Thermophilic Hemicellulases Secreted by Microbial Consortia Selected from an Anaerobic Digester
Abstract
:1. Introduction
2. Results
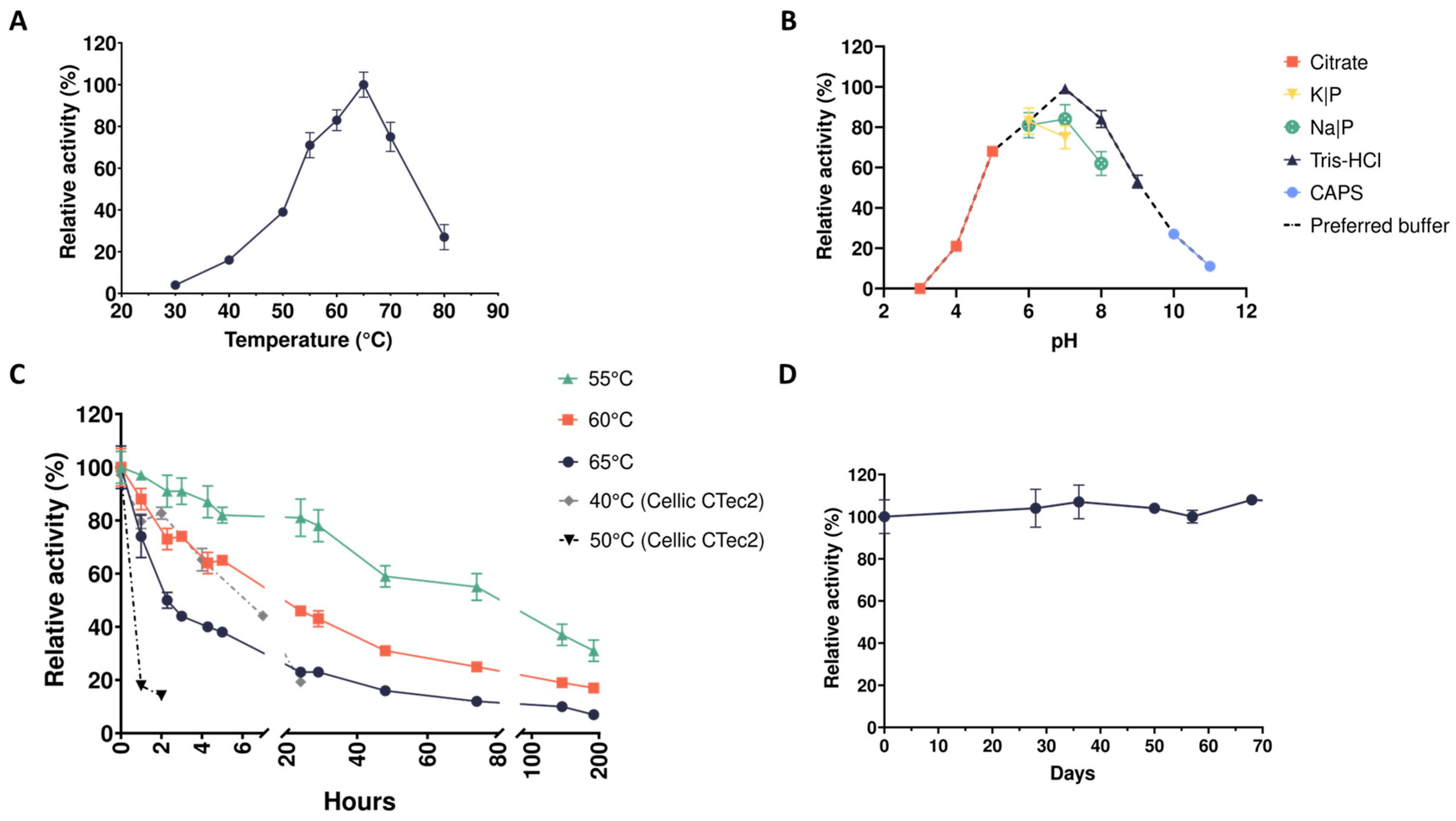
2.1. Thermophilic Microbial Consortia Secrete Robust Xylanases
2.2. Quantification of the