Differential Expression of Mitogen-Activated Protein Kinase Signaling Pathways in the Human Choroid–Retinal Pigment Epithelial Complex Indicates Regional Predisposition to Disease
Abstract
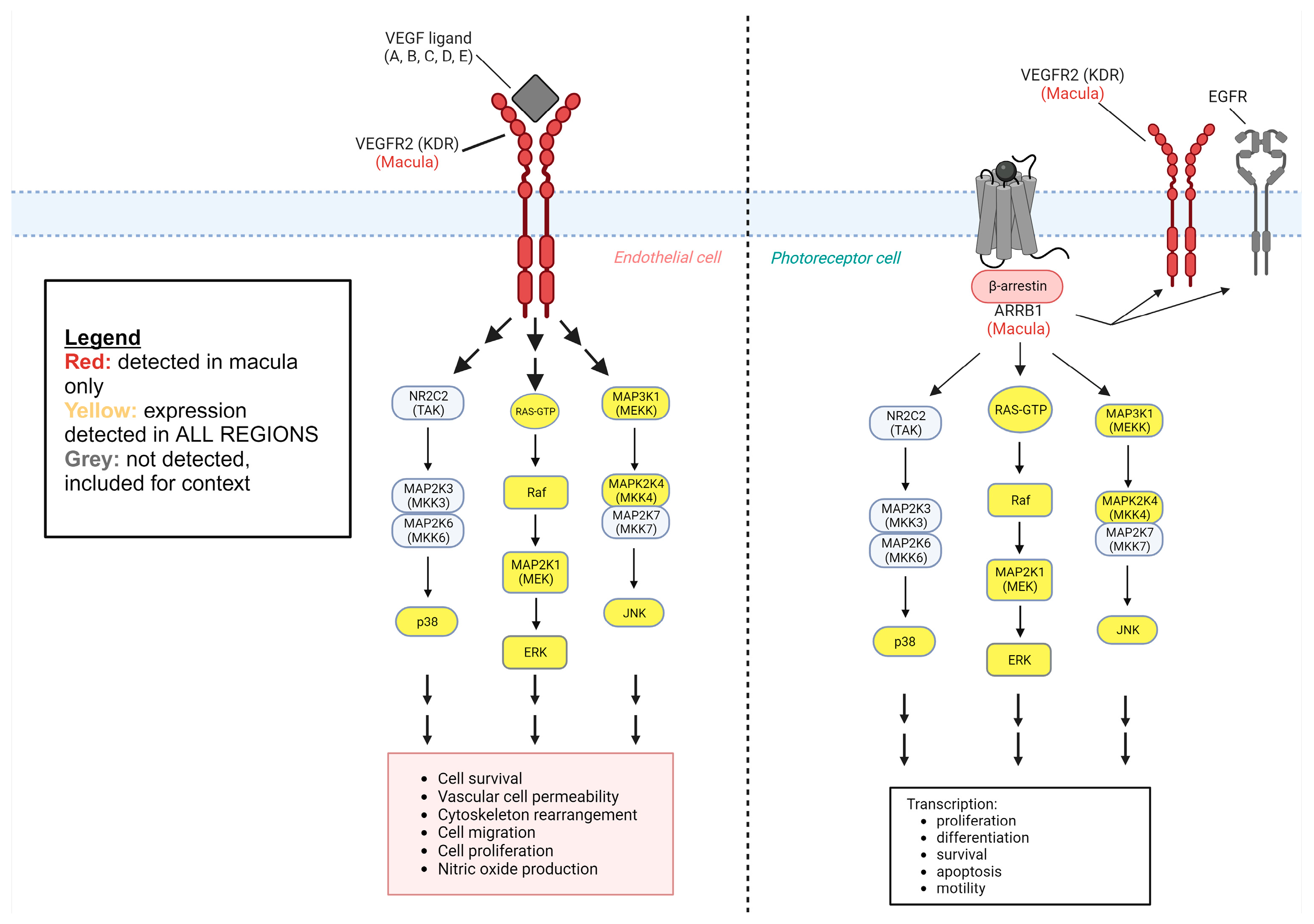
1. Introduction
2. Results
3. Discussion
3.1. Regionally Presenting Retinal Disease
3.2. Downstream Proteins
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Purves, D.; Augustine, G.J.; Fitzpatrick, D. (Eds.) Sunderland (MA). In Neuroscience, 2nd ed.; The Retina; Sinauer Associates: Sunderland, MA, USA, 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK10885/ (accessed on 9 October 2023).
