Towards Targeting Endothelial Rap1B to Overcome Vascular Immunosuppression in Cancer
Abstract
1. Introduction
2. Anti-Angiogenesis to Normalize Vessels, Current Anti-Angiogenic Therapies
3. Combining Immunotherapy with Anti-Angiogenesis (Combination Therapy)
4. Combination Therapy Enhances Clinical Effectiveness
5. Challenges of Combination Therapy, and the Need for New Targets
6. Rap1: A Ras Family Member
7. Ras and Rap1 Effector Domains and Direct Targets
8. Rap1 Is a Positive Regulator of VEGF Signaling
9. Role of Rap1B in VEGF-A-Induced Immunosuppression in the TME
10. RAS: Now Druggable—Paving the Way for Targeting Rap1?
11. Strategies for Targeting RAS-Mutant Tumors and Use of Drugs in Cancer Treatment
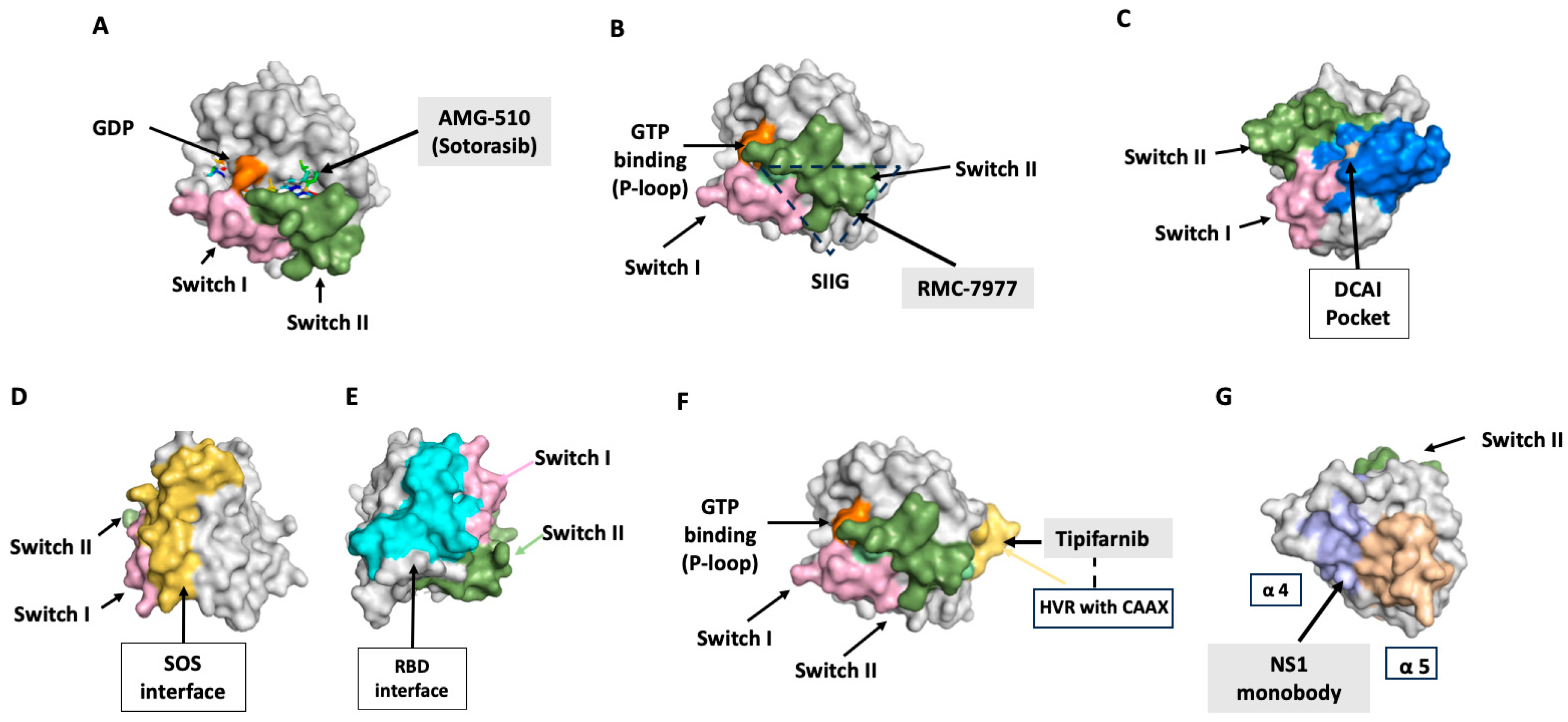
11.1. Switch II Pocket Inhibitors, KRAS-G12C
11.2. Direct Targeting of WT, G12C KRAS
11.3. DCAI Pocket Binders: Inhibiting Ras–Effector Binding
11.4. Indirect Targeting of Ras
11.5. SOS Inhibitors
11.6. SHP2 Inhibitors
11.7. RAS-Mimetics: Inhibition of Ras–Effector Interactions
12. Targeting Post-Translational Modification of RAS
12.1. Farnesyltransferase Inhibitors (FTIs)
12.2. Postprenylation CAAX Box Processing
12.3. Prenyl-Binding Protein Inhibitors
12.4. Geranylgeranyltransferase Inhibitors (GGTIs)
12.5. Ras Oligomerization
13. Inhibition of RAS Signaling Pathways
13.1. Synthetic Lethal Screens
13.2. EGFR Inhibition
13.3. MAPK Pathway Inhibitors
14. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Aird, W.C. (Ed.) Endothelial Cells in Health and Disease; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Cao, Y.; Langer, R.; Ferrara, N. Targeting angiogenesis in oncology, ophthalmology and beyond. Nat. Rev. Drug Discov. 2023, 22, 476–495. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.M.; Choyke, P.L. Imaging of angiogenesis: From microscope to clinic. Nat. Med. 2003, 9, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Vestweber, D. How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 2015, 15, 692–704. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pagès, F.; Sauts-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Ager, A.; Watson, H.A.; Wehenkel, S.C.; Mohammed, R.N. Homing to solid cancers: A vascular checkpoint in adoptive cell therapy using CAR T-cells. Biochem. Soc. Trans. 2016, 44, 377–385. [Google Scholar] [CrossRef]
- Leone, P.; Malerba, E.; Susca, N.; Favoino, E.; Perosa, F.; Brunori, G.; Prete, M.; Racanelli, V. Endothelial cells in tumor microenvironment: Insights and perspectives. Front. Immunol. 2024, 15, 1367875. [Google Scholar] [CrossRef]
- Huinen, Z.R.; Huijbers, E.J.M.; van Beijnum, J.R.; Nowak-Sliwinska, P.; Griffioen, A.W. Anti-angiogenic agents—Overcoming tumour endothelial cell anergy and improving immunotherapy outcomes. Nat. Rev. Clin. Oncol. 2021, 18, 527–540. [Google Scholar] [CrossRef]
- Chouaib, S.; Noman, M.Z.; Kosmatopoulos, K.; Curran, M.A. Hypoxic stress: Obstacles and opportunities for innovative immunotherapy of cancer. Oncogene 2017, 36, 439–445. [Google Scholar] [CrossRef]
- Lin, Q.; Choyke, P.L.; Sato, N. Visualizing vasculature and its response to therapy in the tumor microenvironment. Theranostics 2023, 13, 5223–5246. [Google Scholar] [CrossRef]
- He, J.; Baum, L.G. Endothelial cell expression of galectin-1 induced by prostate cancer cells inhibits T-cell transendothelial migration. Lab. Investig. 2006, 86, 578–590. [Google Scholar] [CrossRef]
- Norling, L.V.; Sampaio, A.L.; Cooper, D.; Perretti, M. Inhibitory control of endothelial galectin-1 on in vitro and in vivo lymphocyte trafficking. FASEB J. 2008, 22, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, V.L.; Postel, R.; Brandwijk, R.J.; Dings, R.P.; Nesmelova, I.; Satijn, S.; Verhofstad, N.; Nakabeppu, Y.; Baum, L.G.; Bakkers, J.; et al. Galectin-1 is essential in tumor angiogenesis and is a target for antiangiogenesis therapy. Proc. Natl. Acad. Sci. USA 2006, 103, 15975–15980. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; van Beijnum, J.R.; Griffioen, C.J.; Huinen, Z.R.; Sopesens, N.G.; Schulz, R.; Jenkins, S.V.; Dings, R.P.M.; Groenendijk, F.H.; Huijbers, E.J.M.; et al. Proinflammatory activity of VEGF-targeted treatment through reversal of tumor endothelial cell anergy. Angiogenesis 2023, 26, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Motz, G.T.; Santoro, S.P.; Wang, L.P.; Garrabrant, T.; Lastra, R.R.; Hagemann, I.S.; Lal, P.; Feldman, M.D.; Benencia, F.; Coukos, G. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 2014, 20, 607–615. [Google Scholar] [CrossRef]
- Deng, H.; Kan, A.; Lyu, N.; Mu, L.; Han, Y.; Liu, L.; Zhang, Y.; Duan, Y.; Liao, S.; Li, S.; et al. Dual Vascular Endothelial Growth Factor Receptor and Fibroblast Growth Factor Receptor Inhibition Elicits Antitumor Immunity and Enhances Programmed Cell Death-1 Checkpoint Blockade in Hepatocellular Carcinoma. Liver Cancer 2020, 9, 338–357. [Google Scholar] [CrossRef]
- Lee, S.; Schmitt, C.A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 2019, 21, 94–101. [Google Scholar] [CrossRef]
- Wu, Z.; Uhl, B.; Gires, O.; Reichel, C.A. A transcriptomic pan-cancer signature for survival prognostication and prediction of immunotherapy response based on endothelial senescence. J. Biomed. Sci. 2023, 30, 21. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, J.; Jacobs, K.A.; Rizzollo, F.; Lodi, F.; Hua, Y.; Poźniak, J.; Narayanan Srinivasan, A.; Houbaert, D.; Shankar, G.; More, S.; et al. Tumor endothelial cell autophagy is a key vascular-immune checkpoint in melanoma. EMBO Mol. Med. 2023, 15, e18028. [Google Scholar] [CrossRef]
