Computational Screening to Predict MicroRNA Targets in the Flavivirus 3′ UTR Genome: An Approach for Antiviral Development
Abstract
:1. Introduction
2. Results
2.1. Data Filtering of miRNA–Flavivirus Interactions
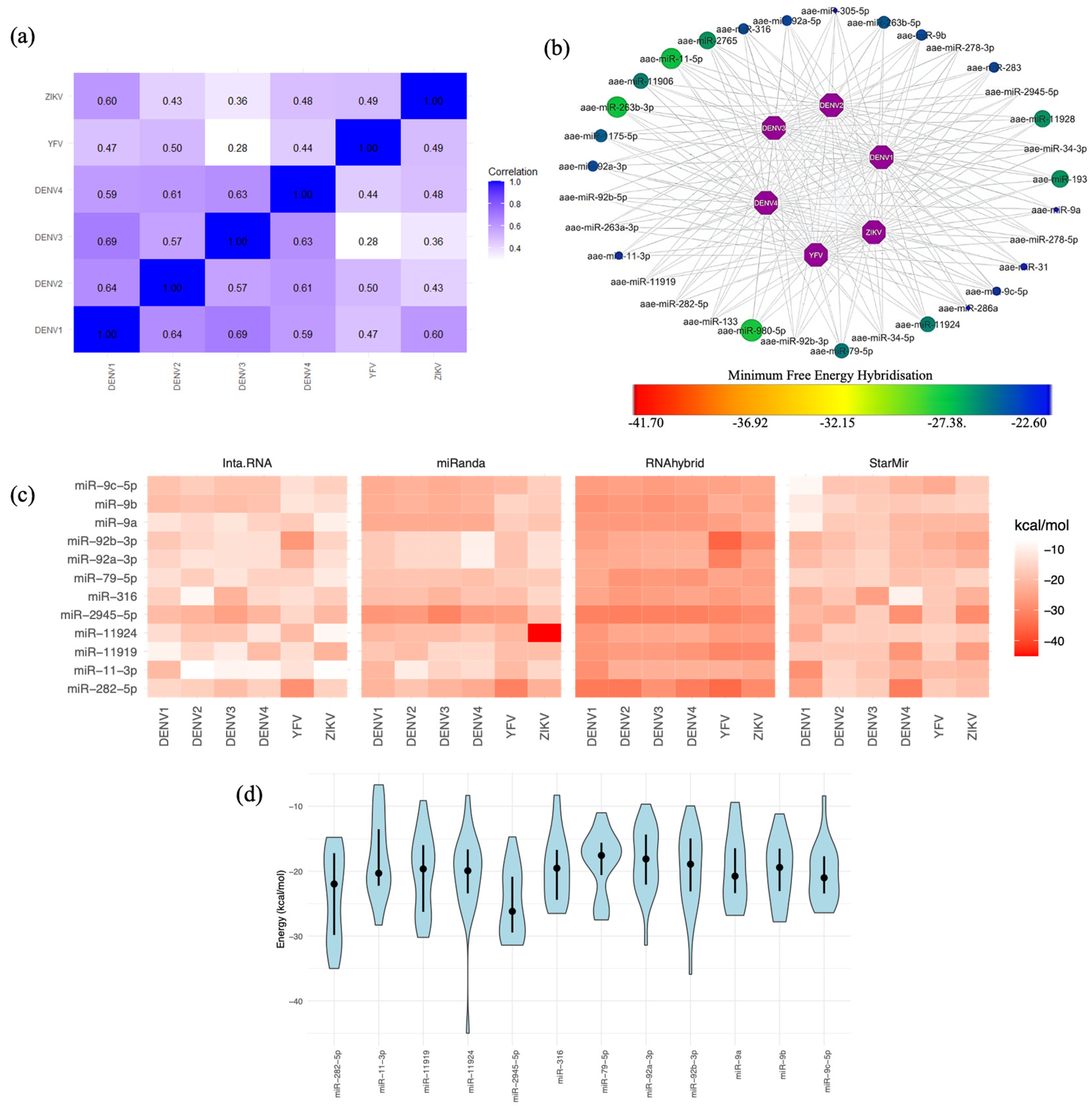
2.2. Human miRNA Interactions with the Flavivirus 3′ UTRs
2.3. Selecting the Optimal Human miRNA That Targets Flavivirus 3′ UTRs
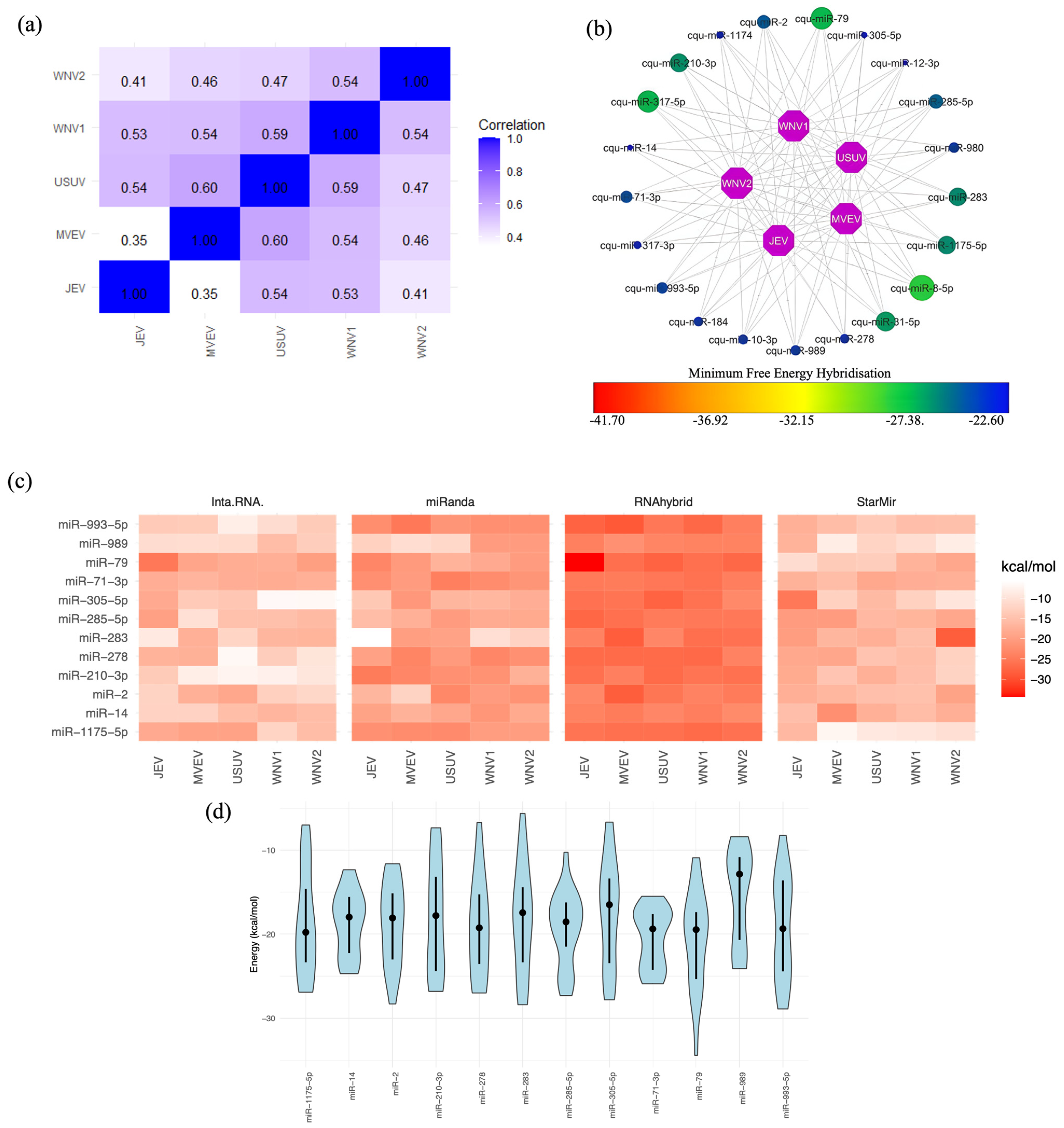
2.4. Mosquito miRNA Interactions with Flavivirus 3′ UTRs
2.5. Selecting the Optimal Mosquito miRNA That Targets Flavivirus 3′ UTRs
3. Discussion
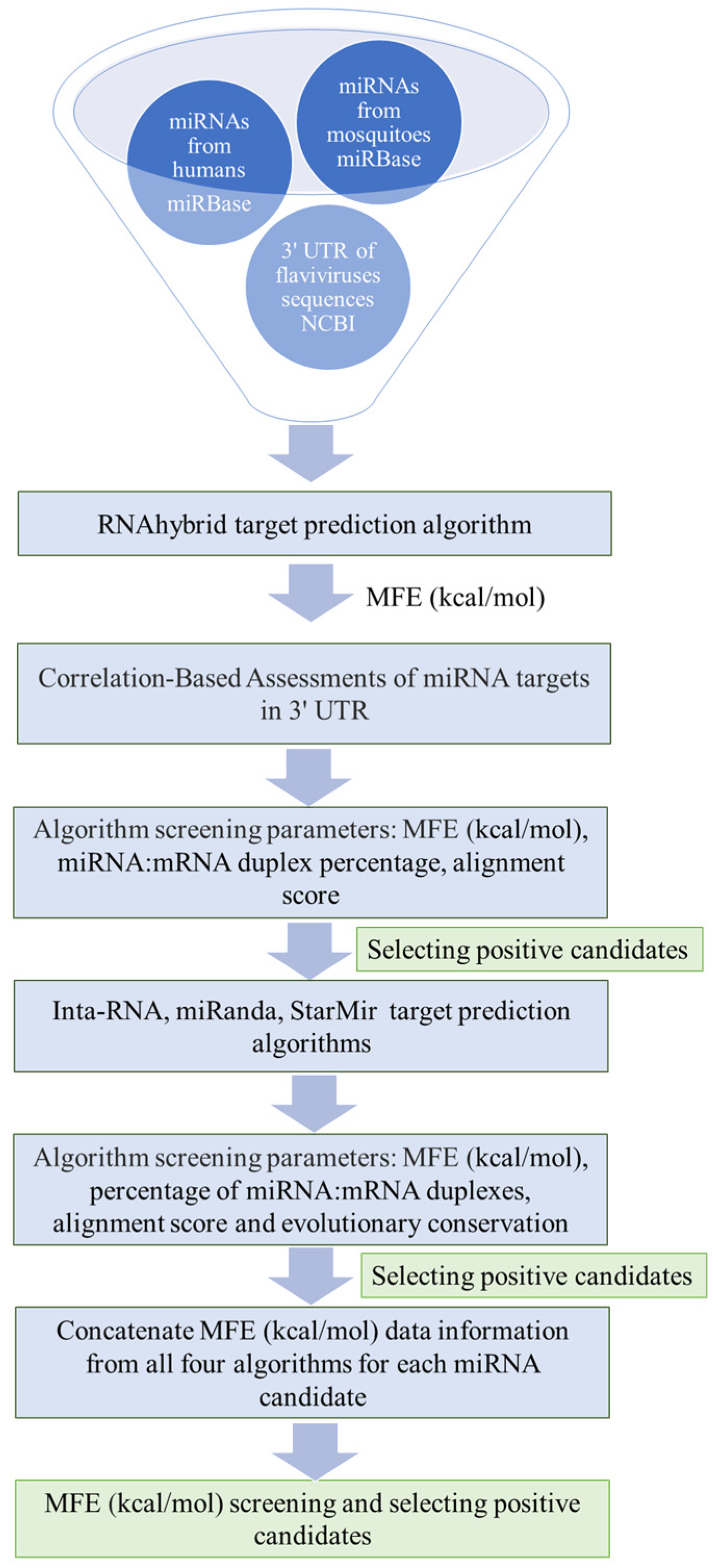
4. Materials and Methods
4.1. Retrieval of the Viral Genome and Mature miRNA Sequences
4.2. miRNA Target Site Algorithms
- RNAhybrid: This algorithm calculates the minimum free energy (MFE) for miRNA–target hybrids using thermodynamic principles. It integrates helix parameters and loop constraints and accounts for G:U wobbles within the seed region. A favourable free energy for hybridisation is typically approximately −20 kcal/mol [43].
- Inta-RNA: Using an enhanced scoring system, this algorithm predicts RNA–RNA interactions. It assesses the thermodynamic stability of interaction duplexes, site accessibility, and seed region attributes. Interactions are predicted when both the total energy and hybridisation energy are less than zero, with scores greater than 140 indicating optimal interactions [44].
- miRanda: This algorithm identifies miRNA–mRNA target duplexes, accommodating mismatches, gaps, and wobble base pairings. It extends beyond the seed region to predict all possible miRNA target sites. We adjusted the threshold binding energy to −20 kcal/mol, set a score threshold of 100, and applied a gap-opening penalty (GOP) of −9 and a gap-extension penalty (GEP) of −4 [45].
- StarMir: Uses miRNA binding data from CLIP studies in non-linear logistic prediction models. It excels at identifying seeded and unseeded target sites by considering thermodynamic, structural, and sequence features from SFold 2.2. The algorithm considers factors, such as the type of seed and site accessibility, and incorporates several parameters, including the Gibbs free energy change of the miRNA–mRNA target hybrid (ΔGhybrid) [43], the miRNA–mRNA target hybridisation (ΔGnucl), the total energy change of the hybridisation (ΔGtotal), and the LogitProb score [46,47].
4.3. Correlation-Based Assessments of miRNA Targets in the 3′ UTR
4.4. Identification of miRNA Binding Sites
4.5. Computational Environments and Software
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van den Elsen, K.; Quek, J.P.; Luo, D. Molecular Insights into the Flavivirus Replication Complex. Viruses 2021, 13, 956. [Google Scholar] [CrossRef] [PubMed]
- Ochsenreiter, R.; Hofacker, I.L.; Wolfinger, M.T. Functional RNA Structures in the 3′ UTR of Tick-Borne, Insect-Specific and No-Known-Vector Flaviviruses. Viruses 2019, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Harapan, H.; Michie, A.; Sasmono, R.T.; Imrie, A. Dengue: A Minireview. Viruses 2020, 12, 829. [Google Scholar] [CrossRef] [PubMed]
- Markoff, L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 2003, 59, 177–228. [Google Scholar] [CrossRef]
- Romero, T.A.; Tumban, E.; Jun, J.; Lott, W.B.; Hanley, K.A. Secondary structure of dengue virus type 4 3′ untranslated region: Impact of deletion and substitution mutations. J. Gen. Virol. 2006, 87, 3291–3296. [Google Scholar] [CrossRef]
- Wei, Y.; Qin, C.; Jiang, T.; Li, X.; Zhao, H.; Liu, Z.; Deng, Y.; Liu, R.; Chen, S.; Yu, M.; et al. Translational regulation by the 3′ untranslated region of the dengue type 2 virus genome. Am. J. Trop. Med. Hyg. 2009, 81, 817–824. [Google Scholar] [CrossRef]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. 2023, 24, 816–833. [Google Scholar] [CrossRef]
- Maurin, T.; Cazalla, D.; Yang, S.; Jr Bortolamiol-Becet, D.; Lai, E.C. RNase III-independent microRNA biogenesis in mammalian cells. RNA 2012, 18, 2166–2173. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microRNA targeting. Science 2014, 346, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Moore, M.J.; Scheel, T.K.; Luna, J.M.; Park, C.Y.; Fak, J.J.; Nishiuchi, E.; Rice, C.M.; Darnell, R.B. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Avila-Bonilla, R.G.; Salas-Benito, J.S. Interactions of host miRNAs in the flavivirus 3´UTR genome: From bioinformatics predictions to practical approaches. Front. Cell. Infect. Microbiol. 2022, 12, 976843. [Google Scholar] [CrossRef]
- Castrillón-Betancur, J.C.; Urcuqui-Inchima, S. Overexpression of miR-484 and miR-744 in Vero cells alters Dengue virus replication. Mem. Inst. Oswaldo Cruz 2017, 112, 281–291. [Google Scholar] [CrossRef]
- Castillo, J.A.; Castrillón, J.C.; Diosa-Toro, M.; Betancur, J.G.; St Laurent, G., 3rd; Smit, J.M.; Urcuqui-Inchima, S. Complex interaction between dengue virus replication and expression of miRNA-133a. BMC Infect. Dis. 2016, 16, 29. [Google Scholar] [CrossRef]
- Cai, W.; Pan, Y.; Cheng, A.; Wang, M.; Yin, Z.; Jia, R. Regulatory Role of Host MicroRNAs in Flaviviruses Infection. Front. Microbiol. 2022, 13, 869441. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.C.; Lin, Y.L.; Liao, J.T.; Su, C.M.; Lin, C.C.; Lin, W.P.; Liao, C.L. Utilizing liver-specific microRNA-122 to modulate replication of dengue virus replicon. Biochem. Biophys. Res. Commun. 2010, 396, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.M.; Langlois, R.A.; TenOever, B.R. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012, 8, e1002465. [Google Scholar] [CrossRef] [PubMed]
- Yen, L.C.; Lin, Y.L.; Sung, H.H.; Liao, J.T.; Tsao, C.H.; Su, C.M.; Lin, C.K.; Liao, C.L. Neurovirulent flavivirus can be attenuated in mice by incorporation of neuron-specific microRNA recognition elements into viral genome. Vaccine 2013, 31, 5915–5922. [Google Scholar] [CrossRef] [PubMed]
