NF-κB Transcription Factors: Their Distribution, Family Expansion, Structural Conservation, and Evolution in Animals
Abstract
1. Introduction
2. Results and Discussion
2.1. p50/p105 Proteins Are Present in Most of the Animals
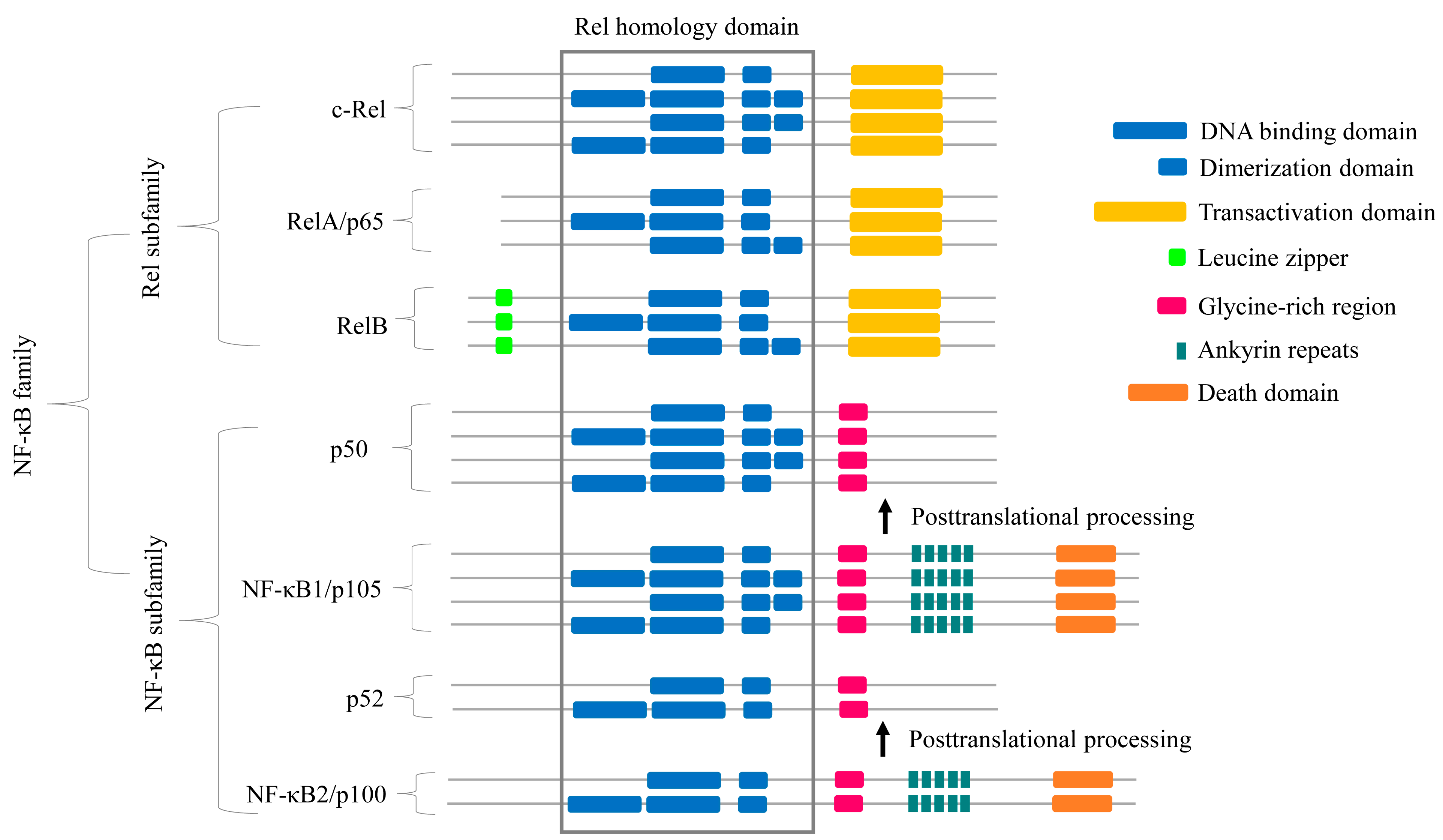
2.2. Some NF-κB Proteins Have Multiple RHD Subdomains
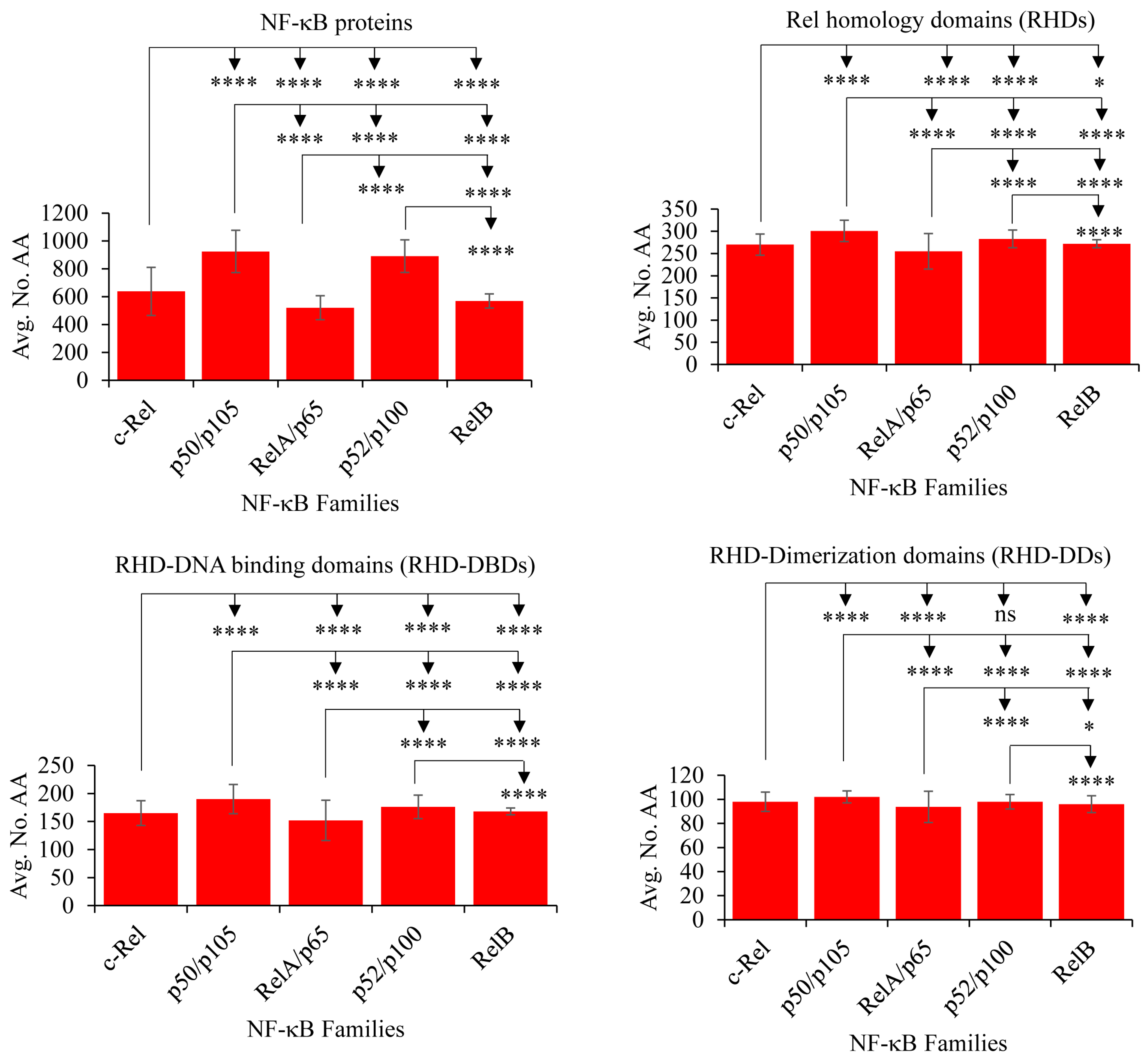
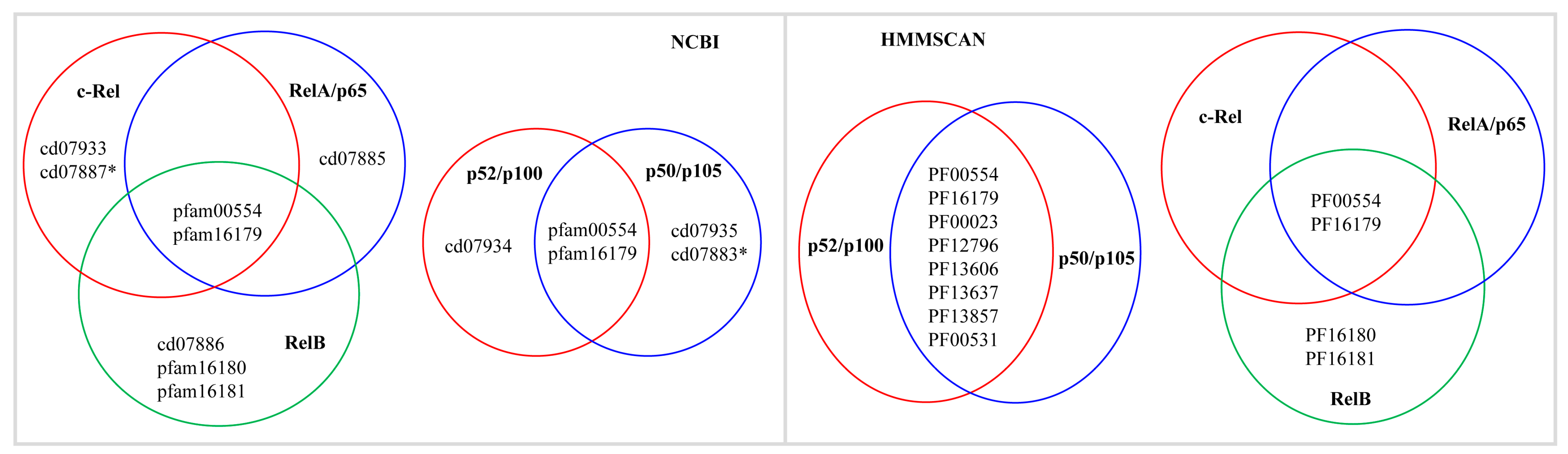
2.3. NF-κB Proteins Conserved Domains and Subdomains Differ in Their Size
2.4. RelB and c-Rel Have the Most and Least Conserved Amino Acids in the RHDs
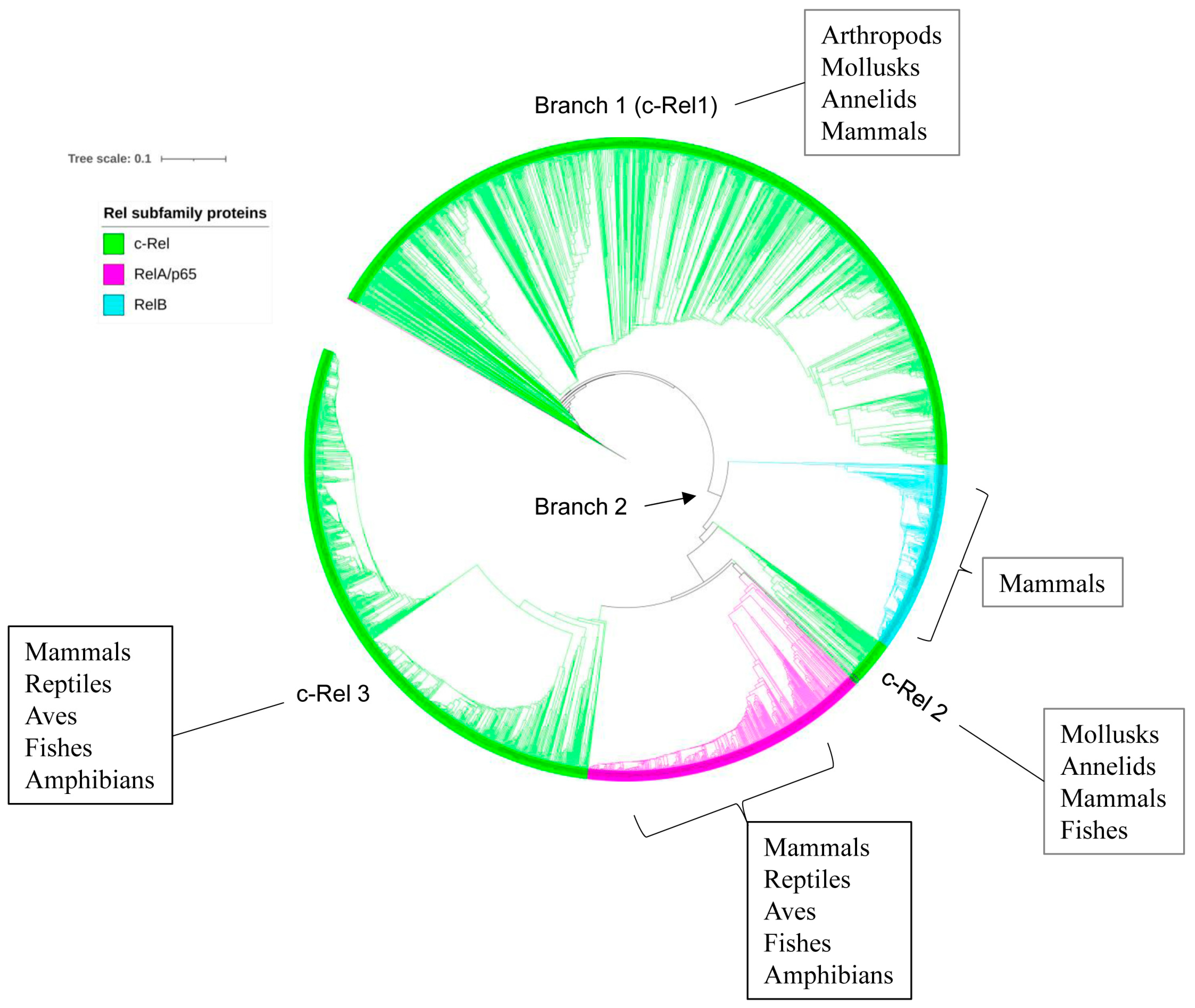
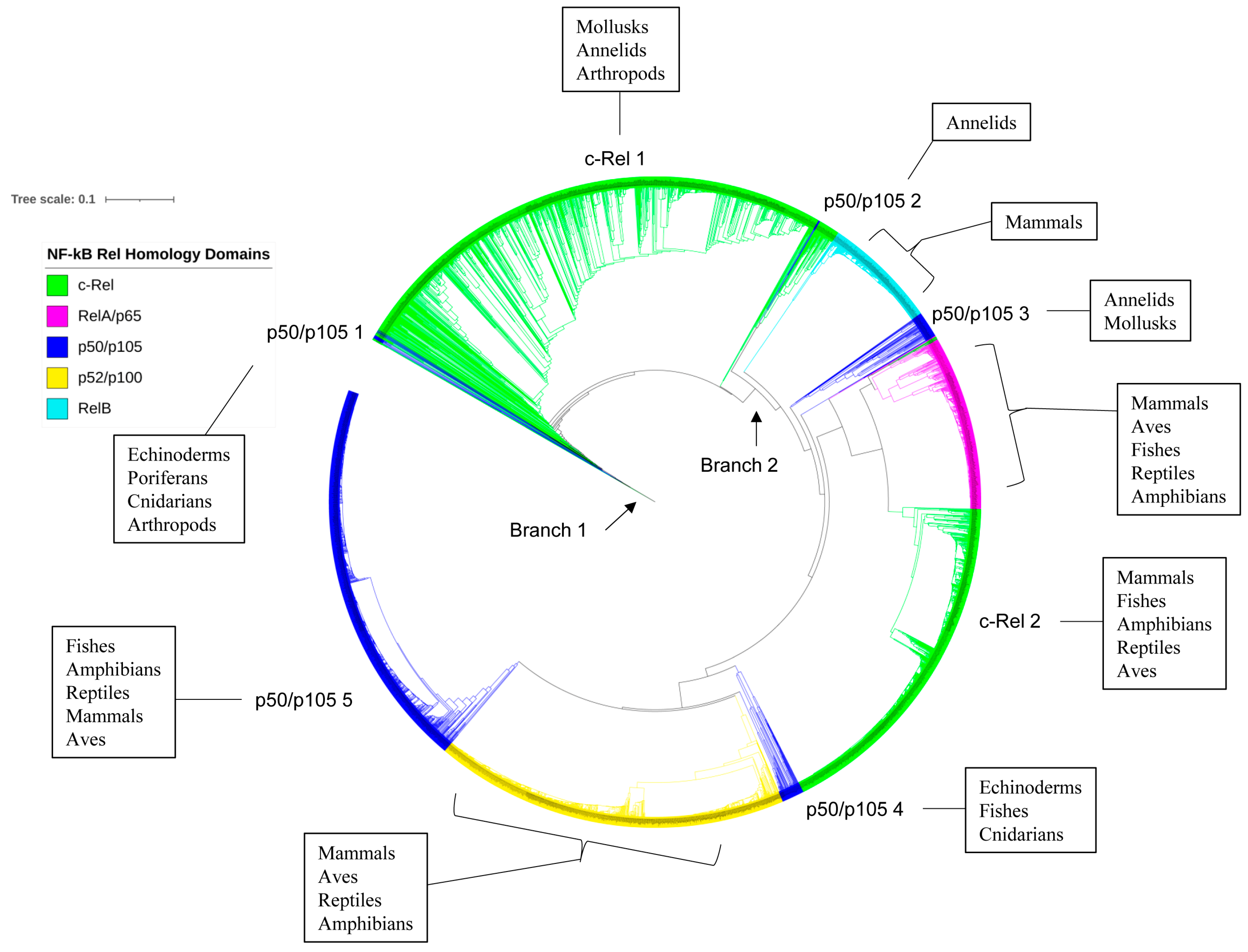
2.5. p50/p105 and c-Rel Might Have Been the Earliest NF-κB Family Members to Emerge
3. Materials and Methods
3.1. Reference NF-κB Family Members Used in This Study
3.2. Selection of Suitable Method for Analyzing NF-κB Family Characteristic Domains
3.3. Genome Data Mining and Annotation of NF-κB Proteins in Animals
3.4. Generation of NF-κB Profile Heat-Map
3.5. Phylogenetic Analysis of NF-κB Proteins
3.6. Identification of RHD, RHD-DBD and RHD-DD in NF-κB Proteins
3.7. Analysis of Amino Acid Conservation
3.8. Statistical Analysis of the NF-κB Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sen, R.; Baltimore, D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986, 46, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D.; Wolenski, F.S. NF-κB: Where did it come from and why? Immunol. Rev. 2012, 246, 14–35. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.T.; Tergaonkar, V. Roles of NF-κB in health and disease: Mechanisms and therapeutic potential. Clin. Sci. 2009, 116, 451–465. [Google Scholar] [CrossRef]
- Ben-Neriah, Y.; Karin, M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 2011, 12, 715–723. [Google Scholar] [CrossRef]
- Tieri, P.; Termanini, A.; Bellavista, E.; Salvioli, S.; Capri, M.; Franceschi, C. Charting the NF-κB pathway interactome map. PLoS ONE 2012, 7, e32678. [Google Scholar] [CrossRef]
- Yu, M.; Chen, J.; Bao, Y.; Li, J. Genomic analysis of NF-κB signaling pathway reveals its complexity in Crassostrea gigas. Fish Shellfish. Immunol. 2018, 72, 510–518. [Google Scholar] [CrossRef]
- Park, M.H.; Hong, J.T. Roles of NF-κB in Cancer and Inflammatory Diseases and Their Therapeutic Approaches. Cells 2016, 5, 15. [Google Scholar] [CrossRef]
- Hoesel, B.; Schmid, J.A. The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef]
