Energy Metabolism Enhance Perylenequinone Biosynthesis in Shiraia sp. Slf14 through Promoting Mitochondrial ROS Accumulation
Abstract
1. Introduction
2. Results
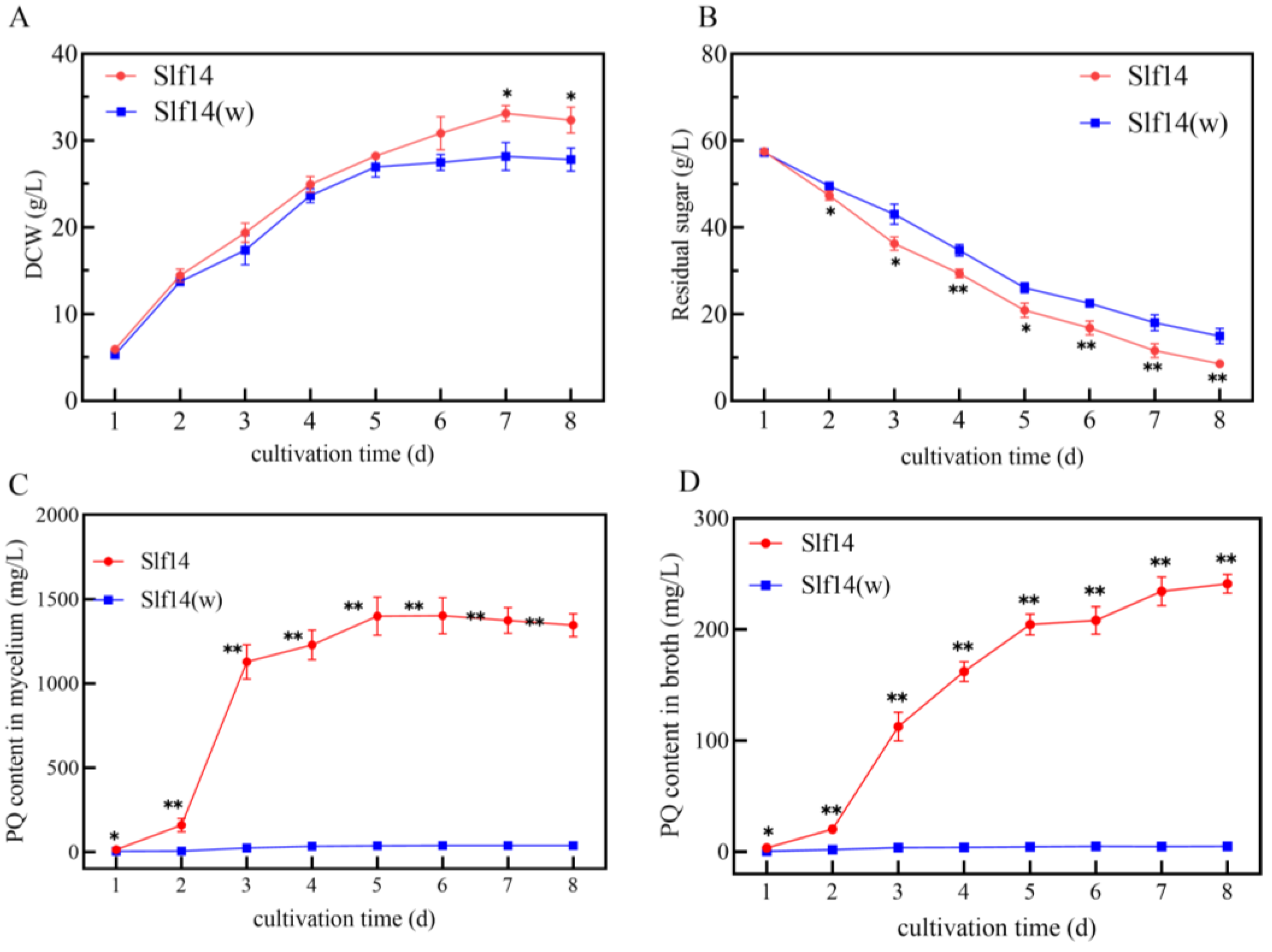
2.1. Growth and PQ Biosynthesis of Strains Slf14 and Slf14(w)
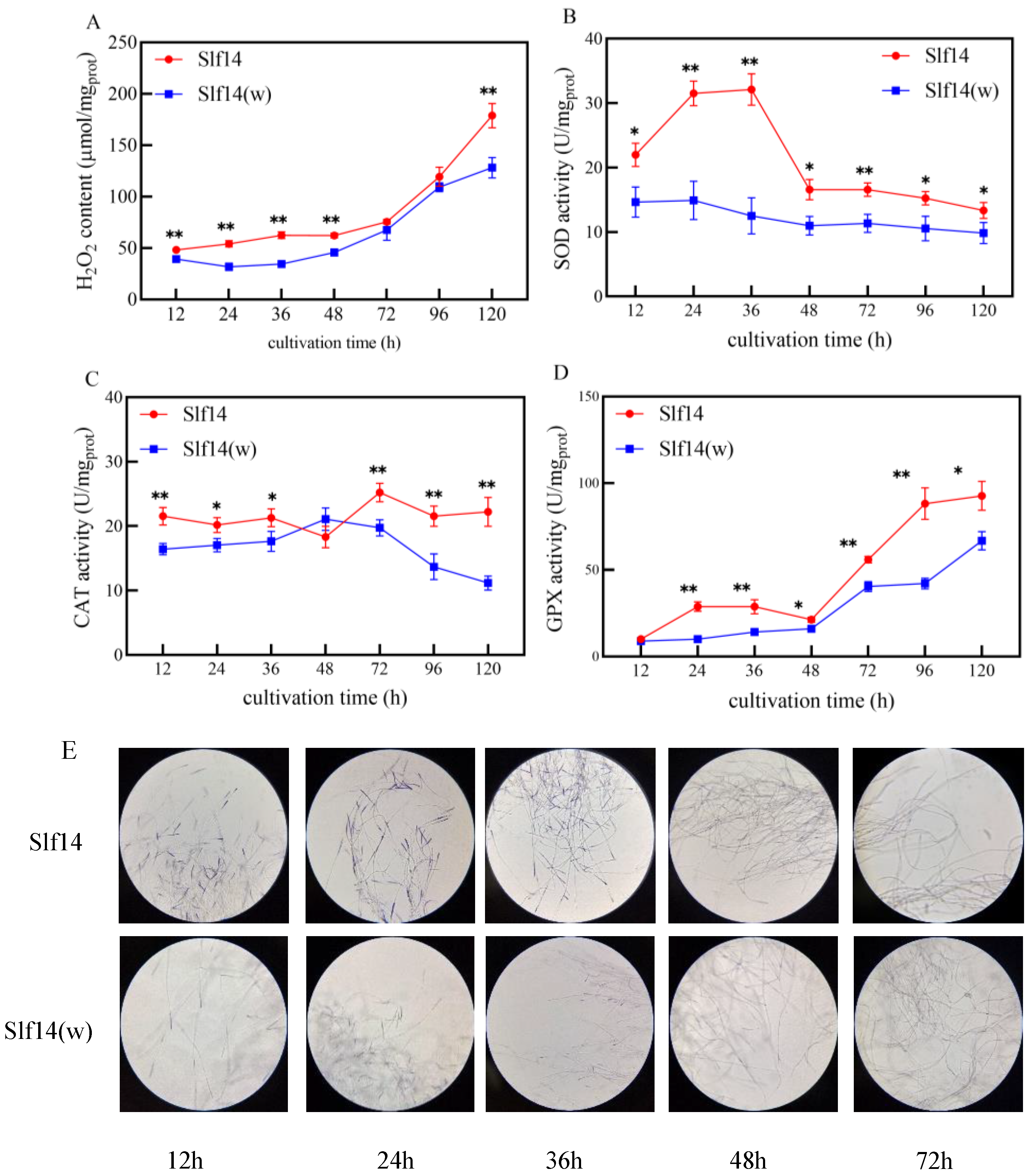
2.2. Redox State of Strains Slf14 and Slf14(w)
2.3. Transcriptome Analysis Results
2.4. qRT-PCR Analysis
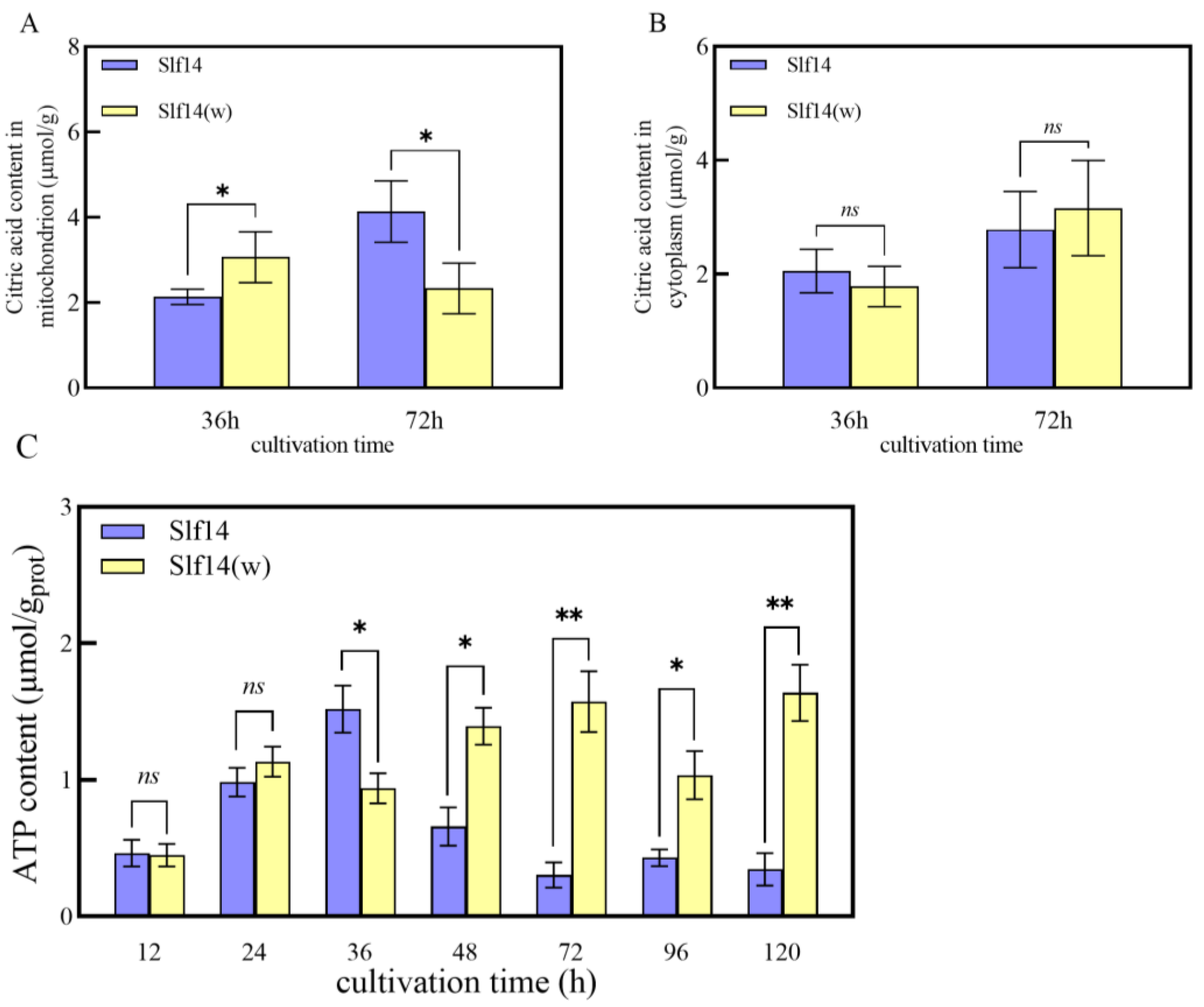
2.5. Measurement of Citric Acid and ATP Content
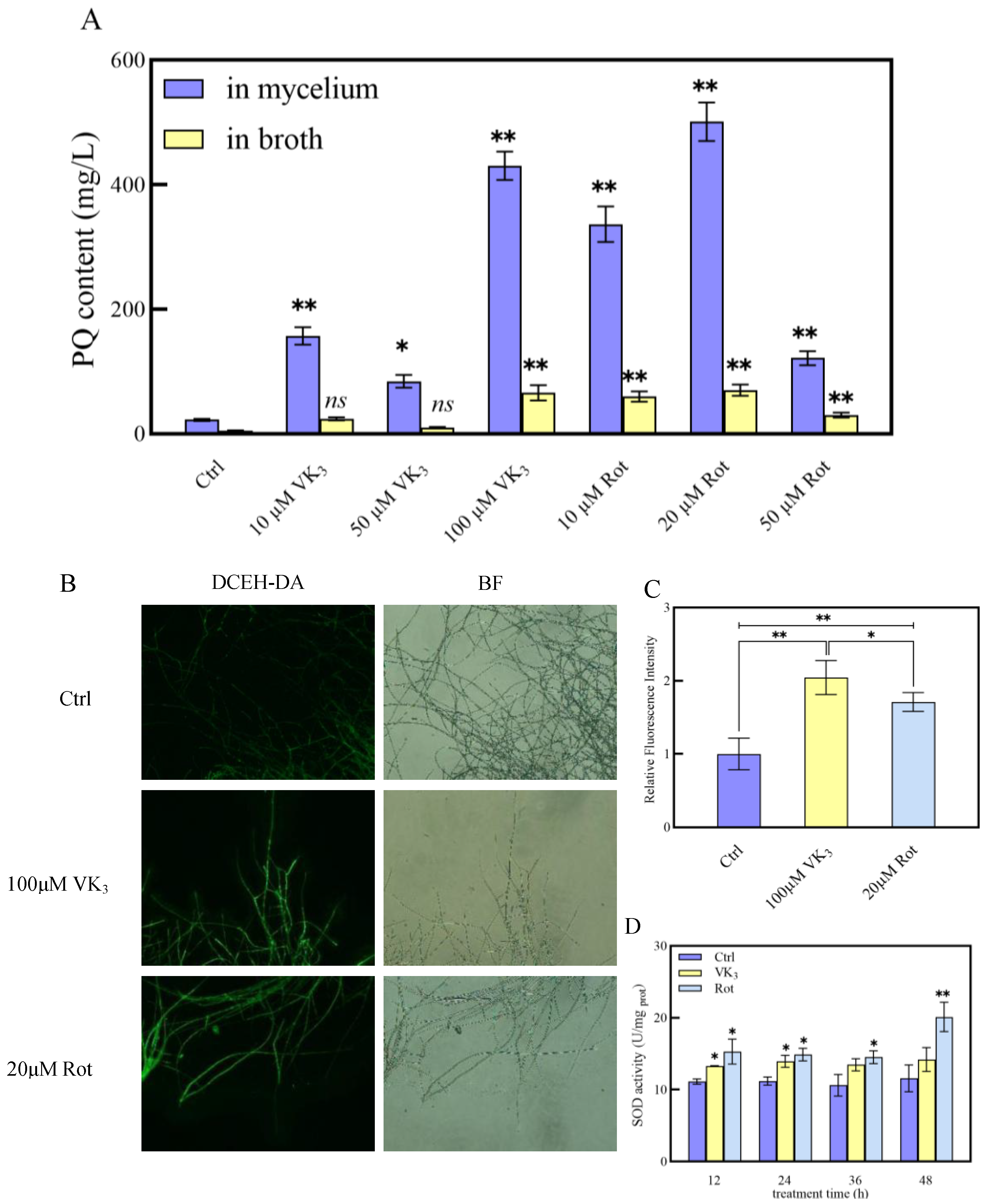
2.6. Effects of Addition of ROS-Related Reagents on PQ Production
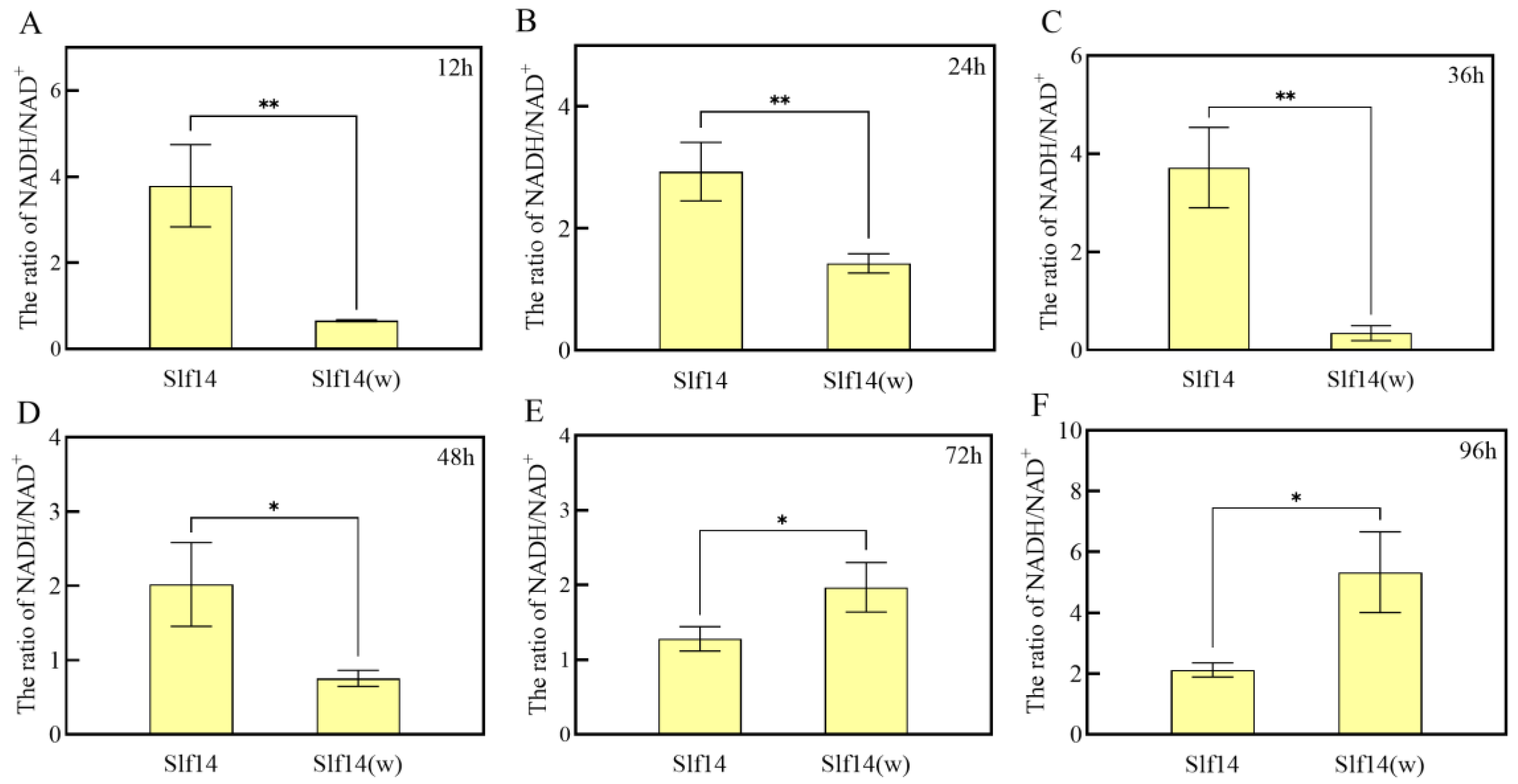
2.7. Mitochondrial NADH/NAD+ Ratio
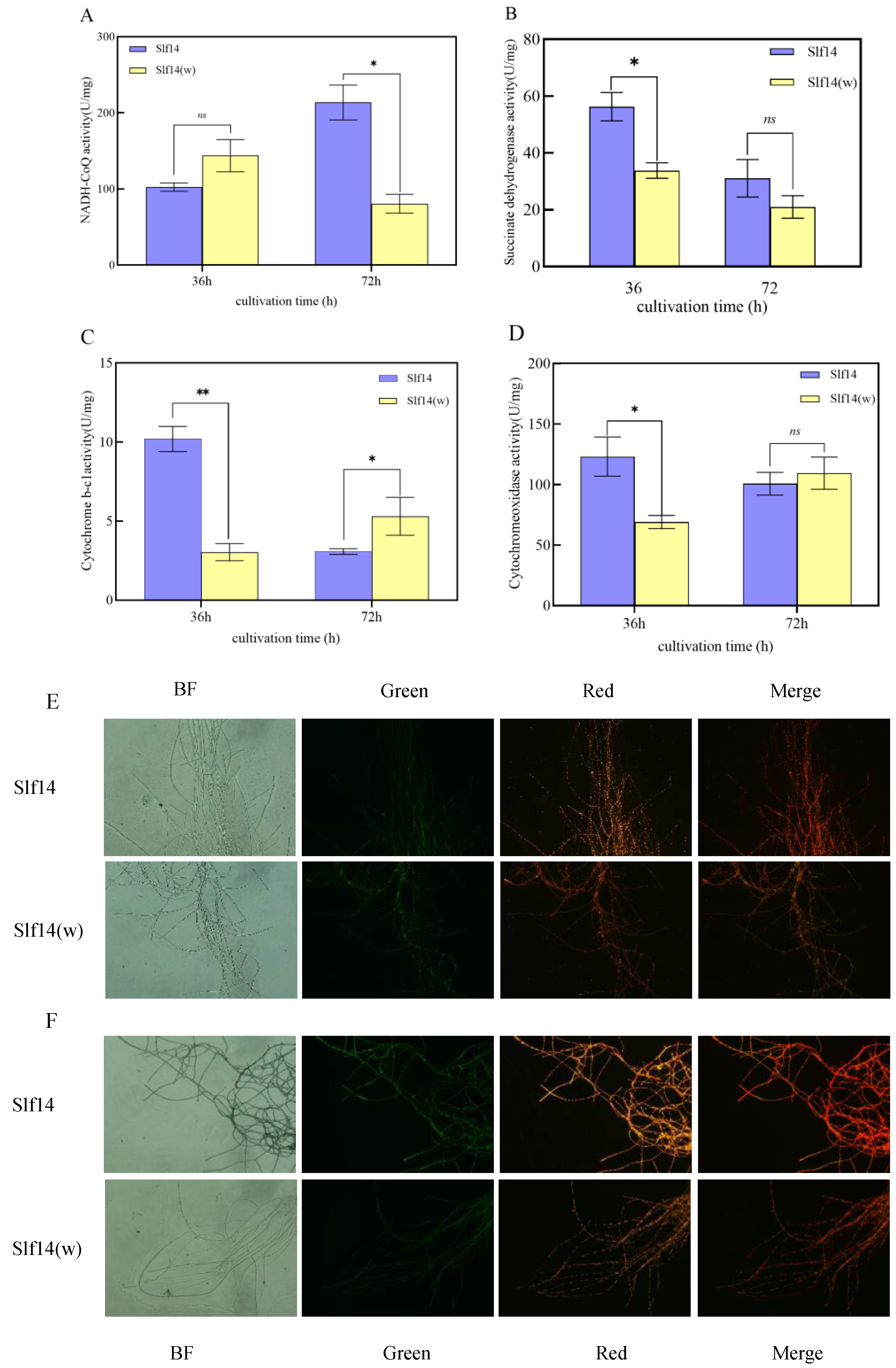
2.8. Mitochondrial Complex Activity and Mitochondrial Membrane Potential Assays
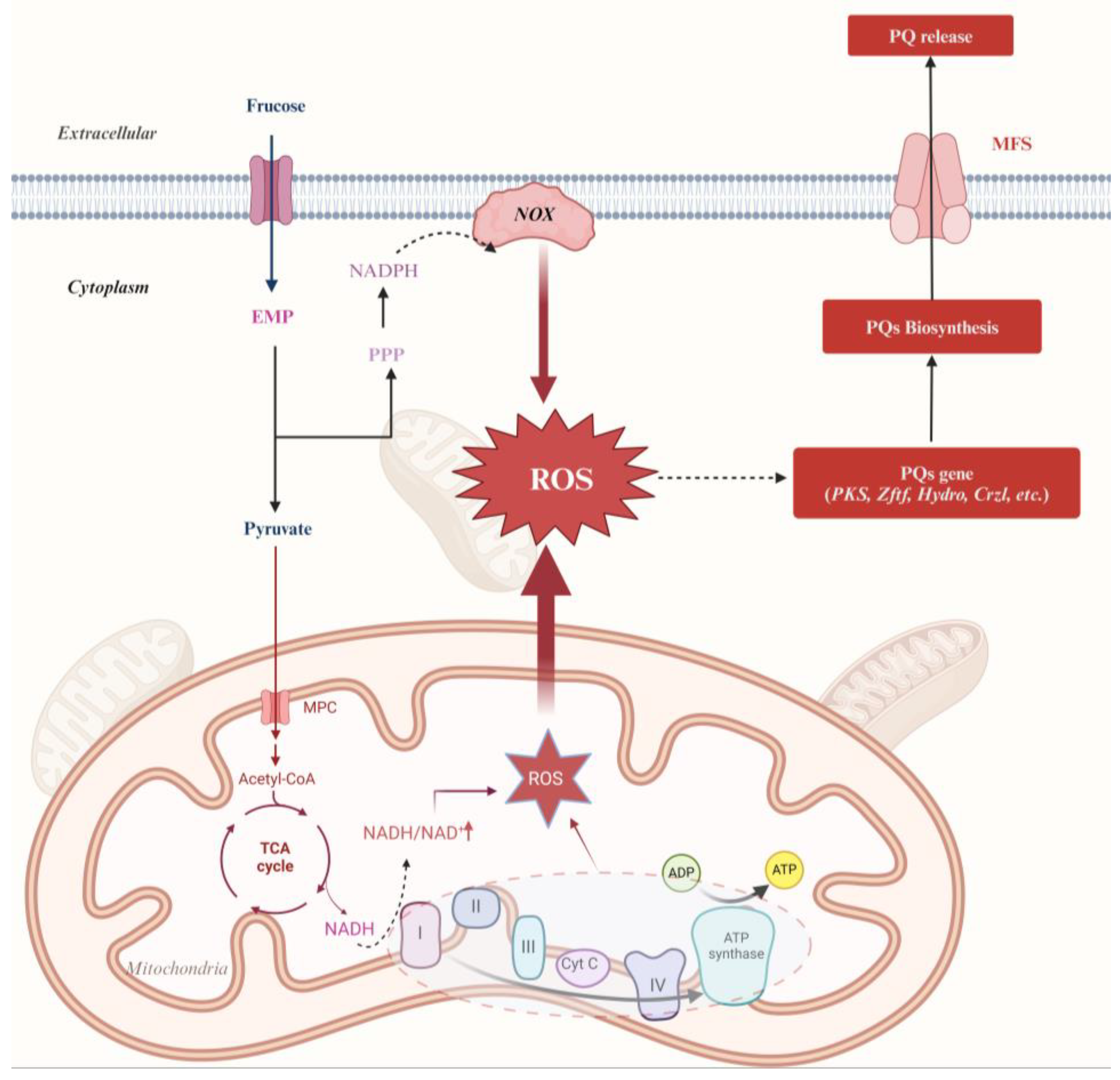
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Culture Conditions
4.2. Analysis of Cell Growth and Residual Sugar
4.3. Extraction and Analysis of Total PQs
4.4. Determination of H2O2 Content and Various Oxidoreductases
4.5. Transcriptome Sequencing, Annotation, and Analysis
4.6. Quantitative Real-Time PCR (qRT-PCR) Analysis of Gene Expression
4.7. Determination of the Contents of Citric Acid and ATP
4.8. Effect of Oxidative Stress Reagents Addition on PQ Biosynthesis
4.9. Determination of the NADH/NAD+ Ratio in Fungi
4.10. Assay of Mitochondrial Complex Activity and Detection of Mitochondrial Membrane Potential
4.11. Bioinformatics Analysis and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| ABC | ATP-binding cassette |
| ACO | Aconitase |
| APX | Aseorbateperoxidase |
| AP1L | AP1-like transcription factor |
| BSA | bovine serum albumin |
| CA | citric acid |
| CAT | Catalase |
| Crzl | Calcineurin-responsive zinc finger transcription factor |
| CS | Citrate synthase |
| DCFH-DA | 2,7-dichlorofluorescein diacetate |
| DCW | Dry cell weight |
| DNS | 3,5-dinitrosalicylic acid |
| ETC | Electron Transport Chain |
| FAD | FAD/FMN-dependent oxidoreductase |
| GR | Glutathione reductase |
| GPX | Glutathione peroxidase |
| HA | Hypocrellin A |
| HK | Hexokinase |
| H2O2 | Hydrogen peroxide |
| Hydro | Hydroxylase |
| MMP | mitochondrial membrane potential |
| MDH | Malate Dehydrogenase |
| MFS | MFS transporters |
| NOX | NADPH oxidase |
| NBT | Nitrotetrazolium blue chloride |
| Omef | O-methytransferase |
| PFA | Potato fructose agar |
| PFB | Potato fructose broth |
| PDH | Pyruvate dehydrogenase |
| PFK | Phosphofructokinase |
| PKS | Polyketide synthase |