- Yang, F.; Akhtar, M.N.; Zhang, D.; El-Mayta, R.; Shin, J.; Dorsey, J.F.; Zhang, L.; Xu, X.; Guo, W.; Bagley, S.J.; et al. An immunosuppressive vascular niche drives macrophage polarization and immunotherapy resistance in glioblastoma. Sci. Adv. 2024, 10, eadj4678. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Smyth, M.J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol. Immunol. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Qian, C.; Liu, C.; Liu, W.; Zhou, R.; Zhao, L. Targeting vascular normalization: A promising strategy to improve immune-vascular crosstalk in cancer immunotherapy. Front. Immunol. 2023, 14, 1291530. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Langenkamp, E.; Georganaki, M.; Loskog, A.; Fuchs, P.F.; Dieterich, L.C.; Kreuger, J.; Dimberg, A. VEGF suppresses T-lymphocyte infiltration in the tumor microenvironment through inhibition of NF-κB-induced endothelial activation. FASEB J. 2015, 29, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Dirkx, A.E.; Oude Egbrink, M.G.; Kuijpers, M.J.; van der Niet, S.T.; Heijnen, V.V.; Bouma-ter Steege, J.C.; Wagstaff, J.; Griffioen, A.W. Tumor angiogenesis modulates leukocyte-vessel wall interactions in vivo by reducing endothelial adhesion molecule expression. Cancer Res. 2003, 63, 2322–2329. [Google Scholar]
- Nagl, L.; Horvath, L.; Pircher, A.; Wolf, D. Tumor Endothelial Cells (TECs) as Potential Immune Directors of the Tumor Microenvironment—New Findings and Future Perspectives. Front. Cell Dev. Biol. 2020, 8, 766. [Google Scholar] [CrossRef]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, H.; Fehrenbacher, L.; Novotny, W.; Cartwright, T.; Hainsworth, J.; Heim, W.; Berlin, J.; Baron, A.; Griffing, S.; Holmgren, E.; et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004, 350, 2335–2342. [Google Scholar] [CrossRef]
- Sandler, A.; Gray, R.; Perry, M.C.; Brahmer, J.; Schiller, J.H.; Dowlati, A.; Lilenbaum, R.; Johnson, D.H. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N. Engl. J. Med. 2006, 355, 2542–2550. [Google Scholar] [CrossRef]
- Yang, J.C.; Haworth, L.; Sherry, R.M.; Hwu, P.; Schwartzentruber, D.J.; Topalian, S.L.; Steinberg, S.M.; Chen, H.X.; Rosenberg, S.A. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N. Engl. J. Med. 2003, 349, 427–434. [Google Scholar] [CrossRef]
- Garon, E.B.; Ciuleanu, T.E.; Arrieta, O.; Prabhash, K.; Syrigos, K.N.; Goksel, T.; Park, K.; Gorbunova, V.; Kowalyszyn, R.D.; Pikiel, J.; et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): A multicentre, double-blind, randomised phase 3 trial. Lancet 2014, 384, 665–673. [Google Scholar] [CrossRef]
- Tabernero, J.; Yoshino, T.; Cohn, A.L.; Obermannova, R.; Bodoky, G.; Garcia-Carbonero, R.; Ciuleanu, T.E.; Portnoy, D.C.; Van Cutsem, E.; Grothey, A.; et al. Ramucirumab versus placebo in combination with second-line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first-line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): A randomised, double-blind, multicentre, phase 3 study. Lancet Oncol. 2015, 16, 499–508. [Google Scholar] [CrossRef]
- Zhu, A.X.; Kang, Y.K.; Yen, C.J.; Finn, R.S.; Galle, P.R.; Llovet, J.M.; Assenat, E.; Brandi, G.; Pracht, M.; Lim, H.Y.; et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019, 20, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.M.; Hurwitz, H.I. Ziv-aflibercept: Binding to more than VEGF-A—Does more matter? Nat. Rev. Clin. Oncol. 2013, 10, 10–11. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A.; Restifo, N.P.; Yang, J.C.; Morgan, R.A.; Dudley, M.E. Adoptive cell transfer: A clinical path to effective cancer immunotherapy. Nat. Rev. Cancer 2008, 8, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Kerbel, R.S. Improving immunotherapy outcomes with anti-angiogenic treatments and vice versa. Nat. Rev. Clin. Oncol. 2018, 15, 310–324. [Google Scholar] [CrossRef]
- Sahu, A.; Kose, K.; Kraehenbuehl, L.; Byers, C.; Holland, A.; Tembo, T.; Santella, A.; Alfonso, A.; Li, M.; Cordova, M.; et al. In vivo tumor immune microenvironment phenotypes correlate with inflammation and vasculature to predict immunotherapy response. Nat. Commun. 2022, 13, 5312. [Google Scholar] [CrossRef]
- Hellebrekers, D.M.; Castermans, K.; Viré, E.; Dings, R.P.; Hoebers, N.T.; Mayo, K.H.; Oude Egbrink, M.G.; Molema, G.; Fuks, F.; van Engeland, M.; et al. Epigenetic regulation of tumor endothelial cell anergy: Silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res. 2006, 66, 10770–10777. [Google Scholar] [CrossRef]
- Griffioen, A.W.; Damen, C.A.; Mayo, K.H.; Barendsz-Janson, A.F.; Martinotti, S.; Blijham, G.H.; Groenewegen, G. Angiogenesis inhibitors overcome tumor induced endothelial cell anergy. Int. J. Cancer 1999, 80, 315–319. [Google Scholar] [CrossRef]
- Tabruyn, S.P.; Sabatel, C.; Nguyen, N.Q.; Verhaeghe, C.; Castermans, K.; Malvaux, L.; Griffioen, A.W.; Martial, J.A.; Struman, I. The angiostatic 16K human prolactin overcomes endothelial cell anergy and promotes leukocyte infiltration via nuclear factor-kappaB activation. Mol. Endocrinol. 2007, 21, 1422–1429. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Issekutz, A.C. Down-modulation of monocyte transendothelial migration and endothelial adhesion molecule expression by fibroblast growth factor: Reversal by the anti-angiogenic agent SU6668. Am. J. Pathol. 2002, 160, 2219–2230. [Google Scholar] [CrossRef]