- Dorrington, M.G.; Fraser, I.D.C. NF-κB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Mazor, M.; Kawano, Y.; Zhu, H.; Waxman, J.; Kypta, R.M. Inhibition of glycogen synthase kinase-3 represses androgen receptor activity and prostate cancer cell growth. Oncogene 2004, 23, 7882–7892. [Google Scholar] [CrossRef] [PubMed]
- Shakoori, A.; Ougolkov, A.; Yu, Z.W.; Zhang, B.; Modarressi, M.H.; Billadeau, D.D.; Mai, M.; Takahashi, Y.; Minamoto, T. Deregulated GSK3beta activity in colorectal cancer: Its association with tumor cell survival and proliferation. Biochem. Biophys. Res. Commun. 2005, 334, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Ougolkov, A.V.; Bone, N.D.; Fernandez-Zapico, M.E.; Kay, N.E.; Billadeau, D.D. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukemia B cells. Blood 2007, 110, 735–742. [Google Scholar] [CrossRef]
- Domoto, T.; Pyko, I.V.; Furuta, T.; Miyashita, K.; Uehara, M.; Shimasaki, T.; Nakada, M.; Minamoto, T. Glycogen synthase kinase-3β is a pivotal mediator of cancer invasion and resistance to therapy. Cancer Sci. 2016, 107, 1363–1372. [Google Scholar] [CrossRef]
- Das, A.; Bhattacharya, B.; Roy, S. Decrypting a path based approach for identifying the interplay between PI3K and GSK3 signaling cascade from the perspective of cancer. Genes Dis. 2022, 9, 868–888. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-κB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Ngo, K.A.; Kishimoto, K.; Davis-Turak, J.; Pimplaskar, A.; Cheng, Z.; Spreafico, R.; Chen, E.Y.; Tam, A.; Ghosh, G.; Mitchell, S. Dissecting the regulatory strategies of NF-κB RelA target genes in the inflammatory response reveals differential transactivation logics. Cell Rep. 2020, 30, 2758–2775.e6. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D.; Kalaitzidis, D.; Liang, M.-C.; Starczynowski, D.T. The c-Rel transcription factor and B-cell proliferation: A deal with the devil. Oncogene 2004, 23, 2275–2286. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, J.; Jenner, R.G.; Murray, H.L.; Gerber, G.K.; Gifford, D.K.; Young, R.A. Coordinated binding of NF-κB family members in the response of human cells to lipopolysaccharide. Proc. Natl. Acad. Sci. USA 2006, 103, 5899–5904. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. Introduction to NF-κB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef]
- Sun, S.C.; Chang, J.H.; Jin, J. Regulation of nuclear factor-κB in autoimmunity. Trends Immunol. 2013, 34, 282–289. [Google Scholar] [CrossRef]