- Hurley, J.B. Retina Metabolism and Metabolism in the Pigmented Epithelium: A Busy Intersection. Annu. Rev. Vis. Sci. 2021, 7, 665–692. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lakkaraju, A. Endo-lysosome function in the retinal pigment epithelium in health and disease. Adv. Exp. Med. Biol. 2012, 723, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, N.; Terasaki, H.; Sonoda, S.; Shiihara, H.; Yamashita, T.; Tomita, M.; Shinohara, Y.; Sakoguchi, T.; Iwata, K.; Sakamoto, T. Regional differences of choroidal structure determined by wide-field optical coherence tomography. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2614–2622. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Notomi, S.; Shiose, S.; Kano, K.; Hashimoto, S.; Fujiwara, K.; Akiyama, M.; Ishikawa, K.; Hisatomi, T.; Sonoda, K.-H. Differences in Central and Peripheral Choroidal Thickness among the Subtypes of Age-Related Macular Degeneration in an Asian Population. J. Clin. Med. 2023, 12, 5364. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, J.; Usategui-Martin, R.; Sanabria, M.R.; Fernandez-Perez, E.; Telleria, J.J.; Coco-Martin, R.M. Pathophysiology of Age-Related Macular Degeneration: Implications for Treatment. Ophthalmic Res. 2022, 65, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Nan, G. The Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway as a Discovery Target in Stroke. J. Mol. Neurosci. 2016, 59, 90–98, Erratum in J. Mol. Neurosci. 2016, 59, 430. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Karin, M. Mammalian MAP kinase signalling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Garg, T.K.; Chang, J.Y. Oxidative stress causes ERK phosphorylation and cell death in cultured retinal pigment epithelium: Prevention of cell death by AG126 and 15-deoxy-delta 12, 14-PGJ2. BMC Ophthalmol. 2003, 3, 5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skeie, J.M.; Mahajan, V.B. Proteomic landscape of the human choroid–retinal pigment epithelial complex. JAMA Ophthalmol 2014, 132, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.-L.; Xue, B.; Jin, D.-Z.; Mao, L.-M.; Wang, J.Q. Interactions and phosphorylation of postsynaptic density 93 (PSD-93) by extracellular signal-regulated kinase (ERK). Brain Res. 2012, 1465, 18–25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Siaw, J.T.; Javanmardi, N.; Eynden, J.V.D.; Lind, D.E.; Fransson, S.; Martinez-Monleon, A.; Djos, A.; Sjöberg, R.