- Dings, R.P.; Vang, K.B.; Castermans, K.; Popescu, F.; Zhang, Y.; Oude Egbrink, M.G.; Mescher, M.F.; Farrar, M.A.; Griffioen, A.W.; Mayo, K.H. Enhancement of T-cell-mediated antitumor response: Angiostatic adjuvant to immunotherapy against cancer. Clin. Cancer Res. 2011, 17, 3134–3145. [Google Scholar] [CrossRef] [PubMed]
- Fukumura, D.; Kloepper, J.; Amoozgar, Z.; Duda, D.G.; Jain, R.K. Enhancing cancer immunotherapy using antiangiogenics: Opportunities and challenges. Nat. Rev. Clin. Oncol. 2018, 15, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Kim, B.Y.S.; Chan, C.K.; Hahn, S.M.; Weissman, I.L.; Jiang, W. Improving immune-vascular crosstalk for cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 195–203. [Google Scholar] [CrossRef]
- Hendry, S.A.; Farnsworth, R.H.; Solomon, B.; Achen, M.G.; Stacker, S.A.; Fox, S.B. The Role of the Tumor Vasculature in the Host Immune Response: Implications for Therapeutic Strategies Targeting the Tumor Microenvironment. Front. Immunol. 2016, 7, 621. [Google Scholar] [CrossRef]
- Duru, G.; van Egmond, M.; Heemskerk, N. A Window of Opportunity: Targeting Cancer Endothelium to Enhance Immunotherapy. Front. Immunol. 2020, 11, 584723. [Google Scholar] [CrossRef]
- Schmittnaegel, M.; Rigamonti, N.; Kadioglu, E.; Cassará, A.; Wyser Rmili, C.; Kiialainen, A.; Kienast, Y.; Mueller, H.J.; Ooi, C.H.; Laoui, D.; et al. Dual angiopoietin-2 and VEGFA inhibition elicits antitumor immunity that is enhanced by PD-1 checkpoint blockade. Sci. Transl. Med. 2017, 9, eaak9670. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Jabouille, A.; Rivera, L.B.; Lodewijckx, I.; Missiaen, R.; Steri, V.; Feyen, K.; Tawney, J.; Hanahan, D.; Michael, I.P.; et al. Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci. Transl. Med. 2017, 9, eaak9679. [Google Scholar] [CrossRef]
- Shigeta, K.; Datta, M.; Hato, T.; Kitahara, S.; Chen, I.X.; Matsui, A.; Kikuchi, H.; Mamessier, E.; Aoki, S.; Ramjiawan, R.R.; et al. Dual Programmed Death Receptor-1 and Vascular Endothelial Growth Factor Receptor-2 Blockade Promotes Vascular Normalization and Enhances Antitumor Immune Responses in Hepatocellular Carcinoma. Hepatology 2020, 71, 1247–1261. [Google Scholar] [CrossRef]
- Shrimali, R.K.; Yu, Z.; Theoret, M.R.; Chinnasamy, D.; Restifo, N.P.; Rosenberg, S.A. Antiangiogenic agents can increase lymphocyte infiltration into tumor and enhance the effectiveness of adoptive immunotherapy of cancer. Cancer Res. 2010, 70, 6171–6180. [Google Scholar] [CrossRef]
- Huang, Y.; Yuan, J.; Righi, E.; Kamoun, W.S.; Ancukiewicz, M.; Nezivar, J.; Santosuosso, M.; Martin, J.D.; Martin, M.R.; Vianello, F.; et al. Vascular normalizing doses of antiangiogenic treatment reprogram the immunosuppressive tumor microenvironment and enhance immunotherapy. Proc. Natl. Acad. Sci. USA 2012, 109, 17561–17566. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ishida, T.; Nadaf, S.; Ohm, J.E.; Carbone, D.P. Antibodies to vascular endothelial growth factor enhance the efficacy of cancer immunotherapy by improving endogenous dendritic cell function. Clin. Cancer Res. 1999, 5, 2963–2970. [Google Scholar] [PubMed]
- Martino, E.C.; Misso, G.; Pastina, P.; Costantini, S.; Vanni, F.; Gandolfo, C.; Botta, C.; Capone, F.; Lombardi, A.; Pirtoli, L.; et al. Immune-modulating effects of bevacizumab in metastatic non-small-cell lung cancer patients. Cell Death Discov. 2016, 2, 16025. [Google Scholar] [CrossRef] [PubMed]
- Ozao-Choy, J.; Ma, G.; Kao, J.; Wang, G.X.; Meseck, M.; Sung, M.; Schwartz, M.; Divino, C.M.; Pan, P.Y.; Chen, S.H. The novel role of tyrosine kinase inhibitor in the reversal of immune suppression and modulation of tumor microenvironment for immune-based cancer therapies. Cancer Res. 2009, 69, 2514–2522. [Google Scholar] [CrossRef]
- Kuo, H.Y.; Khan, K.A.; Kerbel, R.S. Antiangiogenic-immune-checkpoint inhibitor combinations: Lessons from phase III clinical trials. Nat. Rev. Clin. Oncol. 2024, 21, 468–482. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Gettinger, S.N.; Horn, L.; Gandhi, L.; Spigel, D.R.; Antonia, S.J.; Rizvi, N.A.; Powderly, J.D.; Heist, R.S.; Carvajal, R.D.; Jackman, D.M.; et al. Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2015, 33, 2004–2012. [Google Scholar] [CrossRef]
- Hua, Y.; Vella, G.; Rambow, F.; Allen, E.; Antoranz Martinez, A.; Duhamel, M.; Takeda, A.; Jalkanen, S.; Junius, S.; Smeets, A.; et al. Cancer immunotherapies transition endothelial cells into HEVs that generate TCF1(+) T lymphocyte niches through a feed-forward loop. Cancer Cell 2022, 40, 1600–1618.e1610. [Google Scholar] [CrossRef]
- Lee, W.S.; Yang, H.; Chon, H.J.; Kim, C. Combination of anti-angiogenic therapy and immune checkpoint blockade normalizes vascular-immune crosstalk to potentiate cancer immunity. Exp. Mol. Med. 2020, 52, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Hack, S.P.; Zhu, A.X.; Wang, Y. Augmenting Anticancer Immunity Through Combined Targeting of Angiogenic and PD-1/PD-L1 Pathways: Challenges and Opportunities. Front. Immunol. 2020, 11, 598877. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Albiges, L.; Powles, T.; Staehler, M.; Bensalah, K.; Giles, R.H.; Hora, M.; Kuczyk, M.A.; Lam, T.B.; Ljungberg, B.; Marconi, L.; et al. Updated European Association of Urology Guidelines on Renal Cell Carcinoma: Immune Checkpoint Inhibition Is the New Backbone in First-line Treatment of Metastatic Clear-cell Renal Cell Carcinoma. Eur. Urol. 2019, 76, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Liang, X.; Fan, B.; Zhao, C.; Shen, B.; Ji, X.; Liu, Y. Endothelial-Specific Molecule 1 Inhibition Lessens Productive Angiogenesis and Tumor Metastasis to Overcome Bevacizumab Resistance. Cancers 2022, 14, 5681. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Cao, Z.; Ji, H.; Yang, X.; Iwamoto, H.; Wahlberg, E.; Länne, T.; Sun, B.; Cao, Y. Anti-VEGF- and anti-VEGF receptor-induced vascular alteration in mouse healthy tissues. Proc. Natl. Acad. Sci. USA 2013, 110, 12018–12023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Hosaka, K.; Huang, G.; Zang, J.; Chen, F.; Zhang, Y.; Samani, N.J.; Cao, Y. Endocrine vasculatures are preferable targets of an antitumor ineffective low dose of anti-VEGF therapy. Proc. Natl. Acad. Sci. USA 2016, 113, 4158–4163. [Google Scholar] [CrossRef] [PubMed]