- Hum, C.; Loiselle, J.; Ahmed, N.; Shaw, T.A.; Toudic, C.; Pezacki, J.P. MicroRNA Mimics or Inhibitors as Antiviral Therapeutic Approaches Against COVID-19. Drugs 2021, 81, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.N.; Majumdar, N.; Williams, F.; Rajput, S.; Pokhrel, L.R.; Cook, P.P.; Akula, S.M. MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens. Biology 2023, 12, 1334. [Google Scholar] [CrossRef]
- Xu, T.; Li, L.X.; Jia, Y.; Wu, Q.; Zhu, W.; Xu, Z.; Zheng, B.; Lu, X. One microRNA has the potential to target whole viral mRNAs in a given human coronavirus. Front. Microbiol. 2022, 13, 1035044. [Google Scholar] [CrossRef]
- Park, J.H.; Moon, J. Conserved 3′ UTR of Severe Acute Respiratory Syndrome Coronavirus 2: Potential Therapeutic Targets. Front. Genet. 2022, 13, 893141. [Google Scholar] [CrossRef]
- Baig, M.S.; Krishnan, A. A bioinformatics approach to investigate serum and hematopoietic cell-specific therapeutic microRNAs targeting the 3′ UTRs of all four Dengue virus serotypes. Pathog. Dis. 2021, 79, ftab050. [Google Scholar] [CrossRef]
- El-Nabi, S.H.; Elhiti, M.; El-Sheekh, M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses 2020, 143, 110203. [Google Scholar] [CrossRef]
- Hemida, M.G.; Ye, X.; Thair, S.; Yang, D. Exploiting the therapeutic potential of microRNAs in viral diseases: Expectations and limitations. Mol. Diagn. Ther. 2010, 14, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Liu, Z.; Hemida, M.G.; Yang, D. Targeted delivery of mutant tolerant anti-coxsackievirus artificial microRNAs using folate conjugated bacteriophage Phi29 pRNA. PLoS ONE 2011, 6, e21215. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.; Aneja, A.; Ghosh, S.; Devvanshi, H.C.D.; Sahu, R.; Ross, C.; Kshetrapal, P.; Maitra, A.; Das, S. Association of exosomal miR-96-5p and miR-146a-5p with the disease severity in dengue virus infection. J. Med. Virol. 2023, 95, e28614. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; He, L.; Li, Y.; Wang, T.; Feng, L.; Jiang, L.; Zhang, P.; Huang, X. miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J. Infect. 2013, 67, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Rastogi, M.; Singh, S.K. Zika virus NS1 suppresses the innate immune responses via miR-146a in human microglial cells. Int. J. Biol. Macromol. 2021, 193 Pt B, 2290–2296. [Google Scholar] [CrossRef]
- Slonchak, A.; Shannon, R.P.; Pali, G.; Khromykh, A.A. Human MicroRNA miR-532-5p Exhibits Antiviral Activity against West Nile Virus via Suppression of Host Genes SESTD1 and TAB3 Required for Virus Replication. J. Virol. 2015, 90, 2388–2402. [Google Scholar] [CrossRef]
- Ashraf, U.; Zhu, B.; Ye, J.; Wan, S.; Nie, Y.; Chen, Z.; Cui, M.; Wang, C.; Duan, X.; Zhang, H.; et al. MicroRNA-19b-3p Modulates Japanese Encephalitis Virus-Mediated Inflammation via Targeting RNF11. J. Virol. 2016, 90, 4780–4795. [Google Scholar] [CrossRef]
- Sharma, S.; Majumdar, A.; Basu, A. Regulation of Onecut2 by miR-9-5p in Japanese encephalitis virus infected neural stem/progenitor cells. Microbiol. Spectr. 2024, 12, e0323823. [Google Scholar] [CrossRef]
- Zhu, B.; Ye, J.; Nie, Y.; Ashraf, U.; Zohaib, A.; Duan, X.; Fu, Z.F.; Song, Y.; Chen, H.; Cao, S. MicroRNA-15b Modulates Japanese Encephalitis Virus-Mediated Inflammation via Targeting RNF125. J. Immunol. 2015, 195, 2251–2262. [Google Scholar] [CrossRef]
- Escalera-Cueto, M.; Medina-Martínez, I.; del Angel, R.M.; Berumen-Campos, J.; Gutiérrez-Escolano, A.L.; Yocupicio-Monroy, M. Let-7c overexpression inhibits dengue virus replication in human hepatoma Huh-7 cells. Virus Res. 2015, 196, 105–112. [Google Scholar] [CrossRef]
- Su, Y.C.; Huang, Y.F.; Wu, Y.W.; Chen, H.F.; Wu, Y.H.; Hsu, C.C.; Hsu, Y.C.; Lee, J.C. MicroRNA-155 inhibits dengue virus replication by inducing heme oxygenase-1-mediated antiviral interferon responses. FASEB J. 2020, 34, 7283–7294. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Roy, S.; Kumari, B.; Jain, P.; Banerjee, A.; Vrati, S. MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J. Neuroinflamm. 2014, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Ahmed, N.; Pezacki, J.P. miR-383 Regulates Hepatic Lipid Homeostasis and Response to Dengue Virus Infection. ACS Infect. Dis. 2022, 8, 928–941. [Google Scholar] [CrossRef] [PubMed]
- Casseb, S.M.M.; Melo, K.F.L.; Carvalho, C.A.M.; Santos, C.R.D.; Franco, E.C.S.; Vasconcelos, P.F.D.C. Experimental Dengue Virus Type 4 Infection Increases the Expression of MicroRNAs-15/16, Triggering a Caspase-Induced Apoptosis Pathway. Curr. Issues Mol. Biol. 2023, 45, 4589–4599. [Google Scholar] [CrossRef] [PubMed]
- Diosa-Toro, M.; Echavarría-Consuegra, L.; Flipse, J.; Fernández, G.J.; Kluiver, J.; van den Berg, A.; Urcuqui-Inchima, S.; Smit, J.M. MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression. PLoS Negl. Trop. Dis. 2017, 11, e0005981. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, J.; Ashraf, U.; Li, Y.; Wei, S.; Wan, S.; Zohaib, A.; Song, Y.; Chen, H.; Cao, S. MicroRNA-33a-5p Modulates Japanese Encephalitis Virus Replication by Targeting Eukaryotic Translation Elongation Factor 1A1. J. Virol. 2016, 90, 3722–3734. [Google Scholar] [CrossRef]
- Bhagat, R.; Rajpara, P.; Kaur, G.; Gupta, K.; Seth, P. Zika virus E protein dysregulate mir-204/WNT2 signalling in human fetal neural stem cells. Brain Res. Bull. 2021, 176, 93–102. [Google Scholar] [CrossRef]
- Ye, H.; Kang, L.; Yan, X.; Li, S.; Huang, Y.; Mu, R.; Duan, X.; Chen, L. MiR-103a-3p Promotes Zika Virus Replication by Targeting OTU Deubiquitinase 4 to Activate p38 Mitogen-Activated Protein Kinase Signaling Pathway. Front. Microbiol. 2022, 13, 862580. [Google Scholar] [CrossRef]