-M.; Östensson, M.; Carén, H.; et al. 11q Deletion or ALK Activity Curbs DLG2 Expression to Maintain an Undifferentiated State in Neuroblastoma. Cell Rep. 2020, 32, 108171. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Yang, H.; Liu, E.; Wang, H. P38/ERK MAPK signaling pathways are involved in the regulation of filaggrin and involucrin by IL-17. Mol. Med. Rep. 2017, 16, 8863–8867. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meng, X.; Qiu, L.; Song, H.; Dang, N. MAPK Pathway Involved in Epidermal Terminal Differentiation of Normal Human Epidermal Keratinocytes. Open Med. 2018, 13, 189–195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hong, Z.; Cabrera, J.A.; Mahapatra, S.; Kutty, S.; Weir, E.K.; Archer, S.L. Activation of the EGFR/p38/JNK pathway by mitochondrial-derived hydrogen peroxide contributes to oxygen-induced contraction of ductus arteriosus. J. Mol. Med. 2014, 92, 995–1007. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oh, M.-J.; van Agthoven, T.; Choi, J.-E.; Jeong, Y.-J.; Chung, Y.-H.; Kim, C.-M.; Jhun, B.H. BCAR3 regulates EGF-induced DNA synthesis in normal human breast MCF-12A cells. Biochem. Biophys. Res. Commun. 2008, 375, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Sakatsume, M.; Stancato, L.F.; David, M.; Silvennoinen, O.; Saharinen, P.; Pierce, J.; Larner, A.C.; Finbloom, D.S. Interferonγ Activation of Raf-1 Is Jak1-dependent and p21ras-independent. J. Biol. Chem. 1998, 273, 3021–3026. [Google Scholar] [CrossRef] [PubMed]
- Linnekin, D. Early signaling pathways activated by c-Kit in hematopoietic cells. Int. J. Biochem. Cell Biol. 1999, 31, 1053–1074. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Niethammer, M.; Kim, E.; Sheng, M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J. Neurosci. 1996, 16, 2157–2163. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dakoji, S.; Tomita, S.; Karimzadegan, S.; Nicoll, R.A.; Bredt, D.S. Interaction of transmembrane AMPA receptor regulatory proteins with multiple membrane associated guanylate kinases. Neuropharmacology 2003, 45, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Inanobe, A.; Fujita, A.; Ito, M.; Tomoike, H.; Inageda, K.; Kurachi, Y. Inward rectifier K+ channel Kir2.3 is localized at the postsynaptic membrane of excitatory synapses. Am. J. Physiol. Physiol. 2002, 282, C1396–C1403. [Google Scholar] [CrossRef] [PubMed]