- Ahmadieh, H.; Salti, I. Tyrosine kinase inhibitors induced thyroid dysfunction: A review of its incidence, pathophysiology, clinical relevance, and treatment. Biomed. Res. Int. 2013, 2013, 725410. [Google Scholar] [CrossRef]
- Hamnvik, O.P.; Larsen, P.R.; Marqusee, E. Thyroid dysfunction from antineoplastic agents. J. Natl. Cancer Inst. 2011, 103, 1572–1587. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, G.; Chen, J.; Zhuang, L.; Du, Y.; Yu, Q.; Zhuang, W.; Zhao, Y.; Zhou, M.; Zhang, W.; et al. AK112, a novel PD-1/VEGF bispecific antibody, in combination with chemotherapy in patients with advanced non-small cell lung cancer (NSCLC): An open-label, multicenter, phase II trial. eClinicalMedicine 2023, 62, 102106. [Google Scholar] [CrossRef]
- Lai, X.; Friedman, A. How to schedule VEGF and PD-1 inhibitors in combination cancer therapy? BMC Syst. Biol. 2019, 13, 30. [Google Scholar] [CrossRef]
- Pastuskovas, C.V.; Mundo, E.E.; Williams, S.P.; Nayak, T.K.; Ho, J.; Ulufatu, S.; Clark, S.; Ross, S.; Cheng, E.; Parsons-Reponte, K.; et al. Effects of anti-VEGF on pharmacokinetics, biodistribution, and tumor penetration of trastuzumab in a preclinical breast cancer model. Mol. Cancer Ther. 2012, 11, 752–762. [Google Scholar] [CrossRef]
- Arjaans, M.; Schröder, C.P.; Oosting, S.F.; Dafni, U.; Kleibeuker, J.E.; de Vries, E.G. VEGF pathway targeting agents, vessel normalization and tumor drug uptake: From bench to bedside. Oncotarget 2016, 7, 21247–21258. [Google Scholar] [CrossRef]
- Heskamp, S.; Boerman, O.C.; Molkenboer-Kuenen, J.D.; Oyen, W.J.; van der Graaf, W.T.; van Laarhoven, H.W. Bevacizumab reduces tumor targeting of antiepidermal growth factor and anti-insulin-like growth factor 1 receptor antibodies. Int. J. Cancer 2013, 133, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Xu, J.; Bai, Y.; Xu, A.; Cang, S.; Du, C.; Li, Q.; Lu, Y.; Chen, Y.; Guo, Y.; et al. Sintilimab plus a bevacizumab biosimilar (IBI305) versus sorafenib in unresectable hepatocellular carcinoma (ORIENT-32): A randomised, open-label, phase 2-3 study. Lancet Oncol. 2021, 22, 977–990. [Google Scholar] [CrossRef]
- Qin, S.; Chan, S.L.; Gu, S.; Bai, Y.; Ren, Z.; Lin, X.; Chen, Z.; Jia, W.; Jin, Y.; Guo, Y.; et al. Camrelizumab plus rivoceranib versus sorafenib as first-line therapy for unresectable hepatocellular carcinoma (CARES-310): A randomised, open-label, international phase 3 study. Lancet 2023, 402, 1133–1146. [Google Scholar] [CrossRef]
- Moore, K.N.; Bookman, M.; Sehouli, J.; Miller, A.; Anderson, C.; Scambia, G.; Myers, T.; Taskiran, C.; Robison, K.; Mäenpää, J.; et al. Atezolizumab, Bevacizumab, and Chemotherapy for Newly Diagnosed Stage III or IV Ovarian Cancer: Placebo-Controlled Randomized Phase III Trial (IMagyn050/GOG 3015/ENGOT-OV39). J. Clin. Oncol. 2021, 39, 1842–1855. [Google Scholar] [CrossRef] [PubMed]
- Mettu, N.B.; Ou, F.S.; Zemla, T.J.; Halfdanarson, T.R.; Lenz, H.J.; Breakstone, R.A.; Boland, P.M.; Crysler, O.V.; Wu, C.; Nixon, A.B.; et al. Assessment of Capecitabine and Bevacizumab With or Without Atezolizumab for the Treatment of Refractory Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA Netw. Open 2022, 5, e2149040. [Google Scholar] [CrossRef]
- Carmona, G.; Göttig, S.; Orlandi, A.; Scheele, J.; Bäuerle, T.; Jugold, M.; Kiessling, F.; Henschler, R.; Zeiher, A.M.; Dimmeler, S.; et al. Role of the small GTPase Rap1 for integrin activity regulation in endothelial cells and angiogenesis. Blood 2009, 113, 488–497. [Google Scholar] [CrossRef]
- Chrzanowska-Wodnicka, M.; Kraus, A.E.; Gale, D.; White, G.C., 2nd; Vansluys, J. Defective angiogenesis, endothelial migration, proliferation, and MAPK signaling in Rap1b-deficient mice. Blood 2008, 111, 2647–2656. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, F.; Ingram, D.A.; Quilliam, L.A. Rap1a is a key regulator of fibroblast growth factor 2-induced angiogenesis and together with Rap1b controls human endothelial cell functions. Mol. Cell Biol. 2008, 28, 5803–5810. [Google Scholar] [CrossRef] [PubMed]
- Han, C.W.; Jeong, M.S.; Jang, S.B. Structure, signaling and the drug discovery of the Ras oncogene protein. BMB Rep. 2017, 50, 355–360. [Google Scholar] [CrossRef]
- Cox, A.D.; Der, C.J. Ras history: The saga continues. Small GTPases 2010, 1, 2–27. [Google Scholar] [CrossRef]
- Boettner, B.; Van Aelst, L. Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 2009, 21, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.R.; Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef]
- Colicelli, J. Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, 2004, Re13. [Google Scholar] [CrossRef]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef]