| PQ | Perylenequinones |
| PBS | Phosphate-buffered saline |
| ROS | Reactive oxygen species |
| SDH | Succinate dehydrgenase |
| SOD | Superoxide dismutase |
| TEMPO | 2,2,6,6-tetramethylpiperidin-1-oxyl |
| Zftf | Zinc finger transcription |
| α-KGDH | α-ketoglutarate dehydrogenase |
References
- Bao, Z.; Xie, Y.; Xu, C.; Zhang, Z.; Zhu, D. Biotechnological production and potential applications of hypocrellins. Appl. Microbiol. Biotechnol. 2023, 107, 6421–6438. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.-H.; Cai, Y.-J.; Liao, X.-R.; Wu, K.; Wang, L.; Zhang, D.-B.; Meng, Q. Isolation and identification of a new hypocrellin A-producing strain Shiraia sp. SUPER-H168. Microbiol. Res. 2009, 164, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Surup, F.; Pommerehne, K.; Schroers, H.-J.; Stadler, M. Elsinopirins A–D, decalin polyketides from the ascomycete Elsinoё pyri. Biomolecules 2018, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- de Morais, F.A.P.; Gonçalves, R.S.; Vilsinski, B.H.; Lazarin-Bidóia, D.; Balbinot, R.B.; Tsubone, T.M.; Brunaldi, K.; Nakamura, C.V.; Hioka, N.; Caetano, W. Hypericin photodynamic activity in DPPC liposomes—Part II: Stability and application in melanoma B16-F10 cancer cells. Photochem. Photobiol. Sci. 2020, 19, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xu, J. Production of fungal pigments: Molecular Processes and Their Applications. J. Fungi 2022, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Yang, S.; Yang, Y.; Zhang, L.; Cui, L. Synergistic chemotherapy and phototherapy based on red blood cell biomimetic nanomaterials. J. Control. Release 2022, 352, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Bao, J.; Zhang, Z.; Yan, R.; Wang, Y.; Yang, H.; Zhu, D. Enhanced production of perylenequinones in the endophytic fungus Shiraia sp. Slf14 by calcium/calmodulin signal transduction. Appl. Microbiol. Biotechnol. 2017, 102, 153–163. [Google Scholar] [CrossRef]
- Tong, Z.; Mao, L.; Liang, H.; Zhang, Z.; Wang, Y.; Yan, R.; Zhu, D. Simultaneous determination of six perylenequinones in Shiraia sp. Slf14 by HPLC. J. Liq. Chromatogr. Relat. Technol. 2017, 40, 536–540. [Google Scholar] [CrossRef]
- Liu, Z.; Bao, J.; Yang, H.; Zhang, Z.; Yan, R.; Zhu, D. Transcriptome analysis on fructose as the sole carbon source enhancing perylenequinones production of endophytic fungus Shiraia sp. Slf14. 3 Biotech 2020, 10, 190. [Google Scholar] [CrossRef]