- Steinert, P.M.; Marekov, L.N. The Proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J. Biol. Chem. 1995, 270, 17702–17711. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-Y.; Finley, J.C.; Ali, S.S.; Patel, H.H.; Howell, S.B. Copper influx transporter 1 is required for FGF, PDGF and EGF-induced MAPK signaling. Biochem. Pharmacol. 2012, 84, 1007–1013. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Buitenhuis, M.; Verhagen, L.P.; Cools, J.; Coffer, P.J. Molecular mechanisms underlying FIP1L1-PDGFRA–mediated myeloproliferation. Cancer Res. 2007, 67, 3759–3766. [Google Scholar] [CrossRef] [PubMed]
- Mahe, M.; Dufour, F.; Neyret-Kahn, H.; Moreno-Vega, A.; Beraud, C.; Shi, M.; Hamaidi, I.; Sanchez-Quiles, V.; Krucker, C.; Dorland-Galliot, M.; et al. An FGFR 3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol. Med. 2018, 10, e8163. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gee, E.; Milkiewicz, M.; Haas, T.L. p38 MAPK activity is stimulated by vascular endothelial growth factor receptor 2 activation and is essential for shear stress-induced angiogenesis. J. Cell. Physiol. 2009, 222, 120–126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rousseau, S.; Houle, F.; Landry, J.; Huot, J. p38 MAP kinase activation by vascular endothelial growth factor mediates actin reorganization and cell migration in human endothelial cells. Oncogene 1997, 15, 2169–2177. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, N.J.; Banerjee, A.; Kelher, M.R.; Gamboni-Robertson, F.; Hamiel, C.; Sheppard, F.R.; Moore, E.E.; Silliman, C.C. Platelet-activating factor-induced clathrin-mediated endocytosis requires beta-arrestin-1 recruitment and activation of the p38 MAPK signalosome at the plasma membrane for actin bundle formation. J. Immunol. 2006, 176, 7039–7050. [Google Scholar] [CrossRef] [PubMed]
- Gesty-Palmer, D.; Chen, M.; Reiter, E.; Ahn, S.; Nelson, C.D.; Wang, S.; Eckhardt, A.E.; Cowan, C.L.; Spurney, R.F.; Luttrell, L.M.; et al. Distinct beta-arrestin- and G protein-dependent pathways for parathyroid hormone receptor-stimulated ERK1/2 activation. J. Biol. Chem. 2006, 281, 10856–10864. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S.K.; Drake, M.T.; Nelson, C.D.; Houtz, D.A.; Xiao, K.; Madabushi, S.; Reiter, E.; Premont, R.T.; Lichtarge, O.; Lefkowitz, R.J. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J. Biol. Chem. 2006, 281, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Rosanò, L.; Cianfrocca, R.; Tocci, P.; Spinella, F.; Di Castro, V.; Caprara, V.; Semprucci, E.; Ferrandina, G.; Natali, P.G.; Bagnato, A. endothelin A receptor/β-arrestin signaling to the Wnt pathway renders ovarian cancer cells resistant to chemotherapy. Cancer Res. 2014, 74, 7453–7464. [Google Scholar] [CrossRef] [PubMed]