- Hussain, M.; Walker, T.; O’Neill, S.L.; Asgari, S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2013, 43, 146–152. [Google Scholar] [CrossRef]
- Yan, H.; Zhou, Y.; Liu, Y.; Deng, Y.; Chen, X. miR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J. Med. Virol. 2014, 86, 1428–1436. [Google Scholar] [CrossRef]
- Slonchak, A.; Hussain, M.; Torres, S.; Asgari, S.; Khromykh, A.A. Expression of mosquito microRNA Aae-miR-2940-5p is downregulated in response to West Nile virus infection to restrict viral replication. J. Virol. 2014, 88, 8457–8467. [Google Scholar] [CrossRef] [PubMed]
- Procyk, G.; Grodzka, O.; Procyk, M.; Gąsecka, A.; Głuszek, K.; Wrzosek, M. MicroRNAs in Myocarditis-Review of the Preclinical In Vivo Trials. Biomedicines 2023, 11, 2723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.L.; Li, Y.X.; Zheng, S.Q.; Liu, M.; Li, X.; Tang, H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antivir. Res. 2010, 88, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.T.; Pham, A.M.; Lorini, M.H.; Chua, M.A.; Steel, J.; tenOever, B.R. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 2009, 27, 572–576. [Google Scholar] [CrossRef]
- He, F.; Yao, H.; Wang, J.; Xiao, Z.; Xin, L.; Liu, Z.; Ma, X.; Sun, J.; Jin, Q.; Liu, Z. Coxsackievirus B3 engineered to contain microRNA targets for muscle-specific microRNAs displays attenuated cardiotropic virulence in mice. J. Virol. 2015, 89, 908–916. [Google Scholar] [CrossRef]
- Xiao, Z.; He, F.; Feng, M.; Liu, Z.; Liu, Z.; Li, S.; Wang, W.; Yao, H.; Wu, J. Engineered coxsackievirus B3 containing multiple organ-specific miRNA targets showed attenuated viral tropism and protective immunity. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2022, 103, 105316. [Google Scholar] [CrossRef]
- Hassan, M.; Iqbal, M.S.; Naqvi, S.; Alashwal, H.; Moustafa, A.A.; Kloczkowski, A. Prediction of Site Directed miRNAs as Key Players of Transcriptional Regulators Against Influenza C Virus Infection Through Computational Approaches. Front. Mol. Biosci. 2022, 9, 866072. [Google Scholar] [CrossRef]
- Witkos, T.M.; Koscianska, E.; Krzyzosiak, W.J. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar] [CrossRef]
- Saito, T.; Saetrom, P. MicroRNAs--targeting and target prediction. New Biotechnol. 2010, 27, 243–249. [Google Scholar] [CrossRef]
- Maragkakis, M.; Alexiou, P.; Papadopoulos, G.L.; Reczko, M.; Dalamagas, T.; Giannopoulos, G.; Goumas, G.; Koukis, E.; Kourtis, K.; Simossis, V.A.; et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinform. 2009, 10, 295. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Lee, R.; Williams, P.; Chan, C.Y.; Ambros, V.; Ding, Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007, 14, 287–294. [Google Scholar] [CrossRef]
- Rennie, W.; Liu, C.; Carmack, C.S.; Wolenc, A.; Kanoria, S.; Lu, J.; Long, D.; Ding, Y. STarMir: A web server for prediction of microRNA binding sites. Nucleic Acids Res. 2014, 42, W114–W118. [Google Scholar] [CrossRef]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; Pérez-Ishiwara, D.G.; Cerecedo-Mercado, D.A.; Del Ángel, R.M.; Salas-Benito, J.S. miR-927 has pro-viral effects during acute and persistent infection with dengue virus type 2 in C6/36 mosquito cells. J. Gen. Virol. 2020, 101, 825–839. [Google Scholar] [CrossRef]
- Asgari, S. Role of microRNAs in arbovirus/vector interactions. Viruses 2014, 6, 3514–3534. [Google Scholar] [CrossRef]
- Lucas, K.; Raikhel, A.S. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 2013, 43, 24–38. [Google Scholar] [CrossRef]
- Ylla, G.; Fromm, B.; Piulachs, M.D.; Belles, X. The microRNA toolkit of insects. Sci. Rep. 2016, 6, 37736. [Google Scholar] [CrossRef]
- Li, X.; Huang, Y.; Zhang, Y.; He, N. Evaluation of microRNA Expression in Patients with Herpes Zoster. Viruses 2016, 8, 326. [Google Scholar] [CrossRef] [PubMed]
- Busto, G.U.; Guven-Ozkan, T.; Fulga, T.A.; Van Vactor, D.; Davis, R.L. microRNAs That Promote or Inhibit Memory Formation in Drosophila melanogaster. Genetics 2015, 200, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Marco, A. Selection Against Maternal microRNA Target Sites in Maternal Transcripts. G3 2015, 5, 2199–2207. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Igaki, T. Yorkie drives Ras-induced tumor progression by microRNA-mediated inhibition of cellular senescence. Sci. Signal. 2021, 14, eaaz3578. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Jia, X.; Wan, H.; Wang, S.; Zhang, X.; Zhang, Z.; Wang, Y. miR-9 and miR-263 Regulate the Key Genes of the ERK Pathway in the Ovary of Mud Crab Scylla paramamosain. Mar. Biotechnol. 2020, 22, 594–606. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, X.; Han, K.; Jia, X.; Zhou, M.; Zhang, Z.; Wang, Y. The expression regulation of Cyclins and CDKs in ovary via miR-9c and miR-263a of Scylla paramamosain. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2021, 254, 110567. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, B.; Zhao, H.; Li, Y.; Zhang, X.; Wen, J. Downregulation of lncRNA MIR181A2HG by high glucose impairs vascular endothelial cell proliferation and migration through the dysregulation of the miRNAs/AKT2 axis. Int. J. Mol. Med. 2021, 47, 35. [Google Scholar] [CrossRef]
- Pedersen, M.E.; Snieckute, G.; Kagias, K.; Nehammer, C.; Multhaupt, H.A.; Couchman, J.R.; Pocock, R. An epidermal microRNA regulates neuronal migration through control of the cellular glycosylation state. Science 2013, 341, 1404–1408. [Google Scholar] [CrossRef]
- Wang, Z.; Xia, X.; Li, J.; Igaki, T. Tumor elimination by clustered microRNAs miR-306 and miR-79 via noncanonical activation of JNK signaling. eLife 2022, 11, e77340. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, H.; Yang, F.; Wu, C.; Jiang, C.; Yu, W.; Liu, K.; Sheng, Q.; Nie, Z. Bmo-miR-79 downregulates the expression of BmEm4 in the silkworm, Bombyx mori. Gene 2019, 690, 113–119. [Google Scholar] [CrossRef]