- Deng, H.; Liang, X.; Liu, J.; Zheng, X.; Fan, T.-P.; Cai, Y. Advances and perspectives on perylenequinone biosynthesis. Front. Microbiol. 2022, 13, 1070110. [Google Scholar] [CrossRef]
- Ma, Y.J.; Zheng, L.P.; Wang, J.W. Inducing perylenequinone production from a bambusicolous fungus Shiraia sp. S9 through co-culture with a fruiting body-associated bacterium Pseudomonas fulva SB1. Microb. Cell Factories 2019, 18, 121. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Sun, C.; Wang, B.; Wang, Y.; Dong, B.; Liu, J.; Xia, J.; Xie, W.; Wang, J.; Sun, J.; et al. Response mechanism of hypocrellin colorants biosynthesis by Shiraia bambusicola to elicitor PB90. AMB Express 2019, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Guo, B.; Xu, H.; Yuan, Z.; Liu, H.; Guo, T.; Deng, Z.; Zhang, Y.; Yin, D.; Liu, C.; et al. Synthetic biology-based construction of unnatural perylenequinones with improved photodynamic anticancer activities. Angew. Chem. Int. Ed. Engl. 2024, 63, e202317726. [Google Scholar] [CrossRef] [PubMed]
- Rahal, A.; Kumar, A.; Singh, V.; Yadav, B.; Tiwari, R.; Chakraborty, S.; Dhama, K. Oxidative stress, prooxidants, and antioxidants: The interplay. BioMed Res. Int. 2014, 2014, 761264. [Google Scholar] [CrossRef]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef]
- Miller, G.A.D.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R.O.N. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Kwolek-Mirek, M.; Bednarska, S.; Dubicka-Lisowska, A.; Maslanka, R.; Zadrag-Tecza, R.; Kaszycki, P. Unbalance between pyridine nucleotide cofactors in The SOD1 deficient yeast Saccharomyces cerevisiae causes hypersensitivity to alcohols and aldehydes. Int. J. Mol. Sci. 2022, 24, 659. [Google Scholar] [CrossRef]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 3. [Google Scholar] [CrossRef]
- Bibián, M.E.; Pérez-Sánchez, A.; Mejía, A.; Barrios-González, J. Penicillin and cephalosporin biosyntheses are also regulated by reactive oxygen species. Appl. Microbiol. Biotechnol. 2020, 104, 1773–1783. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Espinosa, J.; Shabala, S.; León, J.; Romero-Puertas, M.C.; Aroca, A. Reactive oxygen species- and nitric oxide-dependent regulation of ion and metal homeostasis in plants. J. Exp. Bot. 2023, 74, 5970–5988. [Google Scholar] [CrossRef]
- Heller, J.; Tudzynski, P. Reactive oxygen species in phytopathogenic fungi: Signaling, development, and disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.; Shih, J.A.M. Metabolic flux analysis of secondary metabolism in plants. Metab. Eng. Commun. 2020, 10, 7. [Google Scholar]