- Strissel, K.J.; Sokolov, M.; Trieu, L.H.; Arshavsky, V.Y. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J. Neurosci. 2006, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhutto, I.; Lutty, G. Understanding age-related macular degeneration (AMD): Relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol. Asp. Med. 2012, 33, 295–317. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- García-Quintanilla, L.; Rodríguez-Martínez, L.; Bandín-Vilar, E.; Gil-Martínez, M.; González-Barcia, M.; Mondelo-García, C.; Fernández-Ferreiro, A.; Mateos, J. Recent Advances in Proteomics-Based Approaches to Studying Age-Related Macular Degeneration: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 14759. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- A Perkins, G.; Ellisman, M.H.; A Fox, D. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: Bioenergetic and functional implications. Mol. Vis. 2003, 9, 60–73. [Google Scholar] [PubMed]
- Jarrett, S.G.; Boulton, M.E. Consequences of oxidative stress in age-related macular degeneration. Mol. Asp. Med. 2012, 33, 399–417. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tuo, J.; Grob, S.; Zhang, K.; Chan, C.-C. Genetics of immunological and inflammatory Components in age-related macular degeneration. Ocul. Immunol. Inflamm. 2012, 20, 27–36. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chong, N.V.; Keonin, J.; Luthert, P.J.; Frennesson, C.I.; Weingeist, D.M.; Wolf, R.L.; Mullins, R.F.; Hageman, G.S. decreased thickness and integrity of the macular elastic layer of Bruch’s membrane correspond to the distribution of lesions associated with age-related macular degeneration. Am. J. Pathol. 2005, 166, 241–251. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- A Campochiaro, P.; Soloway, P.; Ryan, S.J.; Miller, J.W. The pathogenesis of choroidal neovascularization in patients with age-related macular degeneration. Mol. Vis. 1999, 5, 34. [Google Scholar] [PubMed]