Xylanolytic Activity in Cell Free Supernatants
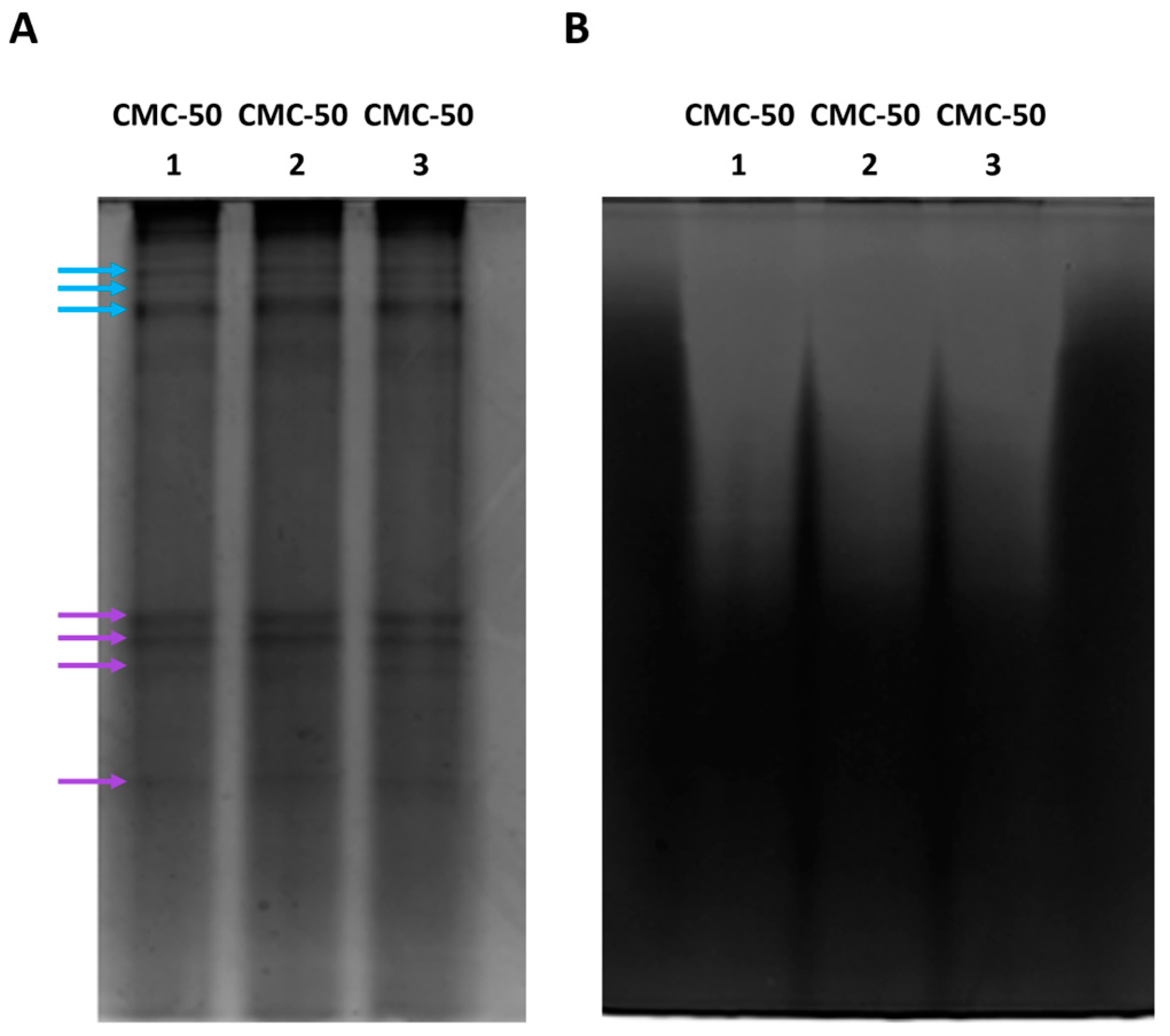
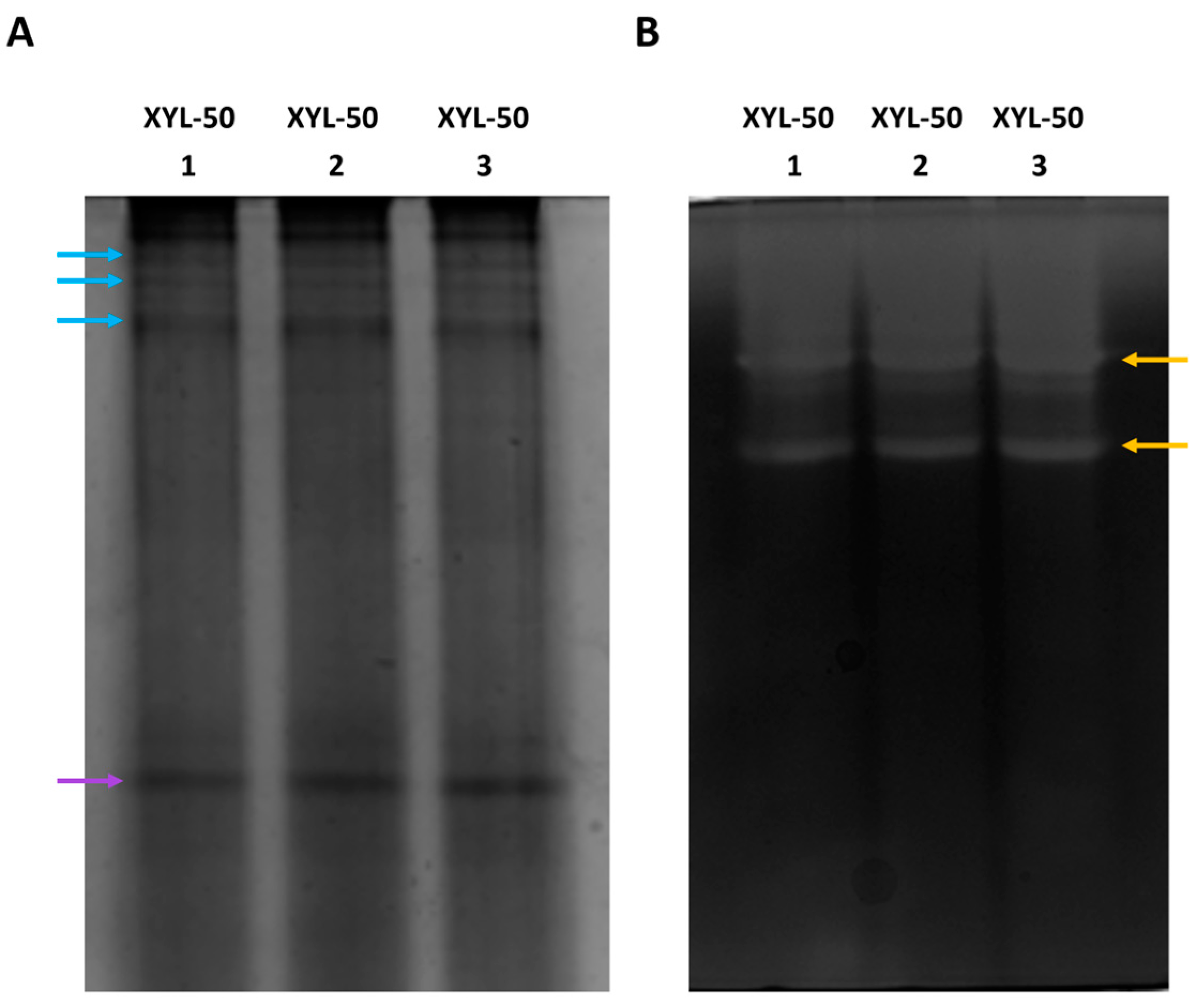