- Pérez-Pérez, W.D.; Carrasco-Navarro, U.; García-Estrada, C.; Kosalková, K.; Gutiérrez-Ruíz, M.C.; Barrios-González, J.; Fierro, F. bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum. Microb. Cell Factories 2022, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, T.; Pan, Y.; Shi, L.; Jin, Y.; Huang, X. The metabolism of reactive oxygen species and their effects on lipid biosynthesis of microalgae. Int. J. Mol. Sci. 2023, 24, 11041. [Google Scholar] [CrossRef]
- Li, C.; Shi, L.; Chen, D.; Ren, A.; Gao, T.; Zhao, M. Functional analysis of the role of glutathione peroxidase (GPx) in the ROS signaling pathway, hyphal branching and the regulation of ganoderic acid biosynthesis in Ganoderma lucidum. Fungal Genet. Biol. 2015, 82, 168–180. [Google Scholar] [CrossRef]
- Fountain, J.C.; Koh, J.; Yang, L.; Pandey, M.K.; Nayak, S.N.; Bajaj, P.; Zhuang, W.-J.; Chen, Z.-Y.; Kemerait, R.C.; Lee, R.D.; et al. Proteome analysis of Aspergillus flavus isolate-specific responses to oxidative stress in relationship to aflatoxin production capability. Sci. Rep. 2018, 8, 3430. [Google Scholar] [CrossRef]
- Hou, L.; Zhou, X.; Gan, F.; Liu, Z.; Zhou, Y.; Qian, G.; Huang, K. Combination of selenomethionine and N-Acetylcysteine alleviates the joint toxicities of Aflatoxin B1 and Ochratoxin A by ERK MAPK signal pathway in porcine Alveolar Macrophages. J. Agric. Food Chem. 2018, 66, 5913–5923. [Google Scholar] [CrossRef]
- Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef]
- Cheng, Z.; Ristow, M. Mitochondria and metabolic homeostasis. Antioxid. Redox Signal 2013, 19, 240–242. [Google Scholar] [CrossRef]
- Poór, P. Effects of salicylic acid on the metabolism of mitochondrial reactive oxygen species in plants. Biomolecules 2020, 10, 341. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A. The role of antioxidants in the chemistry of oxidative stress: A review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Chang, P.; Jin, J.; Yang, Q.; Xing, F. Dimethylformamide Inhibits Fungal Growth and Aflatoxin B1 Biosynthesis in Aspergillus flavus by Down-Regulating Glucose Metabolism and Amino Acid Biosynthesis. Toxins 2020, 12, 683. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Wang, J.; Zeng, Q.; Zhang, Z.; Yan, R. A novel endophytic Huperzine A-producing fungus, Shiraia sp. Slf14, isolated from Huperzia serrata. J. Appl. Microbiol. 2010, 109, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, Y.; Zhang, Z.; Yan, R.; Zhu, D. Whole-Genome shotgun assembly and analysis of the genome of Shiraia sp. Strain Slf14, a novel endophytic fungus producing Huperzine A and Hypocrellin A. Genome Announc. 2014, 2, e00011-14. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Bao, J.; Yan, R.; Du, Z. Effects of different carbon sources on the production of perylenequinone pigments by the endophytic fungus Shiraia sp. Slf14. J. Jiangxi Norm. Univ. (Nat. Sci. Ed.) 2019, 43, 5. [Google Scholar]