- Vilkeviciute, A.; Cebatoriene, D.; Kriauciuniene, L.; Liutkeviciene, R. VEGFA Haplotype and VEGF-A and VEGF-R2 Protein Associations with Exudative Age-Related Macular Degeneration. Cells 2022, 11, 996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hagstrom, S.A.; Ying, G.-S.; Maguire, M.G.; Martin, D.F.; Gibson, J.; Lotery, A.; Chakravarthy, U. VEGFR2 Gene Polymorphisms and Response to Anti–Vascular Endothelial Growth Factor Therapy in Age-Related Macular Degeneration. Ophthalmology 2015, 122, 1563–1568. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Atienzar-Aroca, S.; Serrano-Heras, G.; Valls, A.F.; de Almodovar, C.R.; Muriach, M.; Barcia, J.M.; Garcia-Verdugo, J.M.; Romero, F.J.; Sancho-Pelluz, J. Role of retinal pigment epithelium-derived exosomes and autophagy in new blood vessel formation. J. Cell. Mol. Med. 2018, 22, 5244–5256. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Y.; Tang, Y.; Teng, L.; Wu, Y.; Zhao, X.; Pei, G. Association of β-arrestin and TRAF6 negatively regulates Toll-like receptor–interleukin 1 receptor signaling. Nat. Immunol. 2005, 7, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Alcazar, O.; Cousins, S.W.; Marin-Castan~O, M.E. MMP-14 and TIMP-2 overexpression protects against hydroquinone-induced oxidant injury in RPE: Implications for extracellular matrix turnover. Investig. Opthalmology Vis. Sci. 2007, 48, 5662–5670. [Google Scholar] [CrossRef] [PubMed]
- Usategui-Martín, R.; Pastor-Idoate, S.; Chamorro, A.J.; Fernández, I.; Fernández-Bueno, I.; Marcos-Martín, M.; González-Sarmiento, R.; Pastor, J.C. Meta-analysis of the rs243865 MMP-2 polymorphism and age-related macular degeneration risk. PLoS ONE 2019, 14, e0213624. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kamei, M.; Hollyfield, J.G. TIMP-3 in Bruch’s membrane: Changes during aging and in age-related macular degeneration. Invest. Ophthalmol. Vis Sci. 1999, 40, 2367–2375. [Google Scholar] [PubMed]
- Ball, S.G.; Shuttleworth, C.A.; Kielty, C.M. Vascular endothelial growth factor can signal through platelet-derived growth factor receptors. J. Cell Biol. 2007, 177, 489–500. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Pontén, A.; Aase, K.; Karlsson, L.; Abramsson, A.; Uutela, M.; Bäckström, G.; Hellström, M.; Boström, H.; Li, H.; et al. PDGF-C is a new protease-activated ligand for the PDGF α-receptor. Nat. Cell Biol. 2000, 2, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Kazlauskas, A. A Reactive oxygen species-mediated, self-perpetuating loop persistently activates platelet-derived growth factor receptor α. Mol. Cell. Biol. 2014, 34, 110–122. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cousin, M.A.; Creighton, B.A.; Breau, K.A.; Spillmann, R.C.; Torti, E.; Dontu, S.; Tripathi, S.; Ajit, D.; Edwards, R.J.; Afriyie, S.; et al. Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nat. Genet. 