I.; Wong, A.; Turek, I.; Manallack, D.T.; Irving, H.R. IRAK3 modulates downstream innate immune signalling through its guanylate cyclase activity. Sci. Rep. 2019, 9, 15468. [Google Scholar] [CrossRef]
- Turek, I.; Nguyen, T.H.; Galea, C.; Abad, I.; Freihat, L.; Manallack, D.T.; Velkov, T.; Irving, H. Mutations in the Vicinity of the IRAK3 Guanylate Cyclase Center Impact Its Subcellular Localization and Ability to Modulate Inflammatory Signaling in Immortalized Cell Lines. Int. J. Mol. Sci. 2023, 24, 8572. [Google Scholar] [CrossRef]
- Eigenthaler, M.; Lohmann, S.M.; Walter, U.; Pilz, R.B. Signal transduction by cGMP-dependent protein kinases and their emerging roles in the regulation of cell adhesion and gene expression. In Reviews of Physiology, Biochemistry and Pharmacology, Volume 135; Springer: Berlin/Heidelberg, Germany, 1999; pp. 173–209. [Google Scholar]
- Wong, A.; Chi, W.; Yu, J.; Bi, C.; Tian, X.; Yang, Y.; Gehring, C. Plant adenylate cyclases have come full circle. Nat. Plants 2023, 9, 1389–1397. [Google Scholar] [CrossRef]
- Düner, M.; Lambertz, J.; Mügge, C.; Hemschemeier, A. The soluble guanylate cyclase CYG12 is required for the acclimation to hypoxia and trophic regimes in Chlamydomonas reinhardtii. Plant J. 2018, 93, 311–337. [Google Scholar] [CrossRef] [PubMed]
- Linder, J.U.; Schultz, J.E. Guanylyl cyclases in unicellular organisms. Mol. Cell. Biochem. 2002, 230, 149–158. [Google Scholar] [CrossRef]
- Bianchet, C.; Wong, A.; Quaglia, M.; Alqurashi, M.; Gehring, C.; Ntoukakis, V.; Pasqualini, S. An Arabidopsis thaliana leucine-rich repeat protein harbors an adenylyl cyclase catalytic center and affects responses to pathogens. J. Plant Physiol. 2019, 232, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Man, J.; Harrington, T.A.; Lally, K.; Bartlett, M.E. Asymmetric evolution of protein domains in the leucine-rich repeat receptor-like kinase family of plant signaling proteins. Mol. Biol. Evol. 2023, 40, msad220. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Yang, J.; Peng, M.; Liu, X.; He, H. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Medicago truncatula. Life 2020, 10, 176. [Google Scholar] [CrossRef]
- Wheeler, J.I.; Wong, A.; Marondedze, C.; Groen, A.J.; Kwezi, L.; Freihat, L.; Vyas, J.; Raji, M.R.; Irving, H.R.; Gehring, C. The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant J. 2017, 91, 590–600. [Google Scholar] [CrossRef]
- Clouse, S.D. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 2011, 23, 1219–1230. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef]
- Muleya, V.; Marondedze, C.; Wheeler, J.I.; Thomas, L.; Mok, Y.F.; Griffin, M.W.D.; Manallack, D.T.; Kwezi, L.; Lilley, K.S.; Gehring, C.; et al. Phosphorylation of the dimeric cytoplasmic domain of the phytosulfokine receptor, PSKR1. Biochem. J. 2016, 473, 3081–3098. [Google Scholar] [CrossRef]
- Muleya, V.; Wheeler, J.I.; Ruzvidzo, O.; Freihat, L.; Manallack, D.T.; Gehring, C.; Irving, H.R. Calcium is the switch in the moonlighting dual function of the ligand-activated receptor kinase phytosulfokine receptor 1. Cell Commun. Signal. 2014, 12, 60. [Google Scholar] [CrossRef]
- Sauter, M. Phytosulfokine peptide signalling. J. Exp. Bot. 2015, 66, 5161–5169. [Google Scholar] [CrossRef]
- Qi, Z.; Verma, R.; Gehring, C.; Yamaguchi, Y.; Zhao, Y.; Ryan, C.A.; Berkowitz, G.A. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Nat. Acad. Sci. USA 2010, 107, 21193–21198. [Google Scholar] [CrossRef] [PubMed]