- Pahl, H.L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Mattson, M.P.; Goodman, Y.; Luo, H.; Fu, W.; Furukawa, K. Activation of NF-kappaB protects hippocampal neurons against oxidative stress-induced apoptosis: Evidence for induction of manganese superoxide dismutase and suppression of peroxynitrite production and protein tyrosine nitration. J. Neurosci. Res. 1997, 49, 681–697. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Caamaño, J.; Hunter, C.A. NF-kappaB family of transcription factors: Central regulators of innate and adaptive immune functions. Clin. Microbiol. Rev. 2002, 15, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Signaling to NF-κB. Genes Dev. 2004, 18, 2195–2224. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-C. Non-canonical NF-κB signaling pathway. Cell Res. 2011, 21, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Bonizzi, G.; Karin, M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Baltimore, D. Circuitry of nuclear factor κB signaling. Immunol. Rev. 2006, 210, 171–186. [Google Scholar] [CrossRef]
- Sheppard, K.A.; Rose, D.W.; Haque, Z.K.; Kurokawa, R.; McInerney, E.; Westin, S.; Thanos, D.; Rosenfeld, M.G.; Glass, C.K.; Collins, T. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol. Cell. Biol. 1999, 19, 6367–6378. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-κB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef]
- Jimi, E.; Takakura, N.; Hiura, F.; Nakamura, I.; Hirata-Tsuchiya, S. The Role of NF-κB in Physiological Bone Development and Inflammatory Bone Diseases: Is NF-κB Inhibition “Killing Two Birds with One Stone”? Cells 2019, 8, 1636. [Google Scholar] [CrossRef]
- Dobrzanski, P.; Ryseck, R.P.; Bravo, R. Both N- and C-terminal domains of RelB are required for full transactivation: Role of the N-terminal leucine zipper-like motif. Mol. Cell. Biol. 1993, 13, 1572–1582. [Google Scholar] [CrossRef]
- Williams, L.M.; Gilmore, T.D. Looking down on NF-κB. Mol. Cell. Biol. 2020, 40, e00104-20. [Google Scholar] [CrossRef] [PubMed]
- Leger, M.M.; Ros-Rocher, N.; Najle, S.R.; Ruiz-Trillo, I. Rel/NF-κB transcription factors emerged at the onset of opisthokonts. Genome Biol. Evol. 2022, 14, evab289. [Google Scholar] [CrossRef] [PubMed]
- Dehal, P.; Boore, J.L. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D.; Gerondakis, S. The c-Rel Transcription Factor in Development and Disease. Genes Cancer 2011, 2, 695–711. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.E.; Yeh, G.; Soltero, S.; Smale, S.T. Selective regulation of a defined subset of inflammatory and immunoregulatory genes by an NF-κB p50-IκBζ pathway. Genes Dev. 2024, 38, 536–553. [Google Scholar] [CrossRef]