- Chen, Y.; Xu, C.; Yang, H.; Liu, Z.; Zhang, Z.; Yan, R.; Zhu, D. L-Arginine enhanced perylenequinone production in the endophytic fungus Shiraia sp. Slf14(w) via NO signaling pathway. Appl. Microbiol. Biotechnol. 2022, 106, 2619–2636. [Google Scholar] [CrossRef]
- Anastasiou, D.; Poulogiannis, G.; Asara, J.M.; Boxer, M.B.; Jiang, J.-k.; Shen, M.; Bellinger, G.; Sasaki, A.T.; Locasale, J.W.; Auld, D.S.; et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011, 334, 1278–1283. [Google Scholar] [CrossRef]
- Kuehne, A.; Emmert, H.; Soehle, J.; Winnefeld, M.; Fischer, F.; Wenck, H.; Gallinat, S.; Terstegen, L.; Lucius, R.; Hildebrand, J.; et al. Acute activation of oxidative Pentose Phosphate Pathway as first-line response to oxidative stress in Human skin cells. Mol. Cell 2015, 59, 359–371. [Google Scholar] [CrossRef]
- Lambeth, J.D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef]
- Lu, C.; Ma, Y.; Wang, J. Lanthanum elicitation on hypocrellin A production in mycelium cultures of Shiraia bambusicola is mediated by ROS generation. J. Rare Earth 2019, 37, 895–902. [Google Scholar] [CrossRef]
- Dellomonaco, C.; Clomburg, J.M.; Miller, E.N.; Gonzalez, R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 2011, 476, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Hackett, S.R.; Zanotelli, V.R.T.; Xu, W.; Goya, J.; Park, J.O.; Perlman, D.H.; Gibney, P.A.; Botstein, D.; Storey, J.D.; Rabinowitz, J.D. Systems-level analysis of mechanisms regulating yeast metabolic flux. Science 2016, 354, aaf2786. [Google Scholar] [CrossRef] [PubMed]
- Chubukov, V.; Gerosa, L.; Kochanowski, K.; Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 2014, 12, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Chroumpi, T.; Mäkelä, M.R.; de Vries, R.P. Engineering of primary carbon metabolism in filamentous fungi. Biotechnol. Adv. 2020, 43, 107551. [Google Scholar] [CrossRef]
- Li, X.P.; Zhou, L.L.; Guo, Y.H.; Wang, J.W. The signaling role of extracellular ATP in co-culture of Shiraia sp. S9 and Pseudomonas fulva SB1 for enhancing hypocrellin A production. Microb. Cell Fact. 2021, 20, 144. [Google Scholar] [CrossRef]