- Chen, M.; Storey, K.B. Large-scale identification and comparative analysis of miRNA expression profile in the respiratory tree of the sea cucumber Apostichopus japonicus during aestivation. Mar. Genom. 2014, 13, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Artiagas-Jerónimo, S.; Alberdi, P.; Villar, R.M.; Cabezas-Cruz, A.; Espinosa, P.J.P.; Mateos-Hernámdez, L.; de la Fuente, J. Anaplasma phagocytophilum modifies tick cell microRNA expression and upregulates isc-mir-79 to facilitate infection by targeting the Roundabout protein 2 pathway. Sci. Rep. 2019, 9, 9073. [Google Scholar] [CrossRef]
- He, S.; Wu, J.; Han, D.; Li, Y.; Wang, T.; Wei, H.; Pan, Y.; Zang, H. Differential expression profile of plasma exosomal microRNAs in chronic rhinosinusitis with nasal polyps. Exp. Biol. Med. 2022, 247, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Y.; Wang, X.; Huang, J.; Guo, W.; Wei, P.; Li, G.; Wang, Z.; Huang, Z.; Zhang, L. Putative biomarkers of malignant transformation of sinonasal inverted papilloma into squamous cell carcinoma. J. Int. Med. Res. 2019, 47, 2371–2380. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Wu, B.; Shi, C.; Li, C. MicroRNA-661 promotes non-small cell lung cancer progression by directly targeting RUNX3. Mol. Med. Rep. 2017, 16, 2113–2120. [Google Scholar] [CrossRef]
- Zhou, G.H.; Yang, W.H.; Sun, B. Clinical impact of serum miR-661 in diagnosis and prognosis of non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5696–5701. [Google Scholar] [CrossRef]
- Liu, F.; Cai, Y.; Rong, X.; Chen, J.; Zheng, D.; Chen, L.; Zhang, J.; Luo, R.; Zhao, P.; Ruan, J. MiR-661 promotes tumor invasion and metastasis by directly inhibiting RB1 in non small cell lung cancer. Mol. Cancer 2017, 16, 122. [Google Scholar] [CrossRef]
- Bao, Y.; Yu, Y.; Hong, B.; Lin, Z.; Qi, G.; Zhou, J.; Liu, K.; Zhang, X. Hsa_Circ_0001947/MiR-661/DOK7 Axis Restrains Non-Small Cell Lung Cancer Development. J. Microbiol. Biotechnol. 2021, 31, 1508–1518. [Google Scholar] [CrossRef]
- Ren, C.; Cui, L.; Li, R.; Song, X.; Li, J.; Xi, Q.; Zhang, Z.; Zhao, L. Hsa_circ_0080608 Attenuates Lung Cancer Progression by Functioning as a Competitive Endogenous RNA to Regulate the miR-661/ADRA1A Pathway. Horm. Metab. Res. 2023, 55, 876–884. [Google Scholar] [CrossRef]
- Liu, F.; Gong, R.; He, B.; Chen, F.; Hu, Z. TUSC2P suppresses the tumor function of esophageal squamous cell carcinoma by regulating TUSC2 expression and correlates with disease prognosis. BMC Cancer 2018, 18, 894. [Google Scholar] [CrossRef]
- Long, H.; Li, Y.; Wang, H.; Guo, B.; Song, S.; Zhe, X.; Li, H.; Li, D.; Shao, R.; Pan, Z. C/EBPβ expression decreases in cervical cancer and leads to tumorigenesis. BMC Cancer 2013, 23, 79. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zhu, C.; Qiu, R.; Zan, P.; Zheng, Z.; Xu, T.; Li, G. MicroRNA-661 Enhances TRAIL or STS Induced Osteosarcoma Cell Apoptosis by Modulating the Expression of Cytochrome c1. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 41, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.D.; Pakala, S.B.; Ohshiro, K.; Rayala, S.K.; Kumar, R. MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Res. 2009, 69, 5639–5642. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, Y.; Bublik, D.R.; Pilpel, Y.; Oren, M. miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell Death Differ. 2014, 21, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, L.; He, L.; Wang, J.; Shi, X.; Li, Z.; Shi, S.; Hou, K.; Teng, Y.; Qu, X. MiR-891a-5p as a prognostic marker and therapeutic target for hormone receptor-positive breast cancer. J. Cancer 2020, 11, 3771–3782. [Google Scholar] [CrossRef]
- Sui, C.; Qu, W.; Lian, Y.; Feng, C.; Zhan, Y. Hsa_circ_0069094 knockdown inhibits cell proliferation, migration, invasion and glycolysis, while induces cell apoptosis by miR-661/HMGA1 axis in breast cancer. Anti-Cancer Drugs 2021, 32, 829–841. [Google Scholar] [CrossRef]
- Almohaywi, M.; Sugita, B.M.; Centa, A.; Fonseca, A.S.; Antunes, V.C.; Fadda, P.; Mannion, C.M.; Abijo, T.; Goldberg, S.L.; Campbell, M.C.; et al. Deregulated miRNA Expression in Triple-Negative Breast Cancer of Ancestral Genomic-Characterized Latina Patients. Int. J. Mol. Sci. 2023, 24, 13046. [Google Scholar] [CrossRef]
- Surmiak, M.; Kosałka-Węgiel, J.; Polański, S.; Sanak, M. Endothelial cells response to neutrophil-derived extracellular vesicles miRNAs in anti-PR3 positive vasculitis. Clin. Exp. Immunol. 2021, 204, 267–282. [Google Scholar] [CrossRef]
- Kang, S.H.; Choi, J.S. MicroRNA-661 upregulation in myelodysplastic syndromes induces apoptosis through p53 activation and associates with decreased overall survival. Leuk. Lymphoma 2019, 60, 2779–2786. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Z.; Yang, S. Circ_RUSC2 upregulates the expression of miR-661 target gene SYK and regulates the function of vascular smooth muscle cells. Biochem. Cell Biol. 2019, 97, 709–714. [Google Scholar] [CrossRef]
- Biranvand, A.S.; Khosravi, M.; Esfandiari, G.; Poursaleh, A.; Hosseini-Fard, S.R.; Amirfarhangi, A.; Najafi, M. Associations between miR-661, miR-1202, lncRNA-HOTAIR, lncRNA-GAS5 and MMP9 in differentiated M2-macrophages of patients with varicose veins. Int. Angiol. J. Int. Union Angiol. 2018, 37, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Xie, G. Circular RNA hsa-circ-0012129 Promotes Cell Proliferation and Invasion in 30 Cases of Human Glioma and Human Glioma Cell Lines U373, A172, and SHG44, by Targeting MicroRNA-661 (miR-661). Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 2497–2507. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Liu, M.; Liu, Y.; Li, Y.; Xu, Z.; He, H.; Liu, J.; Zhang, Y.; Ke, Y. Lcn2-derived Circular RNA (hsa_circ_0088732) Inhibits Cell Apoptosis and Promotes EMT in Glioma via the miR-661/RAB3D Axis. Front. Oncol. 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Qi, Y.; Zhang, X.; Guan, Z.; Han, W.; Peng, X. Production and Stabilization of Specific Upregulated Long Noncoding RNA HOXD-AS2 in Glioblastomas Are Mediated by TFE3 and miR-661, Respectively. Int. J. Mol. Sci. 2022, 23, 2828. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Zheng, K.; Yu, J.; Huang, Z. MicroRNA-661 expression is upregulated in pancreatic ductal adenocarcinoma and promotes cell proliferation. Oncol. Lett. 2018, 16, 6293–6298. [Google Scholar] [CrossRef]