- Mitchell, C.D.; Criscitiello, M.F. Comparative study of cartilaginous fish divulges insights into the early evolution of primary, secondary and mucosal lymphoid tissue architecture. Fish Shellfish Immunol. 2020, 107, 435–443. [Google Scholar] [CrossRef]
- Smith, N.C.; Rise, M.L.; Christian, S.L. A Comparison of the Innate and Adaptive Immune Systems in Cartilaginous Fish, Ray-Finned Fish, and Lobe-Finned Fish. Front. Immunol. 2019, 10, 2292. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB in immunobiology. Cell Res. 2011, 21, 223–244. [Google Scholar] [CrossRef]
- Dev, A.; Iyer, S.; Razani, B.; Cheng, G. NF-κB and innate immunity. Curr. Top. Microbiol. Immunol. 2011, 349, 115–143. [Google Scholar] [CrossRef]
- Buchmann, K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef]
- Zanetti, M.; Castiglioni, P.; Schoenberger, S.; Gerloni, M. The role of relB in regulating the adaptive immune response. Ann. N. Y. Acad. Sci. 2003, 987, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Millet, P.; McCall, C.; Yoza, B. RelB: An outlier in leukocyte biology. J. Leukoc. Biol. 2013, 94, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Finnerty, J.R.; Gilmore, T.D. Methods for analyzing the evolutionary relationship of NF-κB proteins using free, web-driven bioinformatics and phylogenetic tools. In NF-kappa B: Methods and Protocols; Humana Press: New York, NY, USA, 2015; pp. 631–646. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Chen, W.; Lee, M.-K.; Jefcoate, C.; Kim, S.-C.; Chen, F.; Yu, J.-H. Fungal cytochrome p450 monooxygenases: Their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol. Evol. 2014, 6, 1620–1634. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. Mev: Multiexperiment viewer. In Biomedical Informatics for Cancer Research; Springer: Boston, MA, USA, 2010; pp. 267–277. [Google Scholar]
- Katoh, K.; Kuma, K.-i.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Zondo, N.M.; Padayachee, T.; Nelson, D.R.; Syed, K. Saprophytic to pathogenic mycobacteria: Loss of cytochrome P450s vis a vis their prominent involvement in natural metabolite biosynthesis. Int. J. Mol. Sci. 2022, 24, 149. [Google Scholar] [CrossRef]
- Dohrmann, M.; Wörheide, G. Dating early animal evolution using phylogenomic data. Sci. Rep. 2017, 7, 3599. [Google Scholar] [CrossRef]
- Gilbert, P.U.; Porter, S.M.; Sun, C.-Y.; Xiao, S.; Gibson, B.M.; Shenkar, N.; Knoll, A.H. Biomineralization by particle attachment in early animals. Proc. Natl. Acad. Sci. USA 2019, 116, 17659–17665. [Google Scholar] [CrossRef]