- Busquets-Hernández, C.; Triola, G. Palmitoylation as a Key Regulator of Ras Localization and Function. Front. Mol. Biosci. 2021, 8, 659861. [Google Scholar] [CrossRef]
- Wright, L.P.; Philips, M.R. Thematic review series: Lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 2006, 47, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.D.; Der, C.J.; Philips, M.R. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin. Cancer Res. 2015, 21, 1819–1827. [Google Scholar] [CrossRef]
- Hancock, J.F.; Parton, R.G. Ras plasma membrane signalling platforms. Biochem. J. 2005, 389, 1–11. [Google Scholar] [CrossRef]
- Ntantie, E.; Gonyo, P.; Lorimer, E.L.; Hauser, A.D.; Schuld, N.; McAllister, D.; Kalyanaraman, B.; Dwinell, M.B.; Auchampach, J.A.; Williams, C.L. An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci. Signal 2013, 6, ra39. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, S.; Henis, Y.I. Interactions of Ras proteins with the plasma membrane and their roles in signaling. Cell Signal 2008, 20, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ahearn, I.; Zhou, M.; Philips, M.R. Posttranslational Modifications of RAS Proteins. Cold Spring Harb. Perspect. Med. 2018, 8, a031484. [Google Scholar] [CrossRef]
- Araya, M.K.; Chen, W.; Ke, Y.; Zhou, Y.; Gorfe, A.A.; Hancock, J.F.; Liu, J. Differential Lipid Binding Specificities of RAP1A and RAP1B are Encoded by the Amino Acid Sequence of the Membrane Anchors. J. Am. Chem. Soc. 2024, 146, 19782–19791. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef]
- Caron, E. Cellular functions of the Rap1 GTP-binding protein: A pattern emerges. J. Cell Sci. 2003, 116, 435–440. [Google Scholar] [CrossRef]
- Simanshu, D.K.; Nissley, D.V.; McCormick, F. RAS Proteins and Their Regulators in Human Disease. Cell 2017, 170, 17–33. [Google Scholar] [CrossRef]
- Anjum, R.; Blenis, J. The RSK family of kinases: Emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008, 9, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.H.; Bos, J.L. Specificity in Ras and Rap signaling. J. Biol. Chem. 2009, 284, 10995–10999. [Google Scholar] [CrossRef]
- Shah, S.; Brock, E.J.; Ji, K.; Mattingly, R.R. Ras and Rap1: A tale of two GTPases. Semin. Cancer Biol. 2019, 54, 29–39. [Google Scholar] [CrossRef]
- Chrzanowska-Wodnicka, M. Regulation of angiogenesis by a small GTPase Rap1. Vascul Pharmacol. 2010, 53, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lakshmikanthan, S.; Sobczak, M.; Chun, C.; Henschel, A.; Dargatz, J.; Ramchandran, R.; Chrzanowska-Wodnicka, M. Rap1 promotes VEGFR2 activation and angiogenesis by a mechanism involving integrin αvβ3. Blood 2011, 118, 2015–2026. [Google Scholar] [CrossRef]
- Lakshmikanthan, S.; Sobczak, M.; Li Calzi, S.; Shaw, L.; Grant, M.B.; Chrzanowska-Wodnicka, M. Rap1B promotes VEGF-induced endothelial permeability and is required for dynamic regulation of the endothelial barrier. J. Cell Sci. 2018, 131, jcs207605. [Google Scholar] [CrossRef] [PubMed]
- Lakshmikanthan, S.; Zheng, X.; Nishijima, Y.; Sobczak, M.; Szabo, A.; Vasquez-Vivar, J.; Zhang, D.X.; Chrzanowska-Wodnicka, M. Rap1 promotes endothelial mechanosensing complex formation, NO release and normal endothelial function. EMBO Rep. 2015, 16, 628–637. [Google Scholar] [CrossRef]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P.; et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020, 30, 745–762. [Google Scholar] [CrossRef]
- Sharma, G.P.; Kosuru, R.; Lakshmikanthan, S.; Zheng, S.; Chen, Y.; Burns, R.; Xin, G.; Cui, W.; Chrzanowska, M. Endothelial Rap1B mediates T-cell exclusion to promote tumor growth: A novel mechanism underlying vascular immunosuppression. Angiogenesis 2023, 26, 265–278. [Google Scholar] [CrossRef]
- Singh, B.; Kosuru, R.; Lakshmikanthan, S.; Sorci-Thomas, M.G.; Zhang, D.X.; Sparapani, R.; Vasquez-Vivar, J.; Chrzanowska, M. Endothelial Rap1 (Ras-Association Proximate 1) Restricts Inflammatory Signaling to Protect From the Progression of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 638–650. [Google Scholar] [CrossRef]
- O’Bryan, J.P. Pharmacological targeting of RAS: Recent success with direct inhibitors. Pharmacol. Res. 2019, 139, 503–511. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Singhal, A.; Li, B.T.; O’Reilly, E.M. Targeting KRAS in cancer. Nat. Med. 2024, 30, 969–983. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Rajalingam, K.; Schreck, R.; Rapp, U.R.; Albert, S. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta 2007, 1773, 1177–1195. [Google Scholar] [CrossRef]
- Qunaj, L.; May, M.S.; Neugut, A.I.; Herzberg, B.O. Prognostic and therapeutic impact of the KRAS G12C mutation in colorectal cancer. Front. Oncol. 2023, 13, 1252516. [Google Scholar] [CrossRef]
- Arcila, M.; Lau, C.; Nafa, K.; Ladanyi, M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J. Mol. Diagn. 2011, 13, 64–73. [Google Scholar] [CrossRef]
- Johnson, L.; Greenbaum, D.; Cichowski, K.; Mercer, K.; Murphy, E.; Schmitt, E.; Bronson, R.T.; Umanoff, H.; Edelmann, W.; Kucherlapati, R.; et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes. Dev. 1997, 11, 2468–2481. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Ichise, H.; Nakao, K.; Hatta, T.; Otani, H.; Sakagami, H.; Kondo, H.; Katsuki, M. Partial functional overlap of the three ras genes in mouse embryonic development. Oncogene 2008, 27, 2961–2968. [Google Scholar] [CrossRef][Green Version]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef]