- Turek, I.; Gehring, C. The Plant Natriuretic Peptide receptor is a guanylyl cyclase and enables cGMP-dependent signaling. Plant Mol. Biol. 2016, 91, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Dhusia, K.; Wu, Y. Understand the functions of scaffold proteins in cell signaling by a mesoscopic simulation method. Biophys. J. 2020, 119, 2116–2126. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Lee, I.-B.; Moon, H.-M.; Hong, S.-C.; Cho, M. Long-term cargo tracking reveals intricate trafficking through active cytoskeletal networks in the crowded cellular environment. Nat. Commun. 2023, 14, 7160. [Google Scholar] [CrossRef]
- Kwiatkowski, M.; Wong, A.; Bi, C.; Gehring, C.; Jaworski, K. Twin cyclic mononucleotide cyclase and phosphodiesterase domain architecture as a common feature in complex plant proteins. Plant Sci. 2022, 325, 111493. [Google Scholar] [CrossRef]
- Kwiatkowski, M.; Wong, A.; Kozakiewicz, A.; Gehring, C.; Jaworski, K. A tandem motif-based and structural approach can identify hidden functional phosphodiesterases. Comput. Struct. Biotechnol. J. 2021, 19, 970–975. [Google Scholar] [CrossRef]
- Isner, J.C.; Olteanu, V.A.; Hetherington, A.J.; Coupel-Ledru, A.; Sun, P.; Pridgeon, A.J.; Jones, G.S.; Oates, M.; Williams, T.A.; Maathuis, F.J.M.; et al. Short- and long-term effects of UVA on Arabidopsis are mediated by a novel cGMP phosphodiesterase. Curr. Biol. 2019, 29, 2580–2585.e4. [Google Scholar] [CrossRef]
- Gehring, C.A.; Irving, H.R.; Parish, R.W. Effects of auxin and abscisic acid on cytosolic calcium and pH in plant cells. Proc. Nat. Acad. Sci. USA 1990, 87, 9645–9649. [Google Scholar] [CrossRef]
- Irving, H.R.; Gehring, C.A.; Parish, R.W. Changes in cytosolic pH and calcium of guard cells precede stomatal movements. Proc. Nat. Acad. Sci. USA 1992, 89, 1790–1794. [Google Scholar] [CrossRef]
- Kader, M.A.; Lindberg, S. Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 2010, 5, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y.; Sakagami, Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 1996, 93, 7623–7627. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Matsubayashi, Y.; Hanai, H.; Sakagami, Y. Phytosulfokine-alpha, a peptide growth factor found in higher plants: Its structure, functions, precursor and receptors. Plant Cell Physiol. 2000, 41, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y.; Sakagami, Y. 120- and 160-kDa receptors for endogenous mitogenic peptide, phytosulfokine-α, in rice plasma membranes. J. Biol. Chem. 2000, 275, 15520–15525. [Google Scholar] [CrossRef]
- Ladwig, F.; Dahlke, R.I.; Stuhrwohdldt, N.; Hartmann, J.; Harter, K.; Sauter, M. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H-ATPase, and BAK1. Plant Cell 2015, 27, 1718–1729. [Google Scholar] [CrossRef]
- He, K.; Xu, S.; Li, J. BAK1 directly regulates brassinosteroid perception and BRI1 activation. J. Integr. Plant Biol. 2013, 55, 1264–1270. [Google Scholar] [CrossRef]
- Suwastika, I.N.; Gehring, C.A. The plasma-membrane H+-ATPase from Tradescantia stem and leaf tissue is modulated in vitro by cGMP. Arch. Biochem. Biophys. 1999, 367, 137–139. [Google Scholar] [CrossRef]
- Hartmann, J.; Linke, D.; Bönniger, C.; Tholey, A.; Sauter, M. Conserved phosphorylation sites in the activation loop of Arabidopsis phytosulokine receptor PSKR1 differentially affect kinase and receptor activity. Biochem. J. 2015, 472, 379–391. [Google Scholar] [CrossRef]
- Al-Younis, I.; Wong, A.; Lemtiri-Chlieh, F.; Schmöckel, S.; Tester, M.; Gehring, C.; Donaldson, L. The Arabidopsis thaliana K+-Uptake Permease 5 (AtKUP5) contains a functional cytosolic adenylate cyclase essential for K+ transport. Front. Plant Sci. 2018, 9, 1645. [Google Scholar] [CrossRef]
- Gehring, C. Adenyl cyclases and cAMP in plant signaling—Past and present. Cell Commun. Signal. 2010, 8, 15. [Google Scholar] [CrossRef]
- Zelman, A.; Dawe, A.; Berkowitz, G.; Gehring, C. Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 2012, 3, 95. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Zhao, C.; Tang, Y.; Wang, J.; Zeng, Y.; Sun, H.; Zheng, Z.; Su, R.; Schneeberger, K.; Parker, J.E.; Cui, H. A mis-regulated cyclic nucleotide-gated channel mediates cytosolic calcium elevation and activates immunity in Arabidopsis. New Phytol. 2021, 230, 1078–1094. [Google Scholar] [CrossRef]