- Hommel, B.; Chapman, C.S.; Cisek, P.; Neyedli, H.F.; Song, J.-H.; Welsh, T.N. No one knows what attention is. Atten. Percept. Psychophys. 2019, 81, 2288–2303. [Google Scholar] [CrossRef]
- Cole, D.B.; Mills, D.B.; Erwin, D.H.; Sperling, E.A.; Porter, S.M.; Reinhard, C.T.; Planavsky, N.J. On the co-evolution of surface oxygen levels and animals. Geobiology 2020, 18, 260–281. [Google Scholar] [CrossRef]


| NF-κB Family Member | Gene Targets | Cellular Functions | Cancer-Related Functions | Reference |
|---|---|---|---|---|
| RelA/p65 | IL-6, TNF-α, Bcl-2, IL-1β, COX-2 | An important player in the canonical NF-κB pathway, it regulates gene expression related to inflammation, immune response, and cell survival. | Promotes the proliferation and survival of cancer cells by inducing anti-apoptotic genes; its overexpression is linked to aggressive cancer types. | [17,18,19,20] |
| RelB | IL-10, Bcl-2, CD40, MMP9, VEGF | A crucial player in the non-canonical NF-κB pathway, it regulates the development of lymphoid organs and the movement of lymphocytes. | Promotes immune evasion. | [17,19,21] |
| c-Rel | IL-2, IL-4, CDOL, Bcl-XL | It regulates T-cell activation and its differentiation, and influences B cell development and its function. | Associated with lymphoid malignancies, it promotes cancer cell survival and contributes to tumor formation. | [17,22,23] |
| p105/p50 (NF-κB1) | IL-1, TNF-α, GM-CSF, MMP9 | Acts as a transcriptional repressor or activator depending on its dimerization partners. It is involved in regulating genes related to inflammation and immune responses. | It dimerizes with RelA or c-Rel to promote oncogenesis. It also promotes tumor growth and metastasis in a signal-dependent manner. | [17,21,24] |
| p100/p52 (NF-κB2) | IL-6, IL-10, lymphotoxin-α, | Involved in the regulation of genes associated with immune responses and lymphocyte development. | Associated with the development of certain lymphomas. | [17,21,24] |
| NF-κB Family Member | Organism Group | Species Names | Protein ID | Protein Size | RHD-DBD Position (% Identity) | RHD-DD Position (% Identity) |
|---|---|---|---|---|---|---|
| c-Rel | Reptilia | Naja naja | KAG8123041.1 * | 447 | 3–46 | 55–83 and 79–130 (17.24%) |
| Mammalia | Microtus ochrogaster | KAH0516101.1 | 566 | 83–135 and 138–192 (21.62%) | 192–261 | |
| Mammalia | Orycteropus afer afer | XP_042636495.1 | 558 | 21–62 and 76–154 (19.51%) | 163–258 | |
| Mammalia | Tupaia chinensis | ELW63349.1 | 458 | 1–77 and 86–133 (26.92%) | 133–202 | |