- Lee, J.; Lee, H.S.; Park, S.B.; Kim, C.; Kim, K.; Jung, D.E.; Song, S.Y. Identification of Circulating Serum miRNAs as Novel Biomarkers in Pancreatic Cancer Using a Penalized Algorithm. Int. J. Mol. Sci. 2021, 22, 1007. [Google Scholar] [CrossRef]
- Hojati, Z.; Omidi, F.; Dehbashi, M.; Mohammad Soltani, B. The Highlighted Roles of Metabolic and Cellular Response to Stress Pathways Engaged in Circulating hsa-miR-494-3p and hsa-miR-661 in Alzheimer’s Disease. Iran. Biomed. J. 2021, 25, 62–67. [Google Scholar] [CrossRef]
- Ali, M.A.; Matboli, M.; El-Khazragy, N.; Saber, O.; El-Nakeep, S.; Abdelzaher, H.M.; Shafei, A.E.; Mostafa, R. Investigating miRNA-661 and ATG4-B mRNA expression as potential biomarkers for hepatocellular carcinoma. Biomark. Med. 2018, 12, 245–256. [Google Scholar] [CrossRef]
- Matboli, M.; Hassan, M.K.; Ali, M.A.; Mansour, M.T.; Elsayed, W.; Atteya, R.; Aly, H.S.; Meteini, M.E.; Elghazaly, H.; El-Khamisy, S.; et al. Impact of circ-0000221 in the Pathogenesis of Hepatocellular via Modulation of miR-661-PTPN11 mRNA Axis. Pharmaceutics 2022, 14, 138. [Google Scholar] [CrossRef]
- Rodosthenous, R.S.; Burris, H.H.; Sanders, A.P.; Just, A.C.; Dereix, A.E.; Svensson, K.; Solano, M.; Téllez-Rojo, M.M.; Wright, R.O.; Baccarelli, A.A. Second trimester extracellular microRNAs in maternal blood and fetal growth: An exploratory study. Epigenetics 2017, 12, 804–810. [Google Scholar] [CrossRef]
- Cuman, C.; Van Sinderen, M.; Gantier, M.P.; Rainczuk, K.; Sorby, K.; Rombauts, L.; Osianlis, T.; Dimitriadis, E. Human Blastocyst Secreted microRNA Regulate Endometrial Epithelial Cell Adhesion. EBioMedicine 2015, 2, 1528–1535. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilivand, M.; Fattahi, A.; Abedelahi, A.; Hamdi, K.; Farzadi, L.; Goharitaban, S.; Niknafs, B. microRNAs in the blastocoel fluid as accessible indicators of chromosomal normality. Reprod. Biol. 2022, 22, 100695. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.P.; Hillen, M.R.; Chouri, E.; Blokland, S.L.M.; Bekker, C.P.J.; Kruize, A.A.; Rossato, M.; van Roon, J.A.G.; Radstake, T.R.D.J. Circulating small non-coding RNAs reflect IFN status and B cell hyperactivity in patients with primary Sjögren’s syndrome. PLoS ONE 2018, 13, e0193157. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; He, H.; Chen, Y.; Zhu, D.; Jiang, T.; Wang, J. CircPDE7B/miR-661 axis accelerates the progression of human keloid fibroblasts by upregulating fibroblast growth factor 2 (FGF2). Mol. Cell. Biochem. 2022, 477, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wan, S.; Yang, T.; Niu, D.; Zhang, A.; Yang, C.; Cai, J.; Wu, J.; Song, J.; Zhang, C.Y.; et al. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci. Rep. 2016, 6, 20032. [Google Scholar] [CrossRef]
- VatanIman, R.; Malekpour, S.H.; Afshari, A.; Zare, M. MiR-770-5p, miR-661 and miR-571 expression level in serum and tissue samples of foot ulcer caused by diabetes mellitus type II in Iranian population. Mol. Biol. Rep. 2021, 48, 7811–7818. [Google Scholar] [CrossRef]
- Zhu, T.; Yuan, J.; Wang, Y.; Gong, C.; Xie, Y.; Li, H. MiR-661 contributed to cell proliferation of human ovarian cancer cells by repressing INPP5J expression. Biomed. Pharmacother. 2015, 75, 123–128. [Google Scholar] [CrossRef]

| miRNA | Flavivirus | Target Position | 3′ UTR Target Sequence |
|---|---|---|---|
| miR-6842-5p | DENV1 | 10,508–10,536 | actagtggttagaggagacccctcccaa |
| DENV2 | 10,587–10,608 | aggttagaggagacccccccga | |
| DENV3 | 10,571–10,590 | aggttagaggagacccccc | |
| DENV4 | 10,513–10,535 | aggttagaggagacccccccaa | |
| YFV | 10,622–10,650 | acctggtttctgggacctcccaccccag | |
| ZIKV | 10,661–10,684 | actagtggttagaggagacccccc | |
| JEV | 10,831–10,852 | aggttagaggagaccccgcat | |
| MVEV | 10,862–10,886 | aggttagaggagaccccactctca | |
| USUV | 10,911–10,934 | agaggttagaggagaccccgcat | |
| WNV1 | 10,807–10,824 | aggttagaggagaccccg | |
| WNV2 | 10,807–10,824 | aggttagaggagaccccg | |
| miR-6791-5p | DENV1 | 10,428–10,455 | gccgtgctgcctgtagctccatcgtgggga |
| DENV2 | 10,344–10,365 | cctgtgagccccgtccaagga | |
| DENV3 | 10,399–10,429 | accgtgctgcctgtagctccgtcgtgggga | |
| DENV4 | 10,327–10,360 | accgtgctgcctgtagctccgccaataatggga | |
| YFV | 10,534–10,554 | gttgtcagcccagaaccccac | |
| ZIKV | 10,397–10,429 | gtcaggcctgctagtcagccacagtttgggga | |
| JEV | 10,655–10,679 | gcggcctgcgcagccccaggagga | |
| MVEV | 10,539–10,564 | gctgcctgcgaccaaccccaggagg | |
| USUV | 10,739–10,761 | agcctgtacggccccaggagga | |
| WNV1 | 10,548–10,572 | gctgcctgcgactcaaccccagga | |
| WNV2 | 10,483–10,510 | gctgcctgcggctcaaccccaggagga | |
| miR-6765-5p | DENV1 | 10,357–10,382 | tatgctgcctgtgagccccgtccaa |
| DENV2 | 10,336–10,362 | ctatgctacctgtgagccccgtccaa | |
| DENV3 | 10,330–10,353 | tgctgcctgtgagccccgtccaa | |
| DENV4 | 10,322–10,351 | agcaaaccgtgctgcctgtagctccgcca | |
| YFV | 10,436–10,467 | ccacggctggagaaccgggctccgcacttaa | |
| ZIKV | 10,745–10,771 | cgctggccgccaggcacagatcgccg | |
| JEV | 10,698–10,721 | agcccccacggcccaagcctcgt | |
| MVEV | 10,797–10,827 | gagaccctgcggaagaaatgagtggcccaa | |
| USUV | 10,766–10,797 | ttaccaaagccgaaaggcccccacggcccaa | |