- Janes, M.R.; Zhang, J.; Li, L.S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e517. [Google Scholar] [CrossRef]
- Zhang, S.S.; Nagasaka, M. Spotlight on Sotorasib (AMG 510) for KRAS (G12C) Positive Non-Small Cell Lung Cancer. Lung Cancer 2021, 12, 115–122. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for Lung Cancers with KRAS p. G12C Mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Jänne, P.A.; Riely, G.J.; Gadgeel, S.M.; Heist, R.S.; Ou, S.I.; Pacheco, J.M.; Johnson, M.L.; Sabari, J.K.; Leventakos, K.; Yau, E.; et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRAS(G12C) Mutation. N. Engl. J. Med. 2022, 387, 120–131. [Google Scholar] [CrossRef] [PubMed]
- McFall, T.; Trogdon, M.; Guizar, A.C.; Langenheim, J.F.; Sisk-Hackworth, L.; Stites, E.C. Co-targeting KRAS G12C and EGFR reduces both mutant and wild-type RAS-GTP. NPJ Precis. Oncol. 2022, 6, 86. [Google Scholar] [CrossRef]
- Gentile, D.R.; Rathinaswamy, M.K.; Jenkins, M.L.; Moss, S.M.; Siempelkamp, B.D.; Renslo, A.R.; Burke, J.E.; Shokat, K.M. Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States. Cell Chem. Biol. 2017, 24, 1455–1466.e1414. [Google Scholar] [CrossRef] [PubMed]
- Holderfield, M.; Lee, B.J.; Jiang, J.; Tomlinson, A.; Seamon, K.J.; Mira, A.; Patrucco, E.; Goodhart, G.; Dilly, J.; Gindin, Y.; et al. Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature 2024, 629, 919–926. [Google Scholar] [CrossRef]
- Schulze, C.J.; Seamon, K.J.; Zhao, Y.; Yang, Y.C.; Cregg, J.; Kim, D.; Tomlinson, A.; Choy, T.J.; Wang, Z.; Sang, B.; et al. Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS. Science 2023, 381, 794–799. [Google Scholar] [CrossRef]
- Wasko, U.N.; Jiang, J.; Dalton, T.C.; Curiel-Garcia, A.; Edwards, A.C.; Wang, Y.; Lee, B.; Orlen, M.; Tian, S.; Stalnecker, C.A.; et al. Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer. Nature 2024, 629, 927–936. [Google Scholar] [CrossRef]
- Maurer, T.; Garrenton, L.S.; Oh, A.; Pitts, K.; Anderson, D.J.; Skelton, N.J.; Fauber, B.P.; Pan, B.; Malek, S.; Stokoe, D.; et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 2012, 109, 5299–5304. [Google Scholar] [CrossRef]
- Cruz-Migoni, A.; Canning, P.; Quevedo, C.E.; Bataille, C.J.R.; Bery, N.; Miller, A.; Russell, A.J.; Phillips, S.E.V.; Carr, S.B.; Rabbitts, T.H. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 2545–2550. [Google Scholar] [CrossRef] [PubMed]
- Kessler, D.; Gmachl, M.; Mantoulidis, A.; Martin, L.J.; Zoephel, A.; Mayer, M.; Gollner, A.; Covini, D.; Fischer, S.; Gerstberger, T.; et al. Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. USA 2019, 116, 15823–15829. [Google Scholar] [CrossRef]
- Leshchiner, E.S.; Parkhitko, A.; Bird, G.H.; Luccarelli, J.; Bellairs, J.A.; Escudero, S.; Opoku-Nsiah, K.; Godes, M.; Perrimon, N.; Walensky, L.D. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc. Natl. Acad. Sci. USA 2015, 112, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Patgiri, A.; Yadav, K.K.; Arora, P.S.; Bar-Sagi, D. An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 2011, 7, 585–587. [Google Scholar] [CrossRef]
- Burns, M.C.; Sun, Q.; Daniels, R.N.; Camper, D.; Kennedy, J.P.; Phan, J.; Olejniczak, E.T.; Lee, T.; Waterson, A.G.; Rossanese, O.W.; et al. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc. Natl. Acad. Sci. USA 2014, 111, 3401–3406. [Google Scholar] [CrossRef] [PubMed]
- Hillig, R.C.; Sautier, B.; Schroeder, J.; Moosmayer, D.; Hilpmann, A.; Stegmann, C.M.; Werbeck, N.D.; Briem, H.; Boemer, U.; Weiske, J.; et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.J.; Anderson, M.; Blades, K.; Brassington, C.; Breeze, A.L.; Chresta, C.; Embrey, K.; Fairley, G.; Faulder, P.; Finlay, M.R.; et al. Small molecule binding sites on the Ras:SOS complex can be exploited for inhibition of Ras activation. J. Med. Chem. 2015, 58, 2265–2274. [Google Scholar] [CrossRef]
- Shi, Z.Q.; Yu, D.H.; Park, M.; Marshall, M.; Feng, G.S. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol. Cell. Biol. 2000, 20, 1526–1536. [Google Scholar] [CrossRef]
- Ruess, D.A.; Heynen, G.J.; Ciecielski, K.J.; Ai, J.; Berninger, A.; Kabacaoglu, D.; Görgülü, K.; Dantes, Z.; Wörmann, S.M.; Diakopoulos, K.N.; et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 2018, 24, 954–960. [Google Scholar] [CrossRef]
- Nichols, R.J.; Haderk, F.; Stahlhut, C.; Schulze, C.J.; Hemmati, G.; Wildes, D.; Tzitzilonis, C.; Mordec, K.; Marquez, A.; Romero, J.; et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 2018, 20, 1064–1073. [Google Scholar] [CrossRef]
- Nakata, S.; Fujita, N.; Kitagawa, Y.; Okamoto, R.; Ogita, H.; Takai, Y. Regulation of platelet-derived growth factor receptor activation by afadin through SHP-2: Implications for cellular morphology. J. Biol. Chem. 2007, 282, 37815–37825. [Google Scholar] [CrossRef]
- Athuluri-Divakar, S.K.; Vasquez-Del Carpio, R.; Dutta, K.; Baker, S.J.; Cosenza, S.C.; Basu, I.; Gupta, Y.K.; Reddy, M.V.; Ueno, L.; Hart, J.R.; et al. A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling. Cell 2016, 165, 643–655. [Google Scholar] [CrossRef]
- Kowalczyk, J.T.; Wan, X.; Hernandez, E.R.; Luo, R.; Lyons, G.C.; Wilson, K.M.; Gallardo, D.C.; Isanogle, K.A.; Robinson, C.M.; Mendoza, A.; et al. Rigosertib Induces Mitotic Arrest and Apoptosis in RAS-Mutated Rhabdomyosarcoma and Neuroblastoma. Mol. Cancer Ther. 2021, 20, 307–319. [Google Scholar] [CrossRef]
- Pallara, C.; Cabot, D.; Rivas, J.; Brun, S.; Seco, J.; Abuasaker, B.; Tarragó, T.; Jaumot, M.; Prades, R.; Agell, N. Peptidomimetics designed to bind to RAS effector domain are promising cancer therapeutic compounds. Sci. Rep. 2022, 12, 15810. [Google Scholar] [CrossRef]