| Aves | Columbina picui | NWQ80171.1 | 289 | 7–50 and 77–159 (22.50%) | 168–263 | |
| Aves | Lamprotornis superbus | KAI1240354.1 | 1426 | 115–259 and 453–500 (25.00%) | 651–746 | |
| Mollusca | Mya arenaria | WAR19259.1 | 412 | 89–193 and 196–233 (23.68%) | 241–341 | |
| Arthropoda | Dermatophagoides pteronyssinus | XP_027194418.1 | 794 | 27–150 and 152–244 (100.00%) | 252–355 | |
| Arthropoda | Drosophila kikkawai | KAH8308414.1 | 1024 | 51–221 | 229–308 and 328–352 (24.00%) | |
| Arthropoda | Drosophila sulfurigaster | KAH8391813.1 | 1056 | 63–233 | 241–321 and 331–362 (17.86%) | |
| Arthropoda | Leptinotarsa decemlineata | XP_023017353.1 | 534 | 122–292 | 3–68 and 299–397 (45.45%) | |
| Arthropoda | Drosophila pandora | KAH8323486.1 | 994 | 53–223 | 231–317 and 320–350 (23.33%) | |
| Arthropoda | Drosophila bipectinata | KAH8278094.1 | 1000 | 55–225 | 233–313 and 325–350 (20.00%) | |
| Arthropoda | Drosophila pseudoananassae | KAH8324519.1 | 998 | 53–223 | 231–313 and 323–348 (22.22%) | |
| Arthropoda | Pseudolycoriella hygida | KAJ6649769.1 | 1011 | 74–244 | 252–348 and 447–469 (88.89%) | |
| Arthropoda | Gonioctena quinquepunctata | KAG5873706.1 | 818 | 76–201 and 207–288 (43.24%) | 296–394 | |
| Arthropoda | Aphidius gifuensis | KAF7994522.1 | 694 | 75–245 | 253–352 and 431–533 (41.00%) | |
| Arthropoda | Phthorimaea operculella | KAI5633865.1 | 804 | 67–193 and 198–274 (94.29%) | 281–389 | |
| Arthropoda | Spodoptera exigua | CAH0702166.1 | 1373 | 17–140 and 467–625 (35.40%) | 633–731 | |
| Arthropoda | Temnothorax longispinosus | TGZ37724.1 * | 1121 | 133–255 and 254–280 (29.17%) | 288–388 | |
| Arthropoda | Timema shepardi | CAD7261384.1 | 722 | 2–89 and 122–165 (25.00%) | 173–273 | |
| Arthropoda | Apis florea | XP_031771860.1 | 460 | 12–91 and 95–134 (17.50%) | 142–242 | |
| Arthropoda | Apis dorsata | XP_006618954.1 * | 466 | 10–91 and 88–134 (22.86%) | 142–242 | |
| Arthropoda | Diaphorina citri | KAI5744658.1 | 480 | 48–146 and 151–188 (15.79%) | 197–293 | |
| Arthropoda | Diaphorina citri | KAI5710357.1 | 512 | 48–146 and 151–188 (15.79%) | 197–293 | |
| Arthropoda | Drosophila birchii | KAH8245463.1 | 1015 | 80–250 | 257–336 and 354–380 (25.93%) | |
| Arthropoda | Rhagoletis zephyria | XP_017461252.1 | 307 | 21–144 and 147–218 (85.71%) | 256–307 | |
| Arthropoda | Choristoneura fumiferana | KAI8422849.1 | 639 | 327–382 and 432–593 (22.64%) | 600–624 | |
| Arthropoda | Timema bartmani | CAD7444423.1 | 1022 | 68–123 and 126–170 (30.56%) | 178–278 | |