- Ma, Y.; Garrido, K.; Ali, R.; Berkowitz, G.A. Phenotypes of cyclic nucleotide-gated cation channel mutants: Probing the nature of native channels. Plant J. 2023, 113, 1223–1236. [Google Scholar] [CrossRef]
- Kipreos, E.T.; Pagano, M. The F-box protein family. Genome Biol. 2000, 1, reviews3002.3001. [Google Scholar] [CrossRef]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Tian, X.; Yang, Y.; Gehring, C. Adenylate cyclase activity of TIR1/AFB links cAMP to auxin-dependent responses. Mol. Plant 2022, 15, 1838–1840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 62, 301–341. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Conde, J.A.; Sans-Coll, G.; Merchante, C. RNA-binding proteins and their role in translational regulation in plants. Essays Biochem. 2022, 66, 87–97. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, M.; Manley, J.L. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 2008, 15, 1040–1048. [Google Scholar] [CrossRef]
- Lipp, J.J.; Marvin, M.C.; Shokat, K.M.; Guthrie, C. SR protein kinases promote splicing of nonconsensus introns. Nat. Struct. Mol. Biol. 2015, 22, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Balagué, C.; Lin, B.; Alcon, C.; Flottes, G.; Malmström, S.; Köhler, C.; Neuhaus, G.; Pelletier, G.; Gaymard, F.; Roby, D. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide–gated channel ion channel family. Plant Cell 2003, 15, 365–379. [Google Scholar] [CrossRef]
- Bassler, J.; Schultz, J.E.; Lupas, A.N. Adenylate cyclases: Receivers, transducers, and generators of signals. Cell Signal. 2018, 46, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Isner, J.-C.; Maathuis, F.J.M. Measurement of cellular cGMP in plant cells and tissues using the endogenous fluorescent reporter FlincG. Plant J. 2011, 65, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Al-Younis, I.; Wong, A.; Gehring, C. The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett. 2015, 589, 3848–3852. [Google Scholar] [CrossRef]
- Kaufmann, C.; Motzkus, M.; Sauter, M. Phosphorylation of the phytosulokine peptide receptor PSKR1 controls receptor activity. J. Exp. Bot. 2017, 68, 1411–1423. [Google Scholar] [CrossRef]
- Sreeramulu, S.; Mostizky, Y.; Sunitha, S.; Shani, E.; Nahum, H.; Salomon, D.; Hayun, L.B.; Gruetter, C.; Rauh, D.; Ori, N.; et al. BSKs are partially redundant positive regulators of brassinosteriod signaling in Arabidopsis. Plant J. 2013, 74, 905–919. [Google Scholar] [CrossRef]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef]
- Donaldson, L.; Meier, S.; Gehring, C. The Arabidopsis cyclic nucleotide interactome. Cell Commun. Signal. 2016, 14, 10. [Google Scholar] [CrossRef]
- Gründling, A.; Lee, V.T. Old concepts, new molecules and current approaches applied to the bacterial nucleotide signalling field. Philos. Trans. R. Soc. B 2016, 371, 20150503. [Google Scholar] [CrossRef]
- Ryu, M.H.; Youn, H.; Kang, I.H.; Gomelsky, M. Identification of bacterial guanylate cyclases. Proteins 2015, 83, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Akhtar, M.; Min, W.; Bai, X.; Ma, T.; Liu, C. Domain of unknown function (DUF) proteins in plants: Function and perspective. Protoplasma 2024, 261, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.J.; Ryjenkov, D.A.; Gomelsky, M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J. Bacteriol. 2005, 187, 4774–4781. [Google Scholar] [CrossRef] [PubMed]
- Schaap, P. Guanylyl cyclases across the tree of life. Front. Biosci. 2005, 10, 1485–1498. [Google Scholar] [CrossRef]