- Whyte, D.B.; Kirschmeier, P.; Hockenberry, T.N.; Nunez-Oliva, I.; James, L.; Catino, J.J.; Bishop, W.R.; Pai, J.K. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997, 272, 14459–14464. [Google Scholar] [CrossRef]
- Gilardi, M.; Wang, Z.; Proietto, M.; Chillà, A.; Calleja-Valera, J.L.; Goto, Y.; Vanoni, M.; Janes, M.R.; Mikulski, Z.; Gualberto, A.; et al. Tipifarnib as a Precision Therapy for HRAS-Mutant Head and Neck Squamous Cell Carcinomas. Mol. Cancer Ther. 2020, 19, 1784–1796. [Google Scholar] [CrossRef]
- Manandhar, S.P.; Hildebrandt, E.R.; Schmidt, W.K. Small-molecule inhibitors of the Rce1p CaaX protease. J. Biomol. Screen. 2007, 12, 983–993. [Google Scholar] [CrossRef]
- Winter-Vann, A.M.; Baron, R.A.; Wong, W.; dela Cruz, J.; York, J.D.; Gooden, D.M.; Bergo, M.O.; Young, S.G.; Toone, E.J.; Casey, P.J. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4336–4341. [Google Scholar] [CrossRef]
- Lau, H.Y.; Ramanujulu, P.M.; Guo, D.; Yang, T.; Wirawan, M.; Casey, P.J.; Go, M.L.; Wang, M. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol. Ther. 2014, 15, 1280–1291. [Google Scholar] [CrossRef] [PubMed]
- Bergo, M.O.; Lieu, H.D.; Gavino, B.J.; Ambroziak, P.; Otto, J.C.; Casey, P.J.; Walker, Q.M.; Young, S.G. On the physiological importance of endoproteolysis of CAAX proteins: Heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J. Biol. Chem. 2004, 279, 4729–4736. [Google Scholar] [CrossRef] [PubMed]
- Belanis, L.; Plowman, S.J.; Rotblat, B.; Hancock, J.F.; Kloog, Y. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol. Biol. Cell 2008, 19, 1404–1414. [Google Scholar] [CrossRef]
- Shalom-Feuerstein, R.; Plowman, S.J.; Rotblat, B.; Ariotti, N.; Tian, T.; Hancock, J.F.; Kloog, Y. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 2008, 68, 6608–6616. [Google Scholar] [CrossRef]
- Chandra, A.; Grecco, H.E.; Pisupati, V.; Perera, D.; Cassidy, L.; Skoulidis, F.; Ismail, S.A.; Hedberg, C.; Hanzal-Bayer, M.; Venkitaraman, A.R.; et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011, 14, 148–158. [Google Scholar] [CrossRef]
- Muratcioglu, S.; Jang, H.; Gursoy, A.; Keskin, O.; Nussinov, R. PDEδ Binding to Ras Isoforms Provides a Route to Proper Membrane Localization. J. Phys. Chem. B 2017, 121, 5917–5927. [Google Scholar] [CrossRef]
- Nancy, V.; Callebaut, I.; El Marjou, A.; de Gunzburg, J. The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J. Biol. Chem. 2002, 277, 15076–15084. [Google Scholar] [CrossRef]
- Zimmermann, G.; Papke, B.; Ismail, S.; Vartak, N.; Chandra, A.; Hoffmann, M.; Hahn, S.A.; Triola, G.; Wittinghofer, A.; Bastiaens, P.I.; et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 2013, 497, 638–642. [Google Scholar] [CrossRef]
- Papke, B.; Murarka, S.; Vogel, H.A.; Martín-Gago, P.; Kovacevic, M.; Truxius, D.C.; Fansa, E.K.; Ismail, S.; Zimmermann, G.; Heinelt, K.; et al. Identification of pyrazolopyridazinones as PDEδ inhibitors. Nat. Commun. 2016, 7, 11360. [Google Scholar] [CrossRef]
- Leung, E.L.; Luo, L.X.; Li, Y.; Liu, Z.Q.; Li, L.L.; Shi, D.F.; Xie, Y.; Huang, M.; Lu, L.L.; Duan, F.G.; et al. Identification of a new inhibitor of KRAS-PDEδ interaction targeting KRAS mutant nonsmall cell lung cancer. Int. J. Cancer 2019, 145, 1334–1345. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Wang, X.; Li, Y.; Zhou, L.; Lu, X.; Dong, G.; Sheng, C. Discovery of novel KRAS–PDEδ inhibitors with potent activity in patient-derived human pancreatic tumor xenograft models. Acta Pharm. Sin. B 2022, 12, 274–290. [Google Scholar] [CrossRef] [PubMed]
- Dumbacher, M.; Van Dooren, T.; Princen, K.; De Witte, K.; Farinelli, M.; Lievens, S.; Tavernier, J.; Dehaen, W.; Wera, S.; Winderickx, J.; et al. Modifying Rap1-signalling by targeting Pde6δ is neuroprotective in models of Alzheimer’s disease. Mol. Neurodegener. 2018, 13, 50. [Google Scholar] [CrossRef]
- Kawata, M.; Farnsworth, C.C.; Yoshida, Y.; Gelb, M.H.; Glomset, J.A.; Takai, Y. Posttranslationally processed structure of the human platelet protein smg p21B: Evidence for geranylgeranylation and carboxyl methylation of the C-terminal cysteine. Proc. Natl. Acad. Sci. USA 1990, 87, 8960–8964. [Google Scholar] [CrossRef]
- Efuet, E.T.; Keyomarsi, K. Farnesyl and geranylgeranyl transferase inhibitors induce G1 arrest by targeting the proteasome. Cancer Res. 2006, 66, 1040–1051. [Google Scholar] [CrossRef]
- Kanda, Y.; Watanabe, Y. Adrenaline increases glucose transport via a Rap1-p38MAPK pathway in rat vascular smooth muscle cells. Br. J. Pharmacol. 2007, 151, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Lee, H.R.; Shin, H.K.; Park, S.Y.; Hong, K.W.; Kim, E.K.; Bae, S.S.; Lee, W.S.; Rhim, B.Y.; Kim, C.D. Cilostazol enhances integrin-dependent homing of progenitor cells by activation of cAMP-dependent protein kinase in synergy with Epac1. J. Neurosci. Res. 2011, 89, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Bustelo, X.R.; Sauzeau, V.; Berenjeno, I.M. GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays News Rev. Mol. Cell. Dev. Biol. 2007, 29, 356–370. [Google Scholar] [CrossRef]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef]
- Lobell, R.B.; Omer, C.A.; Abrams, M.T.; Bhimnathwala, H.G.; Brucker, M.J.; Buser, C.A.; Davide, J.P.; deSolms, S.J.; Dinsmore, C.J.; Ellis-Hutchings, M.S.; et al. Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 2001, 61, 8758–8768. [Google Scholar]