2.3. Activity Profiling of Xylanases via Electrophoresis and Zymography
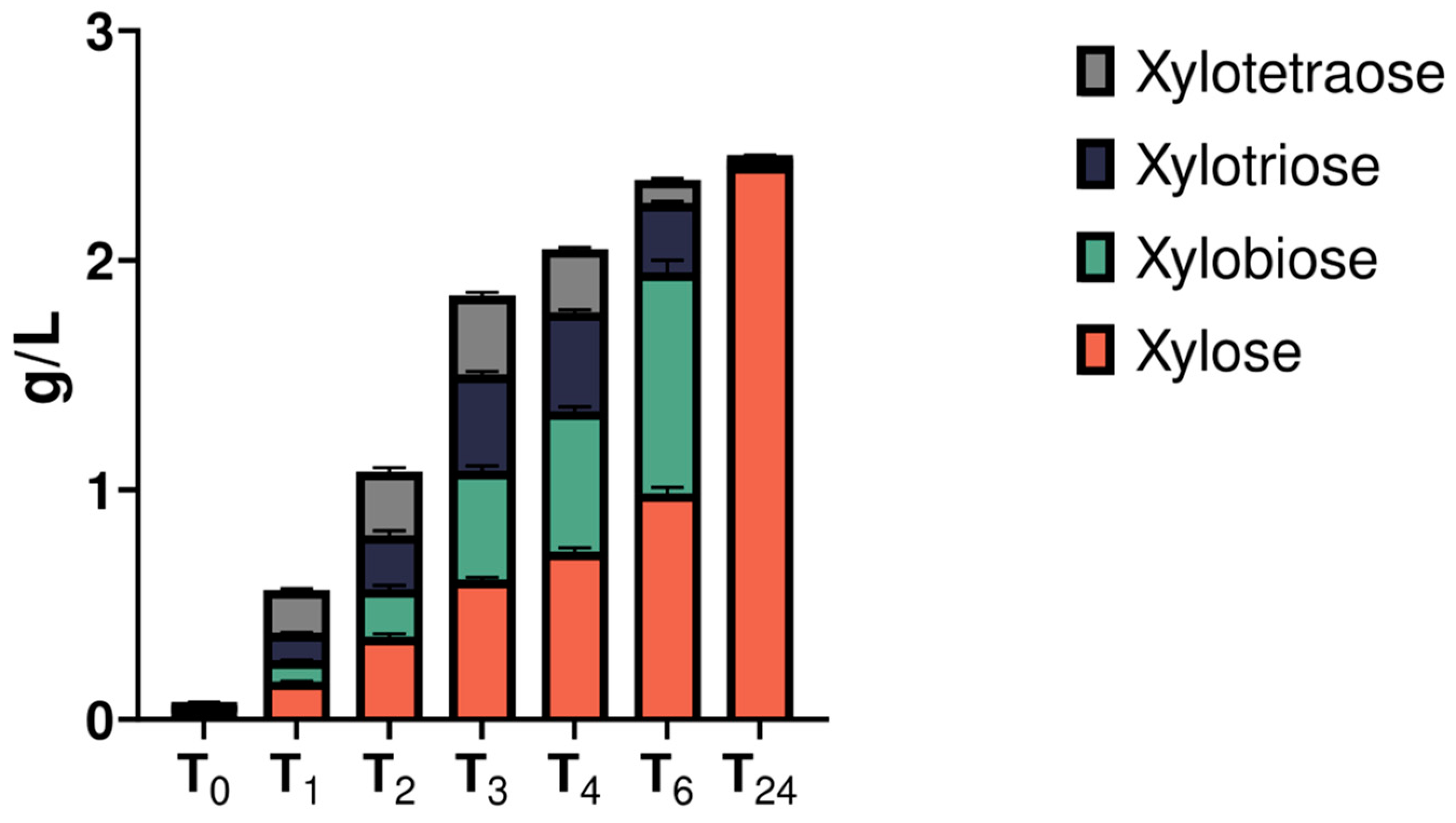
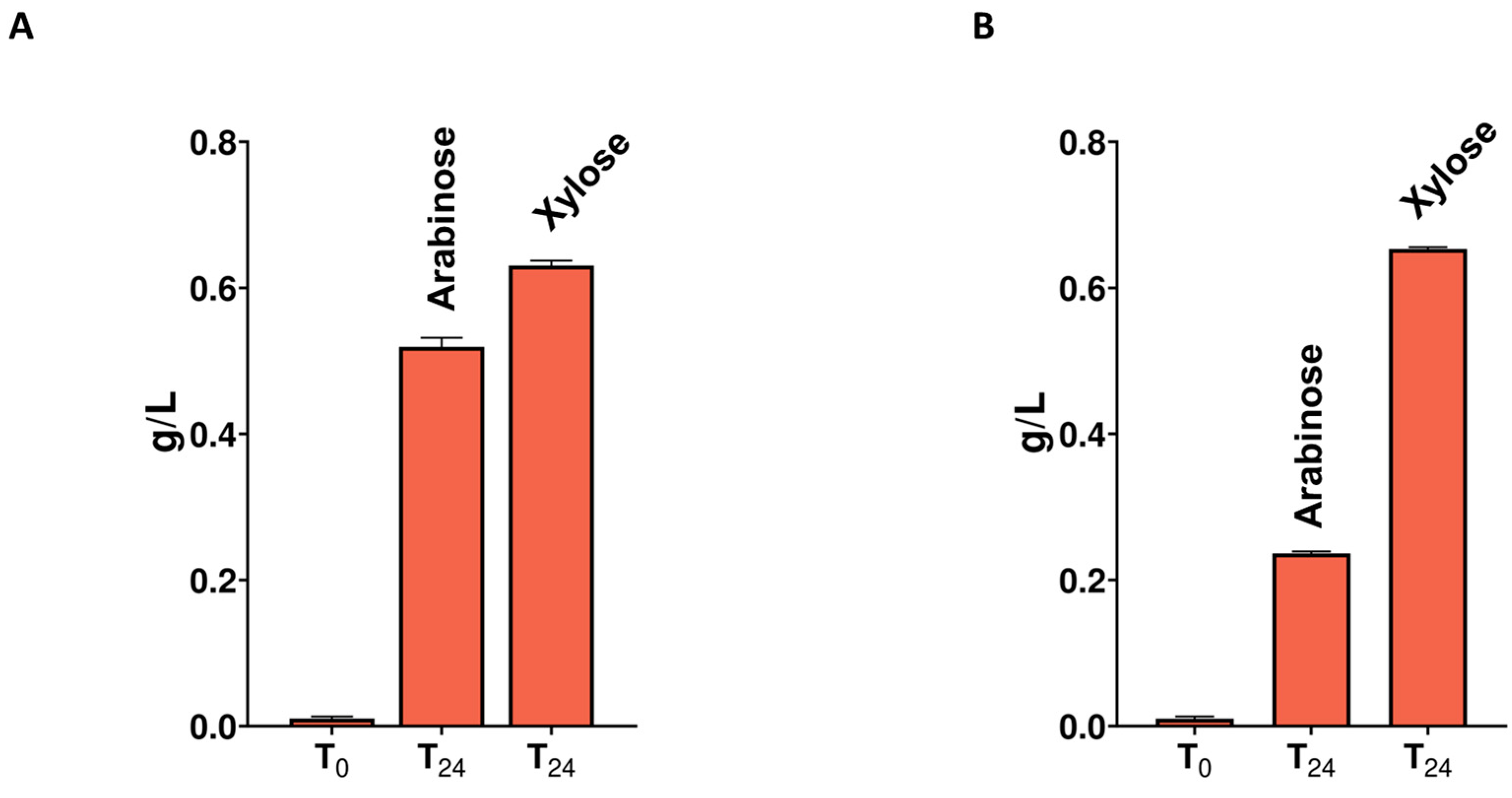