| WNV1 | 10,834–10,868 | tgcacggcccagcctggctgaagctgtaggtcag | |
| WNV2 | 10,537–10,564 | ccacgtaagccctcagaaccgtctcgg | |
| miR-6762-3p | DENV1 | 10,551–10,578 | cggggcccaacaccaggggaagctgta |
| DENV2 | 10,539–10,566 | atgggggcccaaggcgagatgaagctg | |
| DENV3 | 10,458–10,486 | gtggggacgtaaaacctgggaggctgca | |
| DENV4 | 10,355–10,392 | gggaggcgtaataatccccagggaggccatgcgccac | |
| YFV | 10,719–10,753 | aggagaccctccagggaacaaatagtgggaccat | |
| ZIKV | 10,627–10,668 | actggagactagctgtgaatctccagcagagggactagtgg | |
| JEV | 10,760–10,800 | aggttagaggagaccccgtggaaacaacaatatgcggccc | |
| MVEV | 10,795–10,834 | aggagaccctgcggaagaaatgagtggcccaagctcgcc | |
| USUV | 10,844–10,874 | aggagaccccgtggaacttaggtgcggccc | |
| WNV1 | 10,497–10,529 | aggagaaagtcaggccgggaagttcccgccac | |
| WNV2 | 10,869–10,912 | cctgggatagactaggggatcttctgctctgcacaaccagccac | |
| miR-6756-5p | DENV1 | 10,424–10,450 | aagccgtgctgcctgtagctccatcg |
| DENV2 | 10,414–10,435 | tgcagcctgtagctccacctg | |
| DENV3 | 10,325–10,354 | aagctgtgctgcctgtgagccccgtccaa | |
| DENV4 | 10,328–10,350 | cgtgctgcctgtagctccgcca | |
| YFV | 10,663–10,686 | gagcctccgctaccaccctccca | |
| ZIKV | 10,590–10,613 | aggtggcgaccttccccaccctt | |
| JEV | 10,665–10,713 | agccccaggaggactgggttaccaaagccgttgagcccccacggccca | |
| MVEV | 10,701–10,749 | aggccccaggaggactgggtaaacaaagccgtaaggcccccgcagcccg | |
| USUV | 10,747–10,796 | cggccccaggaggactgggttaccaaagccgaaaggcccccacggcccaa | |
| WNV1 | 10,752–10,775 | cgccccacgcggccctagccccg | |
| WNV2 | 10,868–10,598 | aggaccccacgtgctttagcctcaaagccca | |
| miR-661 | DENV1 | 10,518–10,563 | aacgcagcagcggggcccaacaccaggggaagctgtaccctggtg |
| DENV2 | 10,528–10,556 | tcgcagcaacaatgggggcccaaggcga | |
| DENV3 | 10,514–10,535 | aacgcagcagcggggcccgag | |
| DENV4 | 10,552–10,580 | gacgctgggaaagaccagagatcctgct | |
| YFV | 10,584–10,615 | agtgcaggctgggacagccgacctccaggtt | |
| ZIKV | 10,479–10,504 | agtcaggccgagaacgccatggcac | |
| JEV | 10,659–10,685 | ctgcgcagccccaggaggactgggtt | |
| MVEV | 10,743–10,780 | agcccgggccgggaggaggtgatgcaaaccccggcga | |
| USUV | 10,584–10,612 | ggtgctgcctgcgactcaaccccaggcgg | |
| WNV1 | 10,915–10,945 | agctgtaggtcaggggaaggactagaggtt | |
| WNV2 | 10,634–10,662 | agtgcagtctgcgatagtgccccaggtg | |
| miR-608 | DENV1 | 10,549–10,583 | agcggggcccaacaccaggggaagctgtaccctg |
| DENV2 | 10,633–10,676 | gggaaagaccagagatcctgctgtctcctcagcatcattcca | |
| DENV3 | 10,616–10,658 | gggagagaccagagatcctgctgtctcctcagcatcattcca | |
| DENV4 | 10,558–10,600 | gggaaagaccagagatcctgctgtctctgcaacatcaatcca | |
| YFV | 10,659–10,683 | aacggagcctccgctaccaccctc | |
| ZIKV | 10,501–10,560 | cacggaagaagccatgctgcctgtgagcccctcagaggacactgagtcaaaaaacccca | |
| JEV | 10,495–10,522 | gacggtgctgtctgcgtctcagtccca | |
| MVEV | 10,532–10,558 | gacggtgctgcctgcgaccaacccca | |
| USUV | 10,582–10,609 | gacggtgctgcctgcgactcaacccca | |
| WNV1 | 10,543–10,570 | gacggtgctgcctgcgactcaacccca | |
| WNV2 | 10,478–10,505 | gacggtgctgcctgcggctcaacccca | |
| miR-4722-5p | DENV1 | 10,637–10,666 | tgacgctgggagagaccagagatcctgct |
| DENV2 | 10,626–10,656 | tgacgctgggaaagaccagagatcctgctg | |
| DENV3 | 10,609–10,639 | tgacgctgggagagaccagagatcctgctg | |
| DENV4 | 10,551–10,580 | tgacgctgggaaagaccagagatcctgct | |
| YFV | 10,587–10,611 | gcaggctgggacagccgacctcca | |
| ZIKV | 10,494–10,524 | cgccatggcacggaagaagccatgctgcct | |
| JEV | 10,616–10,657 | gcggcctgcgcagccccaggaggactgggttaccaaagccg | |
| MVEV | 10,726–10,750 | aagccgtaaggcccccgcagcccg | |
| USUV | 10,895–10,952 | agaggttagaggagaccccgtggaacttaggtgcggcccaagccgtttccgaagctg | |
| WNV1 | 10,780–10,854 | agaccccgcggtttaaagtgcacggcccagcctggct | |
| WNV2 | 10,865–10,896 | cacctgggatagactaggggatcttctgctc |
| miRNA | Flavivirus | Target Position | 3′ UTR Flavivirus Sequence |
|---|---|---|---|
| miR-9c-5p | DENV1 | 10,635–10,660 | attgacgctgggagagaccagagat |
| DENV2 | 10,624–10,649 | attgacgctgggaaagaccagagat | |
| DENV3 | 10,608–10,633 | attgacgctgggagagaccagagat | |
| DENV4 | 10,550–10,575 | attgacgctgggaaagaccagagat | |
| YFV | 10,437–10,456 | cacggctggagaaccgggc | |
| ZIKV | 10,635–10,661 | aacagcatattgacgctgggaaagac | |
| miR-9b | DENV1 | 10,630–10,660 | agcatattgacgctgggagagaccagagat |
| DENV2 | 10,618–10,648 | agcatattgacgctgggagagaccagagat | |
| DENV3 | 10,603–10,633 | agcatattgacgctgggagagaccagagat | |
| DENV4 | 10,545–10,575 | agcatattgacgctgggaaagaccagagat | |
| YFV | 10,583–10,610 | gcagtgcaggctgggacagccgacctc | |
| ZIKV | 10,638–10,661 | agcatattgacgctgggaaagac | |
| miR-9a | DENV1 | 10,631–10,660 | gcatattgacgctgggagagaccagagat |
| DENV2 | 10,620–10,629 | gcatattgacgctgggaaagaccagagat | |
| DENV3 | 10,604–10,633 | gcatattgacgctgggagagaccagagat | |
| DENV4 | 10,546–10,566 | gcatattgacgctgggaaagaccagagat | |
| YFV | 10,437–10,456 | cacggctggagaaccgggc | |
| ZIKV | 10,635–10,667 | aacagcatattgacgtgggaaagaccagagac | |
| miR-92a-3p | DENV1 | 10,327–10,356 | tcaggccggattaagccatagcacggtaa |
| DENV2 | 10,542–10,571 | ggggcccaaggcgagatgaagctgtagtc | |
| DENV3 | 10,422– 10,453 | cgtggggacgtaaaacctgggaggctgcaaa | |
| DENV4 | 10,603–10,632 | cacagagcgccgcaagatggattggtgtt | |
| YFV | 10,587–10,621 | tgcaggctgggacagccgacctccaggttgcgaa | |
| ZIKV | 10,573–10,600 | cgcaggatgggaaaagaaggtggcgac | |
| miR-92b-3p | DENV1 | 10,326–10,350 | tcaggccggattaagccatagcac |
| DENV2 | 10,506–10,531 | gcatggcgtagtggactagcggtta | |
| DENV3 | 10,421–10,452 | cgtggggacgtaaaacctgggaggctgcaaa | |
| DENV4 | 10,539–10,566 | gagcgccgcaagatggattggtgttgt | |
| YFV | 10,587–10,621 | tgcaggctgggacagccgacctccaggttgcgaa | |
| ZIKV | 10,572–10,599 | cgcaggatgggaaaagaaggtggcgac | |
| miR-79-5p | DENV1 | 10,340–10,375 | ccatagcacggtaagagctatgctgcctgtgagcc |
| DENV2 | 10,528–10,528 | cgcagcaacaatgggggcccaaggcg | |
| DENV3 | 10,516–10,537 | cgcagcagcggggcccgagca | |
| DENV4 | 10,456–10,456 | cgcagcaaaagggggcccgaagcc | |
| YFV | 10,437–10,456 | cacggctggagaaccgggc | |