- Huang, X.; Liu, S.; Wu, L.; Jiang, M.; Hou, Y. High Throughput Single Cell RNA Sequencing, Bioinformatics Analysis and Applications. Adv. Exp. Med. Biol. 2018, 1068, 33–43. [Google Scholar]
- Hong, S.-Y.; Roze, L.; Linz, J. Oxidative stress-related transcription factors in the regulation of secondary metabolism. Toxins 2013, 5, 683–702. [Google Scholar] [CrossRef]
- Jayashree, T.; Subramanyam, C. Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic. Biol. Med. 2000, 29, 5. [Google Scholar]
- Kim, J.H.; Yu, J.; Mahoney, N.; Chan, K.L.; Molyneux, R.J.; Varga, J.; Bhatnagar, D.; Cleveland, T.E.; Nierman, W.C.; Campbell, B.C. Elucidation of the functional genomics of antioxidant-based inhibition of aflatoxin biosynthesis. Int. J. Food Microbiol. 2008, 122, 49–60. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, Y.; Yao, B.; Hu, T.; Ma, Z.; Shi, W.; Ye, Y. Curcumin inhibits Aspergillus flavus infection and aflatoxin production possibly by inducing ROS burst. Food Res. Int. 2023, 167, 112646. [Google Scholar] [CrossRef]
- Liu, R.; Cao, P.; Ren, A.; Wang, S.; Yang, T.; Zhu, T.; Shi, L.; Zhu, J.; Jiang, A.-L.; Zhao, M.-W. SA inhibits complex III activity to generate reactive oxygen species and thereby induces GA overproduction in Ganoderma lucidum. Redox Biol. 2018, 16, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, X.; Ren, A.; Shi, D.-K.; Shi, L.; Zhu, J.; Yu, H.-S.; Zhao, M.-W. Heat stress-induced reactive oxygen species participate in the regulation of HSP expression, hyphal branching and ganoderic acid biosynthesis in Ganoderma lucidum. Microbiol. Res. 2018, 209, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ruan, J.; Chu, P.; Fu, W.; Liang, Z.; Li, Y.; Tong, J.; Xiao, L.; Liu, J.; Li, C.; et al. AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J. 2019, 101, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.X.; Ma, Y.J.; Wang, J.W. Improved hypocrellin A production in Shiraia bambusicola by light-dark shift. J. Photochem. Photobiol. B 2018, 182, 100–107. [Google Scholar] [CrossRef]
- Sun, C.X.; Ma, Y.J.; Wang, J.W. Enhanced production of hypocrellin A by ultrasound stimulation in submerged cultures of Shiraia bambusicola. Ultrason. Sonochem 2017, 38, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Ji, H.Y.; Wang, W.J.; Shen, W.H.; Wang, J.W. Effects of blue light on Hypocrellin A production in Shiraia mycelium cultures. Photochem. Photobiol. 2022, 98, 1343–1354. [Google Scholar] [CrossRef]