| Arthropoda | Aphidius gifuensis | XP_044005943.1 | 340 | 20–93 and 104–164 (34.38%) | 170–268 | |
| Arthropoda | Copidosoma floridanum | XP_023248498.1 | 1289 | 25–199 and 348–522 (29.76%) | 209–308 and 533–629 (32.61%) | |
| p50/p105 | Cnidaria | Orbicella faveolata | XP_020612557.1 | 1121 | 45–260 | 269–295 and 307–410 (100.00%) |
| Reptilia | Thamnophis sirtalis | XP_013917902.1 | 589 | 32–230 and 265–424 (98.11%) | 239–268 and 433–534 (93.33%) | |
| Mammalia | Microtus ochrogaster | KAH0500796.1 | 1000 | 11–49 and 64–226 (20.51%) | 235–336 | |
| Mammalia | Heterocephalus glaber | EHB13915.1 | 1164 | 125–166 and 198–340 (26.19%) | 349–450 | |
| Mollusca | Plakobranchus ocellatus | GFN89059.1 | 1056 | 67–254 | 263–290 and 324–377 (14.29%) | |
| RelA/p65 | Mammalia | Galemys pyrenaicus | KAG8522598.1 | 756 | 204–369 | 378–404 and 424–494 (20.00%) |
| Mammalia | Sousa chinensis | TEA11684.1 | 591 | 17–62 and 102–226 (18.60%) | 235–331 | |
| Aves | Melopsittacus undulatus | XP_033927754.1 | 405 | 13–54 and 72–221 (96.15%) | 230–326 | |
| Aves | Onychostruthus taczanowskii | XP_041269398.1 | 443 | 1–32 and 40–115 (12.90%) | 124–220 | |
| p52/p100 | Mammalia | Galemys pyrenaicus | KAG8523592.1 | 955 | 70–114 and 116–274 (100.00%) | 283–381 |
| Aves | Pygoscelis adeliae | KFW65979.1 | 657 | 30–122 and 172–220 (17.39%) | 229–279 | |
| Aves | Pygoscelis adeliae | XP_009320748.1 | 568 | 9–101 and 116–163 (17.78%) | 172–224 | |
| RelB | Mammalia | Pteropus alecto | XP_024902475.1 | 637 | 154–322 | 331–357 and 401–456 (13.04%) |
| Mammalia | Tupaia chinensis | ELW71096.1 * | 547 | 72–240 | 249–322 and 318–369 (82.76%) | |
| Mammalia | Propithecus coquereli | XP_012508693.1 | 559 | 112–202 and 232–275 (26.32%) | 284–380 | |
| Mammalia | Neotoma lepida | OBS57525.1 | 455 | 44–132 and 138–166 (21.43%) | 175–276 |
| NF-κB Family | Number of NF-κB Protein Sequences | PROMALS3D Conservation Index | Ranking | ||||
|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | |||
| Rel homology domain (RHD) | |||||||
| c-Rel | 1814 | 18 | 10 | 9 | 1 | 2 | V |
| p50/p105 | 934 | 28 | 12 | 25 | 11 | 7 | IV |
| RelA/p65 | 368 | 30 | 0 | 82 | 0 | 92 | III |
| p52/p100 | 741 | 15 | 33 | 69 | 20 | 93 | II |
| RelB | 240 | 23 | 92 | 0 | 0 | 126 | I |
| Rel homology domain-DNA binding domain (RHD-DBD) | |||||||
| c-Rel | 1814 | 8 | 10 | 5 | 0 | 2 | IV |
| p50/p105 | 934 | 16 | 8 | 12 | 6 | 2 | IV |
| RelA/p65 | 368 | 19 | 0 | 47 | 0 | 51 | III |
| p52/p100 | 741 | 10 | 20 | 37 | 8 | 59 | II |
| RelB | 240 | 0 | 59 | 0 | 0 | 72 | I |