| ZIKV | 10,475–10,498 | ctcatagtcaggccgagaacgcc | |
| miR-316 | DENV1 | 10,499–10,527 | ggtagcagactagtggttagaggagacc |
| DENV2 | 10,482–10,513 | atggcgtagtggactagcggttagaggagac | |
| DENV3 | 10,477–10,502 | agcagactagcggttagaggagacc | |
| DENV4 | 10,408–10,438 | tggcatattggactagcggttagaggagac | |
| YFV | 10,579–10,601 | aggcagtgcaggctgggacag | |
| ZIKV | 10,775–10,801 | tcggcggccggtgtggggaaatccat | |
| miR-2945-5p | DENV1 | 10,636–10,667 | ttgacgctgggagagaccagagatcctgctg |
| DENV2 | 10,625–10,656 | ttgacgctgggaaagaccagagatcctgctg | |
| DENV3 | 10,609–10,640 | ttgacgctgggaaagaccagagatcctgctg | |
| DENV4 | 10,551–10,582 | ttgacgctgggaaagaccagagatcctgctg | |
| YFV | 10,440–10,460 | ggctggagaaccgggctccg | |
| ZIKV | 10,466–10,497 | ccatggcacggaagaagccatgctgcctgtg | |
| miR-11924 | DENV1 | 10,401–10,420 | ggccgaaagccacggttcg |
| DENV2 | 10,703–10,723 | ctgttgaatcaacaggttct | |
| DENV3 | 10,687–10,707 | ctgttgaatcaacaggttct | |
| DENV4 | 10,629–10,649 | ttgttgatccaacaggttct | |
| YFV | 10,599–10,616 | agccgacctccaggttg | |
| ZIKV | 10,752–10,788 | cgccaggcacagatcgccgaacttcggcggccggtg | |
| miR-11919-5p | DENV1 | 10,548–10,586 | cagcggggcccaacaccaggggaagctgtaccctggtg |
| DENV2 | 10,472–10,502 | aagctgtacgcatggcgtagtggactagcg | |
| DENV3 | 10,459–10,488 | agctgtacgcacggtgtagcagactagcg | |
| DENV4 | 10,600–10,624 | aggcacagagcgccgcaagatgga | |
| YFV | 10,580–10,632 | aggcagtgcaggctgggacagccgacctccaggttgcgaaaaacctggttt | |
| ZIKV | 10,428–10,465 | aagctgtgcagcctgtaacccccccaggagaagctgg | |
| miR-11-3p | DENV1 | 10,315–10,337 | aggcaagaagtcaggccggatt |
| DENV2 | 10,422–10,472 | aggcacagaacgccagaaaatggaatggtgctgttgaatcaacaggttct | |
| DENV3 | 10,428–10,459 | aggcacagaacgccagaaaatggaatggtgc | |
| DENV4 | 10,286–10,313 | aggctattgaagtcaggccacttgtgc | |
| YFV | 10,820–10,846 | tcaagaataagcagacctttggatga | |
| ZIKV | 10,620–10,647 | gggcctgaactggagactagctgtgaa | |
| miR-282-5p | DENV1 | 10,564–10,600 | ccaggggaagctgtaccctggtggtaaggactagag |
| DENV2 | 10,553–10,588 | gagatgaagctgtagtctcgctggaaggactagag | |
| DENV3 | 10,426–10,449 | ggacgtaaaacctgggaggctgc | |
| DENV4 | 10,457–10,492 | aggaggaagctgtactcctggtggaaggactagag | |
| YFV | 10,698–10,724 | aagacggggtctagaggttagaggag | |
| ZIKV | 10,423–10,468 | tggggaaagctgtgcagcctgtaacccccccaggagaagctggga |
| miRNA | Flavivirus | Target Position | 3′ UTR Flavivirus Sequence |
|---|---|---|---|
| miR-993-5p | JEV | 10,469–10,496 | caaaagctgccaccggatactgggtag |
| MVEV | 10,699–10,723 | tgaggccccaggaggactgggtaa | |
| USUV | 10,748–10,769 | cggccccaggaggactgggtt | |
| WNV1 | 10,844–10,873 | cagcctggctgaagctgtaggtcagggg | |
| WNV2 | 10,504–10,650 | aggaggactgggtgaccaaagctgcgaggtga | |
| miR-989 | JEV | 10,498–10,523 | ggtgctgtctgcgtctcagtcccag |
| MVEV | 10,651–10,670 | agcccgtgtcagatcgcga | |
| USUV | 10,709–10,731 | ggccgcaaagcgccacttcgcc | |
| WNV1 | 10,973–10,994 | agccacacggcacagtgcgcc | |
| WNV2 | 10,906–10,927 | agccacacggcacagtgcgcc | |
| miR-79 | JEV | 10,914–10,945 | acatcagctactaggcacagagcgccgaagt |
| MVEV | 10,653–10,692 | ccgtgtcagatcgcgaaagcgccacttcgccgaggagtg | |
| USUV | 10,865–10,892 | ggtgcggcccaagccgtttccgaagct | |
| WNV1 | 10,975–11,007 | ccacacggcacagtgcgccgacaatggtggct | |
| WNV2 | 10,784–10,807 | tggctgaagctgtaagccaaggg | |
| miR-71-3p | JEV | 10,818–10,846 | ggtggaaggactagaggttagaggagac |
| MVEV | 10,778–10,803 | cgaaggactagaggttagaggagac | |
| USUV | 10,553–10,582 | tcgtggaaggactagaggttagaggagac | |
| WNV1 | 10,870–10,895 | ggaaggactagaggttagtggagac | |
| WNV2 | 10,805–10,830 | ggaaggactagaggttagaggagac | |
| miR-305-5p | JEV | 10,930–10,963 | cagagcgccgaagtatgtagctggtggtgagga |
| MVEV | 10,969–11,002 | cgagcgccgaacactgtgactgatgggggagaa | |
| USUV | 10,553–10,582 | cagggcaacctgccaccggaagttgagta | |
| WNV1 | 10,985–11,018 | cagtgcgccgacaatggtggctggtggtgcgag | |
| WNV2 | 10,618–10,644 | tgtgccactctgcggagagtgcagtc | |
| miR-285-5p | JEV | 10,591–10,609 | cctgctcactggaagttg |
| MVEV | 10,513–10,553 | gctgccaccgaaggttggtagacggtg | |
| USUV | 10,561–10,582 | cctgccaccggaagttgagta | |
| WNV1 | 10,956–10,979 | ttctgctctgcacaaccagccac | |
| WNV2 | 10,889–10,912 | ttctgctctgcacaaccagccac | |
| miR-283 | JEV | 10,669–10,702 | ccccaggaggactgggttaccaaagccgttgag |
| MVEV | 10,554–10,597 | ccccaggaggactgggttaccaaagctgattctccacggttgg | |
| USUV | 10,603–10,646 | ccccaggcggactgggttaacaaagctgaccgctgatgatgg | |
| WNV1 | 10,565–10,585 | ccccaggaggactgggtgaa | |
| WNV2 | 10,500–10,520 | ccccaggaggactgggtgac | |
| miR-278 | JEV | 10,578–10,597 | tcggaagtaggtccctgct |
| MVEV | 10,616–10,637 | tcggaagaggagtccctgcca | |
| USUV | 10,607–10,641 | caggcggactgggttaacaaagctgaccgctgat | |
| WNV1 | 10,498–10,517 | aggagaaagtcaggccggg | |
| WNV2 | 10,560–10,579 | tcggaaggaggaccccacg | |
| miR-210-3p | JEV | 10,693–10,720 | aagccgttgagcccccacggcccaagc |
| MVEV | 10,649–10,673 | aagcccgtgtcagatcgcgaaagc | |
| USUV | 10,697–10,718 | cagcccgtgtcaggccgcaaa | |
| WNV1 | 10,657–10,684 | aagcccaatgtcagaccacgctacggc | |
| WNV2 | 10,579–10,600 | aagcccagtgtcagaccacac | |
| miR-2 | JEV | 10,936–10,962 | cgccgaagtatgtagctggtggtgag |
| MVEV | 10,827–10,855 | agctcgccgaagctgtaaggcgggtgga | |
| USUV | 10,623–10,646 | acaaagctgaccgctgatgatgg | |
| WNV1 | 10,990–11,015 | cgccgacaatggtggctggtggtgc | |
| WNV2 | 10,923–10,948 | cgccgacataggtggctggtggtgc | |
| miR-14 | JEV | 10,809–10,834 | tgtagaggaggtggaaggactagag |
| MVEV | 10,684–10,720 | gaggagtgcaatctgtgaggccccaggaggactggg | |
| USUV | 10,731–10,767 | aaggagtgcagcctgtacggccccaggaggactggg | |
| WNV1 | 10,859–10,883 | tgtaggtcaggggaaggactagag | |
| WNV2 | 10,726–10,757 | agggagaagggactagaggttagaggagacc | |
| miR-1175-5p | JEV | 10,886–10,911 | agactgggagatcttctgctctatc |
| MVEV | 10,923–10,949 | agactaggagatcttctgctctattc | |
| USUV | 10,975–11,001 | agactaggagatcttctgctctattc | |
| WNV1 | 10,943–10,968 | agactaggagatcttctgctctgca | |