- Li, X.P.; Wang, Y.; Ma, Y.J.; Wang, J.W.; Zheng, L.P. Nitric oxide and hydrogen peroxide signaling in extractive Shiraia fermentation by Triton X-100 for Hypocrellin A production. Int. J. Mol. Sci. 2020, 21, 882. [Google Scholar] [CrossRef]
- Murphy, M.P.; Holmgren, A.; Larsson, N.-G.; Halliwell, B.; Chang, C.J.; Kalyanaraman, B.; Rhee, S.G.; Thornalley, P.J.; Partridge, L.; Gems, D.; et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011, 13, 361–366. [Google Scholar] [CrossRef]
- Singh, A.; Mehta, S.; Yadav, S.; Nagar, G.; Ghosh, R.; Roy, A.; Chakraborty, A.; Singh, I.K. How to cope with the challenges of environmental stresses in the era of global climate change: An update on ROS stave off in plants. Int. J. Mol. Sci. 2022, 23, 1995. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2008, 417, 1–13. [Google Scholar] [CrossRef]
- Mishra, R.K.; Kumar, V.; Ghose, D.; Kalita, A.; Dhiman, P.; Prakash, A.; Thakur, N.; Mitra, G.; Chaudhari, V.D.; Arora, A.; et al. Free spermidine evokes superoxide radicals that manifest toxicity. eLife 2022, 11, e77704. [Google Scholar]
- Ray, R.; Shah, A.M. NADPH oxidase and endothelial cell function. Clin. Sci. 2005, 109, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M. NAD+/NADH homeostasis affects metabolic adaptation to hypoxia and secondary metabolite production infilamentous fungi. Biosci. Biotech. Biochem. 2018, 82, 216–224. [Google Scholar] [CrossRef]
- Karamanlidis, G.; Lee, C.F.; Garcia-Menendez, L.; Kolwicz, S.C.; Suthammarak, W.; Gong, G.; Sedensky, M.M.; Morgan, P.G.; Wang, W.; Tian, R. Mitochondrial Complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013, 18, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Lopes, L.D.; Shaffer, B.T.; Kidarsa, T.A.; Vining, O.; Philmus, B.; Song, C.; Stockwell, V.O.; Raaijmakers, J.M.; McPhail, K.L.; et al. Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. mBio 2018, 9, e01845-17. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P. Regulatory aspects of the tricarboxylic acid cycle in filamentous fungi—A review Trans. Brit. Mycol. 1988, 90, 11. [Google Scholar]
- Chen, J.; Xu, W.; Wang, Z.; Tian, L.; Liu, W.; Ren, A.; Liu, R.; Zhu, J.; Shi, L. Mitochondrial pyruvate carrier influences ganoderic acid biosynthesis in Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2023, 107, 1361–1371. [Google Scholar] [CrossRef]
- Furukawa, T.; Katayama, H.; Oikawa, A.; Negishi, L.; Ichikawa, T.; Suzuki, M.; Murase, K.; Takayama, S.; Sakuda, S. Dioctatin activates clpP to degrade mitochondrial components and inhibits aflatoxin production. Cell Chem. Biol. 2020, 27, 1396–1409.e10. [Google Scholar] [CrossRef]