- Spencer-Smith, R.; Li, L.; Prasad, S.; Koide, A.; Koide, S.; O’Bryan, J.P. Targeting the α4-α5 interface of RAS results in multiple levels of inhibition. Small GTPases 2019, 10, 378–387. [Google Scholar] [CrossRef]
- Spencer-Smith, R.; Koide, A.; Zhou, Y.; Eguchi, R.R.; Sha, F.; Gajwani, P.; Santana, D.; Gupta, A.; Jacobs, M.; Herrero-Garcia, E.; et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 2017, 13, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Spencer-Smith, R.; O’Bryan, J.P. Targeting the α4-α5 dimerization interface of K-RAS inhibits tumor formation in vivo. Oncogene 2019, 38, 2984–2993. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Koide, A.; Zuberi, M.; Ketavarapu, G.; Denbaum, E.; Teng, K.W.; Rhett, J.M.; Spencer-Smith, R.; Hobbs, G.A.; Camp, E.R.; et al. Identification of the nucleotide-free state as a therapeutic vulnerability for inhibition of selected oncogenic RAS mutants. Cell Rep. 2022, 38, 110322. [Google Scholar] [CrossRef]
- Whaby, M.; Nair, R.S.; O’Bryan, J.P. Probing RAS Function Using Monobody and NanoBiT Technologies. Methods Mol. Biol. 2024, 2797, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Shoari, A.; Tahmasebi, M.; Khodabakhsh, F.; Cohan, R.A.; Oghalaie, A.; Behdani, M. Angiogenic biomolecules specific nanobodies application in cancer imaging and therapy; review and updates. Int. Immunopharmacol. 2022, 105, 108585. [Google Scholar] [CrossRef]
- Wang, T.; Yu, H.; Hughes, N.W.; Liu, B.; Kendirli, A.; Klein, K.; Chen, W.W.; Lander, E.S.; Sabatini, D.M. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 2017, 168, 890–903.e815. [Google Scholar] [CrossRef]
- Jones, G.G.; Del Río, I.B.; Sari, S.; Sekerim, A.; Young, L.C.; Hartig, N.; Areso Zubiaur, I.; El-Bahrawy, M.A.; Hynds, R.E.; Lei, W.; et al. SHOC2 phosphatase-dependent RAF dimerization mediates resistance to MEK inhibition in RAS-mutant cancers. Nat. Commun. 2019, 10, 2532. [Google Scholar] [CrossRef]
- Sulahian, R.; Kwon, J.J.; Walsh, K.H.; Pailler, E.; Bosse, T.L.; Thaker, M.; Almanza, D.; Dempster, J.M.; Pan, J.; Piccioni, F.; et al. Synthetic Lethal Interaction of SHOC2 Depletion with MEK Inhibition in RAS-Driven Cancers. Cell Rep. 2019, 29, 118–134.e118. [Google Scholar] [CrossRef]
- Hatzivassiliou, G.; Song, K.; Yen, I.; Brandhuber, B.J.; Anderson, D.J.; Alvarado, R.; Ludlam, M.J.; Stokoe, D.; Gloor, S.L.; Vigers, G.; et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010, 464, 431–435. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Zhang, C.; Bollag, G.; Shokat, K.M.; Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010, 464, 427–430. [Google Scholar] [CrossRef]
- Yen, I.; Shanahan, F.; Merchant, M.; Orr, C.; Hunsaker, T.; Durk, M.; La, H.; Zhang, X.; Martin, S.E.; Lin, E.; et al. Pharmacological Induction of RAS-GTP Confers RAF Inhibitor Sensitivity in KRAS Mutant Tumors. Cancer Cell 2018, 34, 611–625.e617. [Google Scholar] [CrossRef] [PubMed]
- Engelman, J.A.; Chen, L.; Tan, X.; Crosby, K.; Guimaraes, A.R.; Upadhyay, R.; Maira, M.; McNamara, K.; Perera, S.A.; Song, Y.; et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008, 14, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Hoeflich, K.P.; Merchant, M.; Orr, C.; Chan, J.; Den Otter, D.; Berry, L.; Kasman, I.; Koeppen, H.; Rice, K.; Yang, N.Y.; et al. Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res. 2012, 72, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Tabernero, J.; Janku, F.; Wainberg, Z.A.; Paz-Ares, L.; Vansteenkiste, J.; Van Cutsem, E.; Pérez-García, J.; Stathis, A.; Britten, C.D.; et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin. Cancer Res. 2015, 21, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, G.I.; LoRusso, P.; Kwak, E.; Pandya, S.; Rudin, C.M.; Kurkjian, C.; Cleary, J.M.; Pilat, M.J.; Jones, S.; de Crespigny, A.; et al. Phase Ib study of the MEK inhibitor cobimetinib (GDC-0973) in combination with the PI3K inhibitor pictilisib (GDC-0941) in patients with advanced solid tumors. Investig. New Drugs 2020, 38, 419–432. [Google Scholar] [CrossRef]
- Ebi, H.; Corcoran, R.B.; Singh, A.; Chen, Z.; Song, Y.; Lifshits, E.; Ryan, D.P.; Meyerhardt, J.A.; Benes, C.; Settleman, J.; et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J. Clin. Investig. 2011, 121, 4311–4321. [Google Scholar] [CrossRef]
- Molina-Arcas, M.; Hancock, D.C.; Sheridan, C.; Kumar, M.S.; Downward, J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 2013, 3, 548–563. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghadrdoost Nakhchi, B.; Kosuru, R.; Chrzanowska, M. Towards Targeting Endothelial Rap1B to Overcome Vascular Immunosuppression in Cancer. Int. J. Mol. Sci. 2024, 25, 9853. https://doi.org/10.3390/ijms25189853
Ghadrdoost Nakhchi B, Kosuru R, Chrzanowska M. Towards Targeting Endothelial Rap1B to Overcome Vascular Immunosuppression in Cancer. International Journal of Molecular Sciences. 2024; 25(18):9853. https://doi.org/10.3390/ijms25189853
Chicago/Turabian StyleGhadrdoost Nakhchi, Behshid, Ramoji Kosuru, and Magdalena Chrzanowska. 2024. "Towards Targeting Endothelial Rap1B to Overcome Vascular Immunosuppression in Cancer" International Journal of Molecular Sciences 25, no. 18: 9853. https://doi.org/10.3390/ijms25189853
APA StyleGhadrdoost Nakhchi, B., Kosuru, R., & Chrzanowska, M. (2024). Towards Targeting Endothelial Rap1B to Overcome Vascular Immunosuppression in Cancer. International Journal of Molecular Sciences, 25(18), 9853. https://doi.org/10.3390/ijms25189853