| WNV2 | 10,876–10,901 | agactaggggatcttctgctctgca |
| miRNA | Organism | Function | Reference |
|---|---|---|---|
| miR-9c | Drosophila melanogaster | Highly expressed in brain. Participates in memory reduction. | [58] |
| Drosophila melanogaster | Highly expressed in eggs. Participates in transcript clearance in the maternal-to-zygotic transition process during development. | [59] | |
| Drosophila melanogaster | Induction of cellular proliferation by inhibition of PntP1, an inductor of Dap. | [60] | |
| Scylla paramamosain | Negative regulation of the ERK pathway that is important in ovarian development. | [61] | |
| Scylla paramamosain | Regulation of the cell cycle in ovarian development by inhibition of cyclin A and CDK1. | [68] | |
| miR-282-5p | Bombyx mori | Inhibition of chitinase 5 during the moulting process. | [69] |
| miR-79 | Ixodes scapularis | Enhances Anaplasma phagocytophilum infection in ISE6 cells through the inhibition of Roundabout protein 2. | [74] |
| Drosophila melanogaster | Suppresses tumour growth through activation of the JNK signaling pathway by inhibition of the RNF146 protein. | [71] | |
| Drosophila melanogaster | Induction of cellular proliferation by inhibition of PntP1, an inductor of Dap. | [60] | |
| Bombyx mori | Inhibits BmEm4, a gene involved in development and metamorphosis. | [72] | |
| Apostichopus japonicus | Participates in metabolic rate suppression during aestivation. | [73] | |
| Caenorhabditis elegans | Controls the hermaphrodite-specific neuron migration during embryogenesis by targeting SQV-5 and SQV-7, enzymes involved in glycosaminoglycan biosynthesis | [70] |
| miRNA | Function | Reference |
|---|---|---|
| miR-6842-5p | Reduction of proliferation, migration, and the formation of capillary-like structures in HUVEC cells by suppression of AKT2. | [77] |
| miR-661 | Upregulated in squamous cell carcinoma. | [85] |
| Upregulated in non-small-cell lung cancer and promotes proliferation and invasion through the RUNX3 and RB1/E2F pathways but inhibits apoptosis through DOK7. Biomarker for diagnosis and prognosis. | [86,87,88] | |
| Promotes migration and proliferation of lung cancer cells by targeting ADRA1A. | [89] | |
| Binds to Tumour suppressor candidate-2 pseudogene (TSC2P) in esophageal squamous cell carcinoma. | [90] | |
| Downregulated in cervical carcinoma tissues. | [91] | |
| Antitumoural effect by promoting apoptosis in osteosarcoma cells through binding to Cytochrome C1. | [92] | |
| Downregulated in breast cancer tissues and a breast epithelial cell line. Inhibits proliferation and glycolysis by targeting HMGA1, regulates the expression of metastatic tumour antigen 1, and has antitumoural effect by binding to Mdm2 and Mdm4, thus stabilising p53. However, it is upregulated in breast cancer tissues from patients with metastasis and in triple-negative breast cancer from LatinAmerican patients. | [93,94,95,96,97] | |
| Downregulated in HUVEC cells stimulated with extracellular vesicles from anti-PR3-activated neutrophils (granulomatosis with polyangiitis). | [98] | |
| Upregulated in patients with myelodysplastic syndrome. It induces apoptosis through the p53 pathway. | [99] | |
| Inhibits proliferation and migration of vascular smooth muscle cells by targeting SYK mRNA. | [100] | |
| Downregulated in macrophages from patients with varicose veins. | [101] | |
| Downregulated in glioma tissues. Inhibits metastasis and promotes apoptosis in glioma cells by targeting RAB3D, and it degrades HOXD-AS2, a lncRNA that promotes cell proliferation and cell cycle progression. | [102,103,104] | |
| Upregulated in pancreatic ductal adenocarcinoma and associated with a bad prognosis but downregulated in patients with other pancreatic cancers. It promotes cell proliferation through activation of the Wnt signalling pathway. | [105,106] | |
| Downregulated in sera from patients with Alzheimer’s disease. | [107] | |
| Overexpressed in patients with hepatocellular carcinoma. It inhibits PTPN11, a tumour suppressive tyrosine phosphatase. | [108,109] | |
| Present in high levels in blood samples of large-for-gestational age mothers during the second trimester of pregnancy. | [110] | |
| Overexpressed in non-implanted blastocysts. It inhibits embryo–endometrial adhesion by downregulation of poliovirus receptor-related 1. | [111] | |
| Overexpressed in the blastocoel fluid of aneuploid embryos. | [112] | |
| Overexpressed in serum from patients with incomplete Sjögren’s syndrome. | [113] | |
| Downregulated in human keloid tissues. It inhibits expression of FGF2, a factor involved in keloid progression. | [114] | |
| Increased in patients with diabetes mellitus type 2 with microvascular complications or foot ulcers. Putative biomarker. | [115,116] | |
| Upregulated in ovarian cancer tissues acting as a tumour promoter through the NPP5J-induced AKT pathway. | [117] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila-Bonilla, R.G.; Salas-Benito, J.S. Computational Screening to Predict MicroRNA Targets in the Flavivirus 3′ UTR Genome: An Approach for Antiviral Development. Int. J. Mol. Sci. 2024, 25, 10135. https://doi.org/10.3390/ijms251810135
Avila-Bonilla RG, Salas-Benito JS. Computational Screening to Predict MicroRNA Targets in the Flavivirus 3′ UTR Genome: An Approach for Antiviral Development. International Journal of Molecular Sciences. 2024; 25(18):10135. https://doi.org/10.3390/ijms251810135
Chicago/Turabian StyleAvila-Bonilla, Rodolfo Gamaliel, and Juan Santiago Salas-Benito. 2024. "Computational Screening to Predict MicroRNA Targets in the Flavivirus 3′ UTR Genome: An Approach for Antiviral Development" International Journal of Molecular Sciences 25, no. 18: 10135. https://doi.org/10.3390/ijms251810135