- Fan, W.; Li, B.; Du, N.; Hui, T.; Cao, Y.; Li, X.; Ren, H. Energy metabolism as the target of 3-phenyllactic acid against Rhizopus oryzae. Int. J. Food Microbiol. 2022, 369, 109606. [Google Scholar] [CrossRef]
- Venkitasubramanian, T.A.; Gupta, S.K. Biosynthesis of aflatoxins. Ann. Nutr. L’alimentation 1970, 31, 635–642. [Google Scholar]
- Xu, C.; Lin, W.; Cheng, Y.; Gao, B.; Zhang, Z.; Du, Z. Heat stress enhanced perylenequinones biosynthesis of Shiraia sp. Slf14(w) through nitric oxide formation. Appl. Microbiol. Biotechnol. 2023, 107, 3745–3761. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Mitchell, J.B.; Samuni, J.A.; Krishna, M.C.; DeGraff, W.G.; Ahn, M.S.; Samuni, U.; Russo, A. Biologically active metal-independent superoxide dismutase mimics. Biochemistry 1990, 29, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xie, L.; Li, Q.L.; Xu, Q.Y.; Lin, L.R.; Liu, L.L.; Yang, T.C. Treponema pallidum membrane protein Tp47 promotes angiogenesis through ROS-induced autophagy. J. Eur. Acad. Dermatol. 2022, 37, 558–572. [Google Scholar] [CrossRef]
- Liu, R.; Zhu, T.; Chen, X.; Wang, Z.; Yang, Z.; Ren, A.; Shi, L.; Yu, H.; Zhao, M. GSNOR regulates ganoderic acid content in Ganoderma lucidum under heat stress through S-nitrosylation of catalase. Commun. Biol. 2022, 5, 32. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Meng, X.; Xiao, Y.; Yang, H.; Zhang, Z.; Zhu, D. Energy Metabolism Enhance Perylenequinone Biosynthesis in Shiraia sp. Slf14 through Promoting Mitochondrial ROS Accumulation. Int. J. Mol. Sci. 2024, 25, 10113. https://doi.org/10.3390/ijms251810113
Wu X, Meng X, Xiao Y, Yang H, Zhang Z, Zhu D. Energy Metabolism Enhance Perylenequinone Biosynthesis in Shiraia sp. Slf14 through Promoting Mitochondrial ROS Accumulation. International Journal of Molecular Sciences. 2024; 25(18):10113. https://doi.org/10.3390/ijms251810113
Chicago/Turabian StyleWu, Xueyi, Xuan Meng, Yiwen Xiao, Huilin Yang, Zhibin Zhang, and Du Zhu. 2024. "Energy Metabolism Enhance Perylenequinone Biosynthesis in Shiraia sp. Slf14 through Promoting Mitochondrial ROS Accumulation" International Journal of Molecular Sciences 25, no. 18: 10113. https://doi.org/10.3390/ijms251810113
APA StyleWu, X., Meng, X., Xiao, Y., Yang, H., Zhang, Z., & Zhu, D. (2024). Energy Metabolism Enhance Perylenequinone Biosynthesis in Shiraia sp. Slf14 through Promoting Mitochondrial ROS Accumulation. International Journal of Molecular Sciences, 25(18), 10113. https://doi.org/10.3390/ijms251810113