- Mulaudzi, T.; Ludidi, N.; Ruzvidzo, O.; Morse, M.; Hendricks, N.; Iwuoha, E.; Gehring, C. Identification of a novel Arabidopsis thaliana nitric oxide-binding molecule with guanylate cyclase activity in vitro. FEBS Lett. 2011, 585, 2693–2697. [Google Scholar] [CrossRef]
- Ooi, A.; Lemtiri-Chlieh, F.; Wong, A.; Gehring, C. Direct modulation of the guard cell outward-rectifying potassium channel (GORK) by abscisic acid. Mol. Plant 2017, 10, 1469–1472. [Google Scholar] [CrossRef]
- Wong, A.; Bi, C.; Pasqualini, S.; Gehring, C. Abscisic acid (ABA) signaling: Finding novel components off the beaten track. Plant Growth Regul. 2022, 97, 585–592. [Google Scholar] [CrossRef]
- Wong, A.; Donaldson, L.; Portes, M.T.; Eppinger, J.; Feijó, J.A.; Gehring, C. Arabidopsis DIACYLGLYCEROL KINASE4 is involved in nitric oxide-dependent pollen tube guidance and fertilization. Development 2020, 147, dev183715. [Google Scholar] [CrossRef]
- Zarban, R.; Vogler, M.; Wong, A.; Eppinger, J.; Al-Babili, S.; Gehring, C. Discovery of a nitric oxide-Responsive protein in Arabidopsis thaliana. Molecules 2019, 24, 2691. [Google Scholar] [CrossRef]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijó, J.A. Nitric oxide: A multitasked signalling gas in plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [CrossRef]
- Wong, A.; Tian, X.; Yang, Y.; Gehring, C. Identification of potential nitric oxide-sensing proteins using the H-NOX motif. Mol. Plant 2021, 14, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Boon, E.M.; Huang, S.H.; Marletta, M.A. A molecular basis for NO selectivity in soluble guanylate cyclase. Nat. Chem. Biol. 2005, 1, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Liu, R.; Wu, J.-X.; Chen, L. Structural insights into the mechanism of human soluble guanylate cyclase. Nature 2019, 574, 206–210. [Google Scholar] [CrossRef]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.-M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Magnone, M.; Sturla, L.; Guida, L.; Spinelli, S.; Begani, G.; Bruzzone, S.; Fresia, C.; Zocchi, E. Abscisic Acid: A Conserved Hormone in Plants and Humans and a Promising Aid to Combat Prediabetes and the Metabolic Syndrome. Nutrients 2020, 12, 1724. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Cackowski, F.C.; Yumoto, K.; Decker, A.M.; Wang, Y.; Hotchkin, M.; Lee, E.; Buttitta, L.; Taichman, R.S. Abscisic acid regulates dormancy of prostate cancer disseminated tumor cells in the bone marrow. Neoplasia 2021, 23, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Lievens, L.; Pollier, J.; Goossens, A.; Beyaert, R.; Staal, J. Abscisic acid as pathogen effector and immune regulator. Front. Plant Sci. 2017, 8, 587. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turek, I.; Wong, A.; Domingo, G.; Vannini, C.; Bracale, M.; Irving, H.; Gehring, C. Moonlighting Crypto-Enzymes and Domains as Ancient and Versatile Signaling Devices. Int. J. Mol. Sci. 2024, 25, 9535. https://doi.org/10.3390/ijms25179535
Turek I, Wong A, Domingo G, Vannini C, Bracale M, Irving H, Gehring C. Moonlighting Crypto-Enzymes and Domains as Ancient and Versatile Signaling Devices. International Journal of Molecular Sciences. 2024; 25(17):9535. https://doi.org/10.3390/ijms25179535
Chicago/Turabian StyleTurek, Ilona, Aloysius Wong, Guido Domingo, Candida Vannini, Marcella Bracale, Helen Irving, and Chris Gehring. 2024. "Moonlighting Crypto-Enzymes and Domains as Ancient and Versatile Signaling Devices" International Journal of Molecular Sciences 25, no. 17: 9535. https://doi.org/10.3390/ijms25179535
APA StyleTurek, I., Wong, A., Domingo, G., Vannini, C., Bracale, M., Irving, H., & Gehring, C. (2024). Moonlighting Crypto-Enzymes and Domains as Ancient and Versatile Signaling Devices. International Journal of Molecular Sciences, 25(17), 9535. https://doi.org/10.3390/ijms25179535