2021, 53, 1006–1021. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yano, M.; Koumoto, Y.; Kanesaki, Y.; Wu, X.; Kido, H. 20S Proteasome prevents aggregation of heat-denatured proteins without PA700 regulatory subcomplex like a molecular chaperone. Biomacromolecules 2004, 5, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Strickland, E.; Demartino, G.N.; Thomas, P.J. Recognition and processing of misfolded proteins by PA700, the 19S regulatory complex of the 26S proteasome. Methods Mol. Biol. 2005, 301, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Glavey, S.V.; Naba, A.; Manier, S.; Clauser, K.; Tahri, S.; Park, J.; Reagan, M.R.; Moschetta, M.; Mishima, Y.; Gambella, M.; et al. Proteomic characterization of human multiple myeloma bone marrow extracellular matrix. Leukemia 2017, 31, 2426–2434. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.N.A.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.D.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006, 38, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.J.D.; Irvine, A.D.; Terron-Kwiatkowski, A.; Sandilands, A.; E Campbell, L.; Zhao, Y.; Liao, H.; Evans, A.T.; Goudie, D.R.; Lewis-Jones, S.; et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006, 38, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Pietruszyńska, M.; Zawadzka-Krajewska, A.; Duda, P.; Rogowska, M.; Grabska-Liberek, I.; Kulus, M. Ophthalmic manifestations of atopic dermatitis. Adv. Dermatol. Allergol. 2020, 37, 174–179. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, Y.; Park, W.K.; Kim, R.-Y.; Kim, M.; Park, Y.-H. Characteristics of retinal detachment associated with atopic dermatitis. BMC Ophthalmol. 2021, 21, 359. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kushima, I.; Aleksic, B.; Nakatochi, M.; Shimamura, T.; Okada, T.; Uno, Y.; Morikawa, M.; Ishizuka, K.; Shiino, T.; Kimura, H.; et al. Comparative Analyses of Copy-Number Variation in Autism Spectrum Disorder and Schizophrenia Reveal Etiological Overlap and Biological Insights. Cell Rep. 2018, 24, 2838–2856. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Hoven, A.; Dress, R.J.; Schaal, H.; Alferink, J.; Scheu, S. Identification of a novel DIg2 isoform differentially expressed in IFNβ-producing plasmacytoid dendritic cells. BMC Genom. 2018, 19, 194. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Naisbitt, S.; Valtschanoff, J.; Allison, D.W.; Sala, C.; Kim, E.; Craig, A.M.; Weinberg, R.J.; Sheng, M. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J. Neurosci. 2000, 20, 4524–4534. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jing-Ping, Z.; Tian, Q.-B.; Sakagami, H.; Kondo, H.; Endo, S.; Suzuki, T. p55 protein is a member of PSD scaffold proteins in the rat brain and interacts with various PSD proteins. Mol. Brain Res. 2005, 135, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, S.M.; Fradkin, S.I.; Demmin, D.L. Schizophrenia and the retina: Towards a 2020 perspective. Schizophr. Res. 2020, 219, 84–94. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhardwaj, A.; Yadav, A.; Yadav, M.; Tanwar, M. Genetic dissection of non-syndromic retinitis pigmentosa. Indian J. Ophthalmol. 2022, 70, 2355–2385. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alexovič, M.; Sabo, J.; Longuespée, R. Microproteomic sample preparation. Proteomics 2021, 21, e2000318. [Google Scholar] [CrossRef] [PubMed]
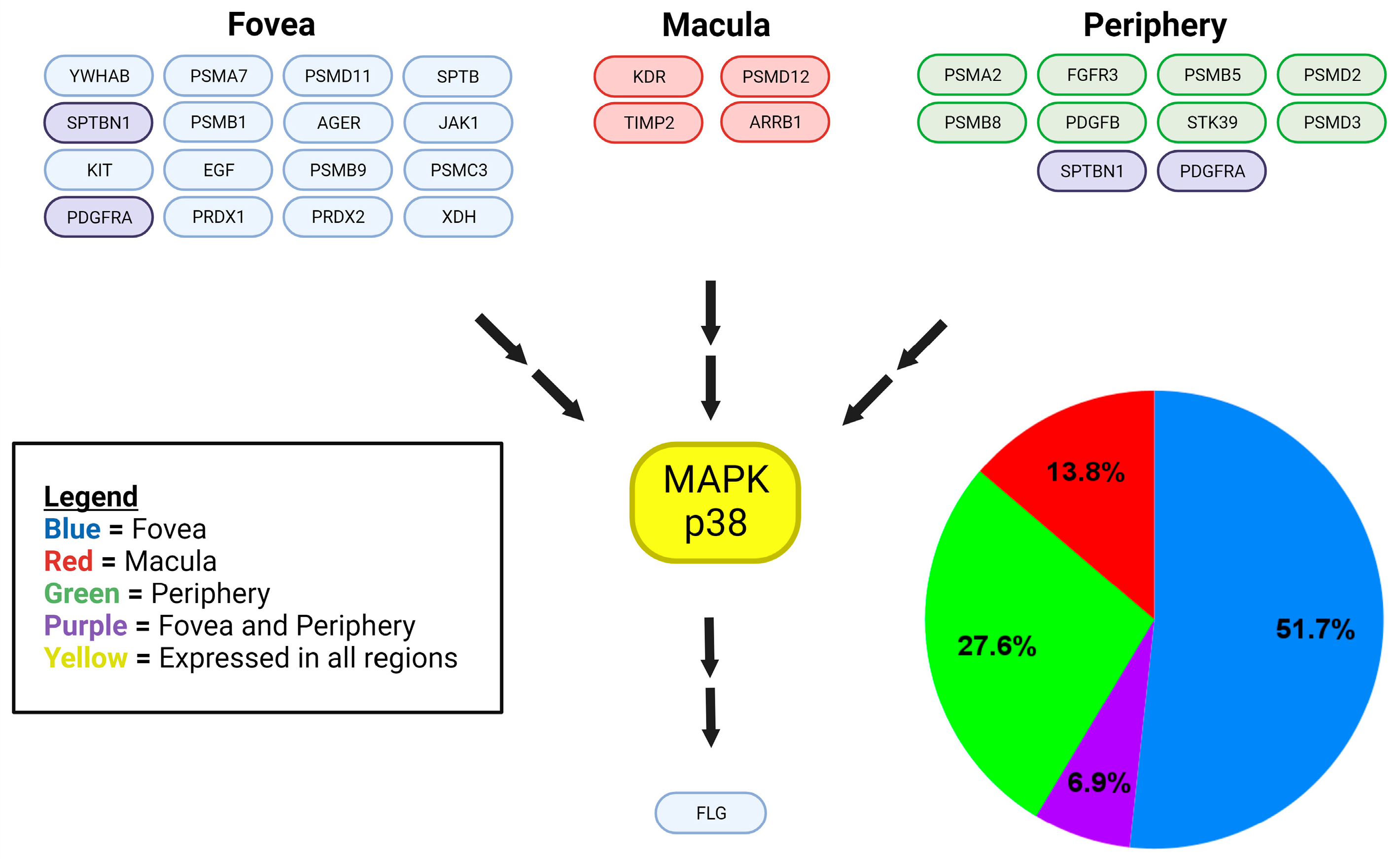
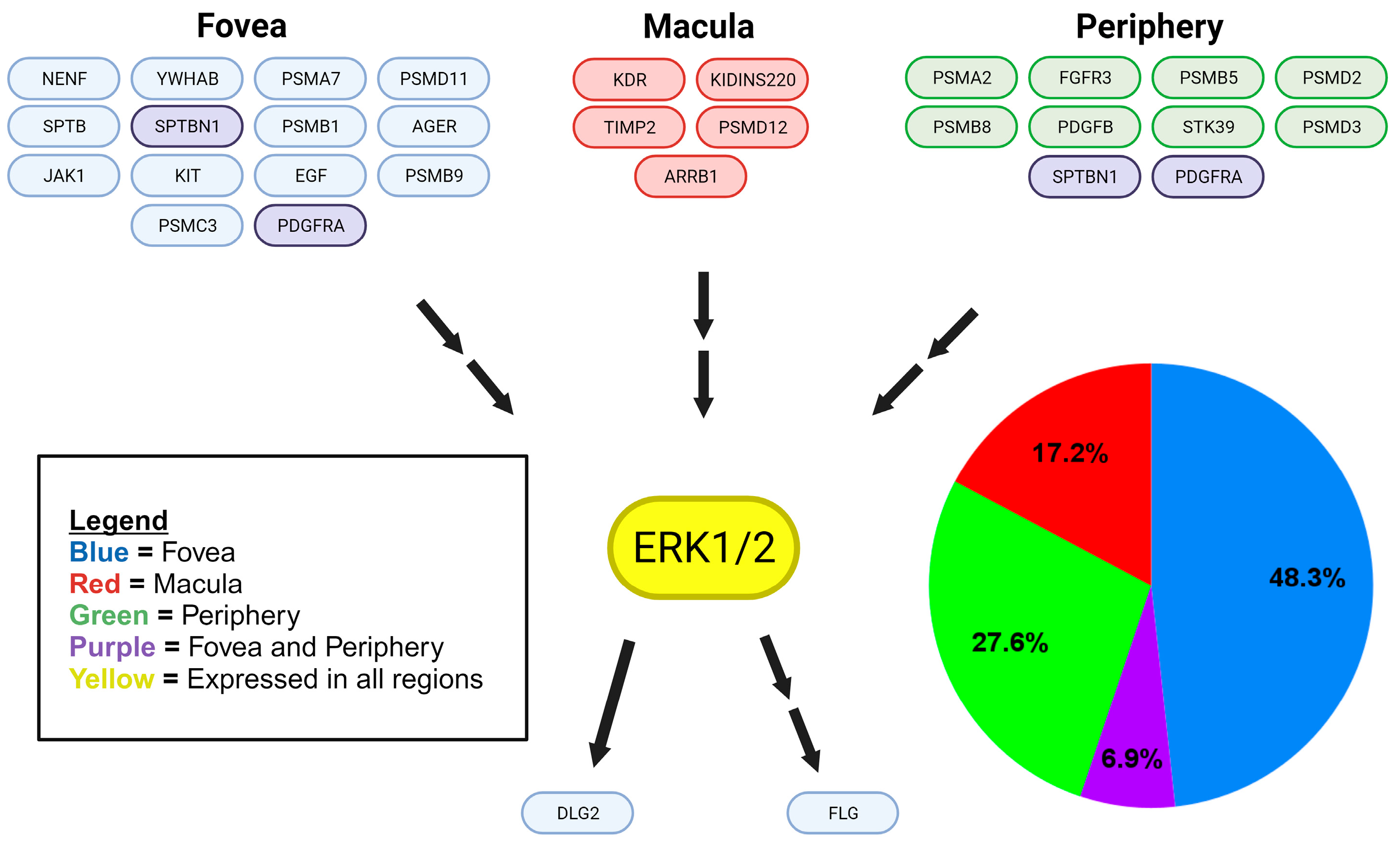
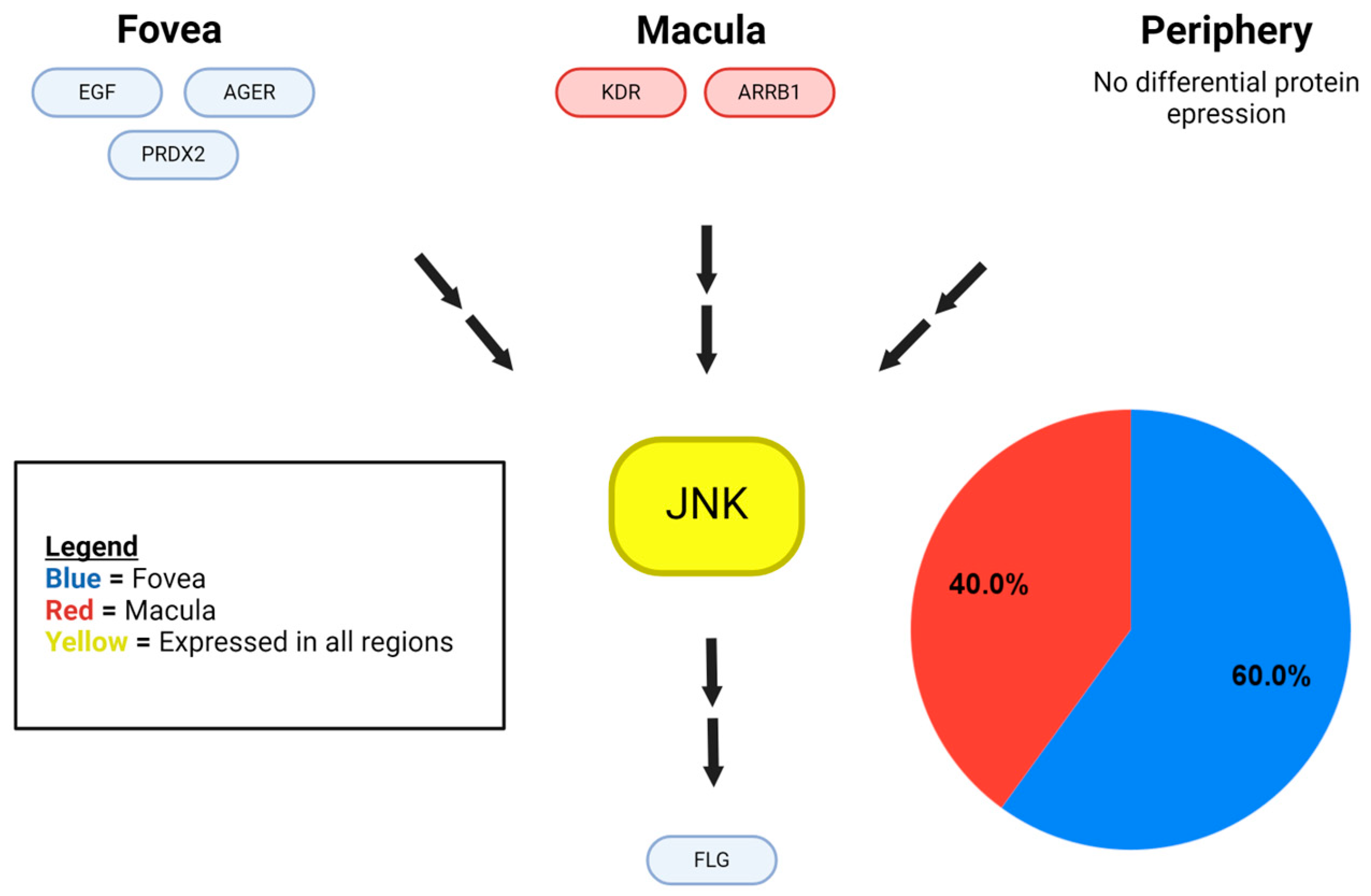
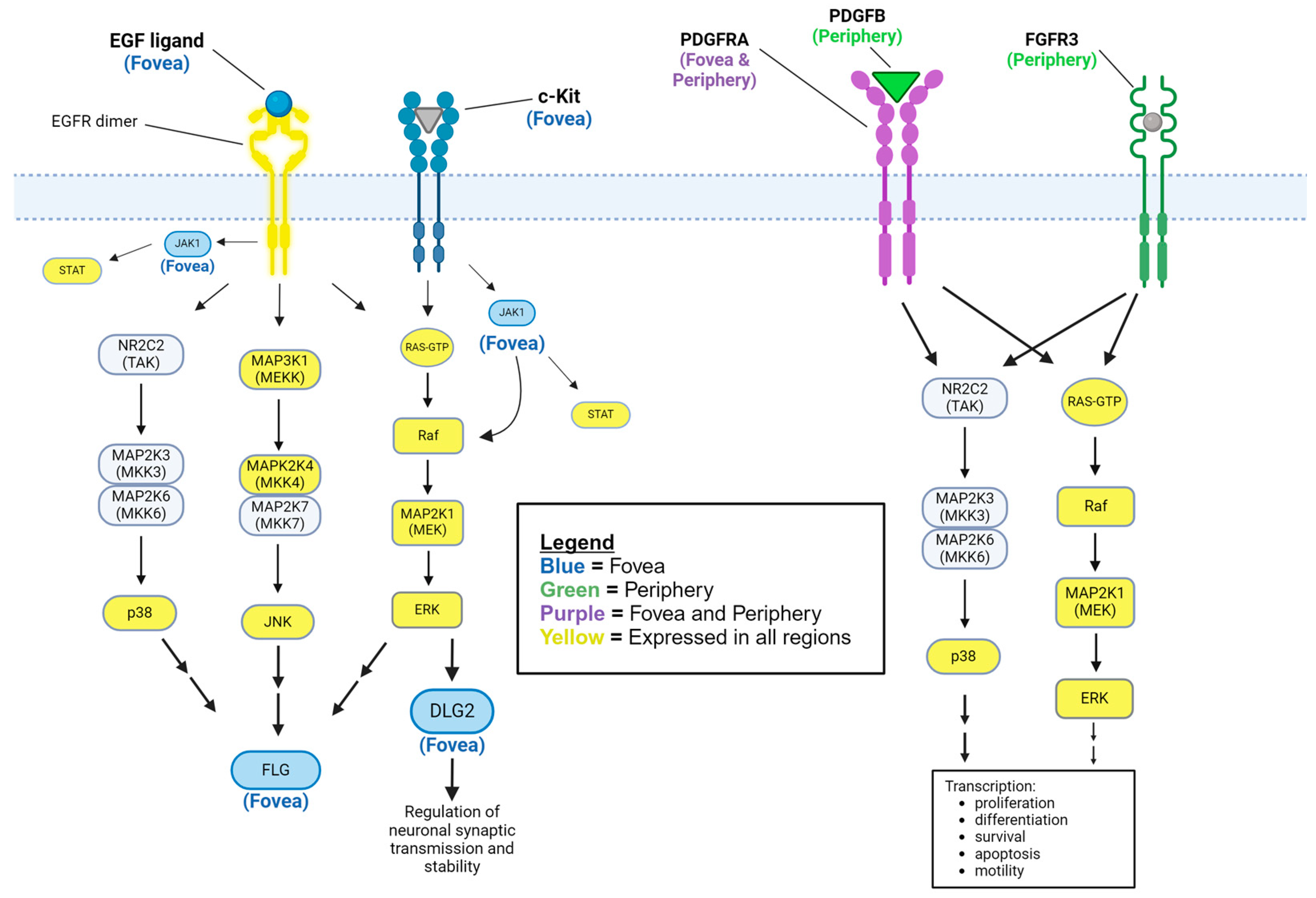
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hailey, D.R.; Kanjilal, D.; Koulen, P. Differential Expression of Mitogen-Activated Protein Kinase Signaling Pathways in the Human Choroid–Retinal Pigment Epithelial Complex Indicates Regional Predisposition to Disease. Int. J. Mol. Sci. 2024, 25, 10105. https://doi.org/10.3390/ijms251810105
Hailey DR, Kanjilal D, Koulen P. Differential Expression of Mitogen-Activated Protein Kinase Signaling Pathways in the Human Choroid–Retinal Pigment Epithelial Complex Indicates Regional Predisposition to Disease. International Journal of Molecular Sciences. 2024; 25(18):10105. https://doi.org/10.3390/ijms251810105
Chicago/Turabian StyleHailey, Dylan R., Debolina Kanjilal, and Peter Koulen. 2024. "Differential Expression of Mitogen-Activated Protein Kinase Signaling Pathways in the Human Choroid–Retinal Pigment Epithelial Complex Indicates Regional Predisposition to Disease" International Journal of Molecular Sciences 25, no. 18: 10105. https://doi.org/10.3390/ijms251810105
APA StyleHailey, D. R., Kanjilal, D., & Koulen, P. (2024). Differential Expression of Mitogen-Activated Protein Kinase Signaling Pathways in the Human Choroid–Retinal Pigment Epithelial Complex Indicates Regional Predisposition to Disease. International Journal of Molecular Sciences, 25(18), 10105. https://doi.org/10.3390/ijms251810105