| Rel homology domain-dimerization domain (RHD-DD) | |||||||
| c-Rel | 1814 | 3 | 3 | 5 | 1 | 2 | V |
| p50/p105 | 934 | 14 | 11 | 7 | 6 | 7 | IV |
| RelA/p65 | 368 | 10 | 5 | 22 | 0 | 40 | II |
| p52/p100 | 741 | 26 | 20 | 21 | 0 | 32 | III |
| RelB | 240 | 0 | 16 | 0 | 0 | 72 | I |
| NF-κB Family | NCBI Protein ID | Organism | Protein Size | NCBI Batch Web CD-Search Domain | |
|---|---|---|---|---|---|
| Accession | Short name | ||||
| c-Rel | CAA52954.1 | Homo sapiens | 619 | c07933 | RHD-n_c-Rel |
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
| c-Rel | AAK72690.1 | Crassostrea gigas | 615 | cd07887 | RHD-n_Dorsal_Dif |
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
| RelA/p65 | AAA36408.1 | Homo sapiens | 551 | cd07885 | RHD-n_RelA |
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
| RelB | NP_006500.2 | Homo sapiens | 579 | cd07886 | RHD-n_RelB |
| pfam16181 | RelB_transactiv | ||||
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
| pfam16180 | RelB_leu_zip | ||||
| p50/p105 | AAA36361.1 | Homo sapiens | 969 | cd07935 | RHD-n_NFkB1 |
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
| p50/p105 | NP_999819.1 | Strongylocentrotus purpuratus | 1125 | cd07883 | RHD-n_NFkB |
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
| p52/p100 | NP_002493.3 | Homo sapiens | 899 | cd07934 | RHD-n_NFkB2 |
| pfam00554 | RHD_DNA_bind | ||||
| pfam16179 | RHD_dimer | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Msweli, S.; Pakala, S.B.; Syed, K. NF-κB Transcription Factors: Their Distribution, Family Expansion, Structural Conservation, and Evolution in Animals. Int. J. Mol. Sci. 2024, 25, 9793. https://doi.org/10.3390/ijms25189793
Msweli S, Pakala SB, Syed K. NF-κB Transcription Factors: Their Distribution, Family Expansion, Structural Conservation, and Evolution in Animals. International Journal of Molecular Sciences. 2024; 25(18):9793. https://doi.org/10.3390/ijms25189793
Chicago/Turabian StyleMsweli, Siphesihle, Suresh B. Pakala, and Khajamohiddin Syed. 2024. "NF-κB Transcription Factors: Their Distribution, Family Expansion, Structural Conservation, and Evolution in Animals" International Journal of Molecular Sciences 25, no. 18: 9793. https://doi.org/10.3390/ijms25189793
APA StyleMsweli, S., Pakala, S. B., & Syed, K. (2024). NF-κB Transcription Factors: Their Distribution, Family Expansion, Structural Conservation, and Evolution in Animals. International Journal of Molecular Sciences, 25(18), 9793. https://doi.org/10.3390/ijms25189793