2.4. Hydrolytic Potential of Cell Free Supernatants towards Lignocellulose-Derived Polysaccharides and Spent Mushroom Substrate
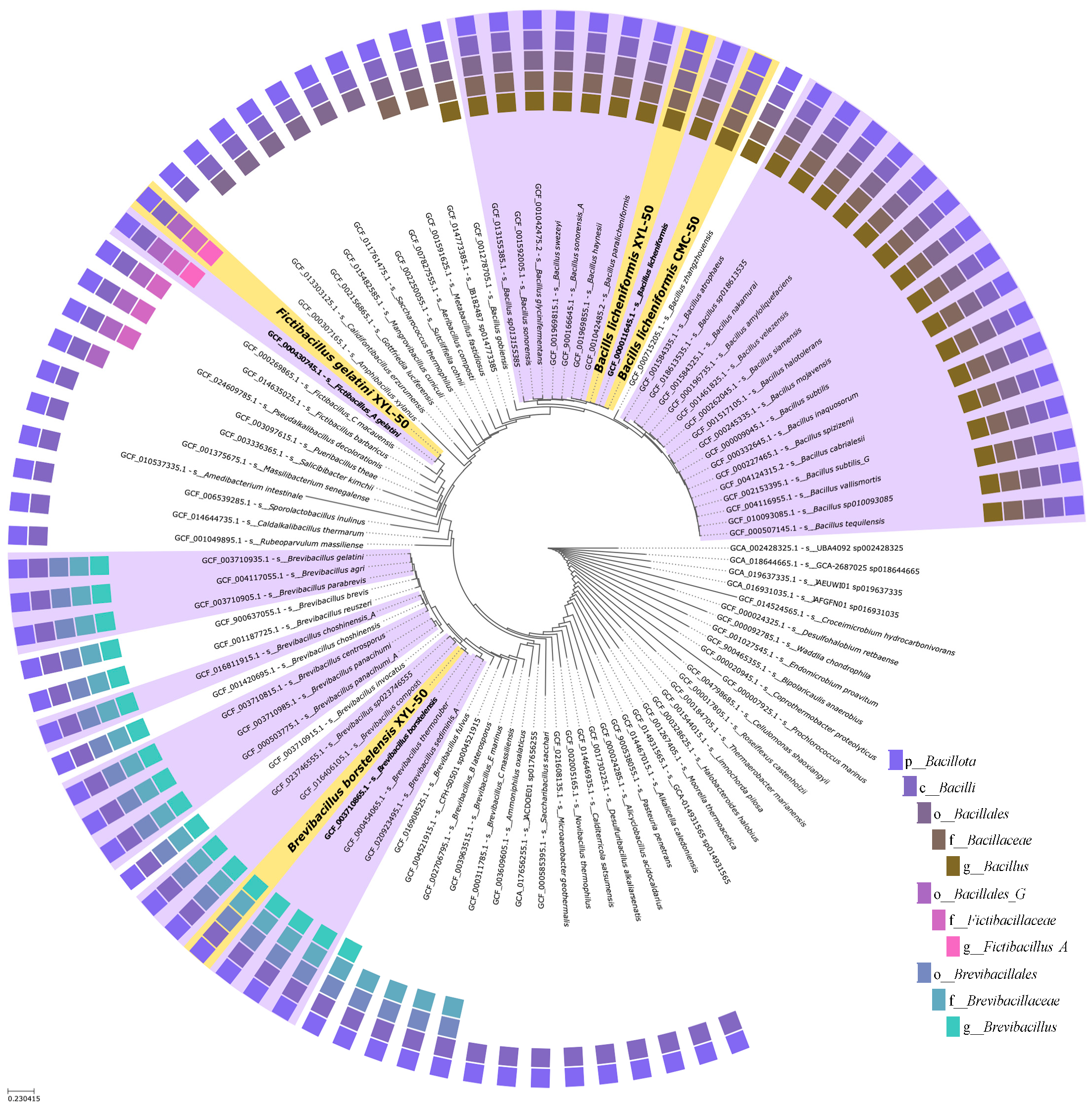
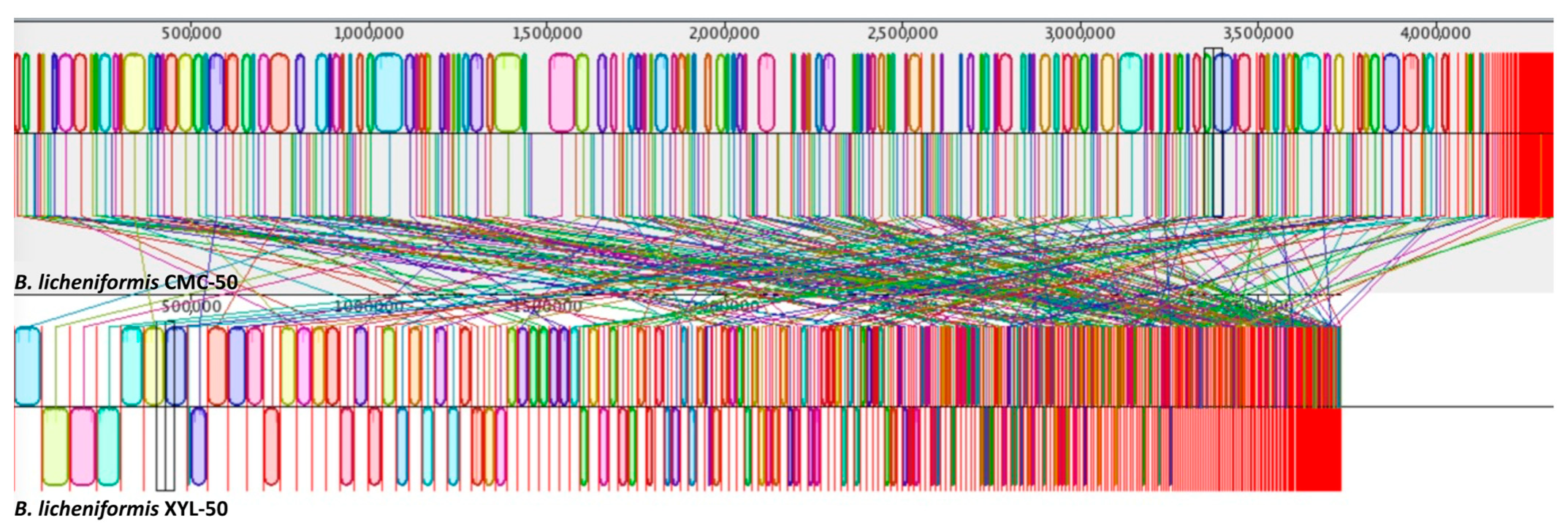
2.5. Phylogenetic and Comparative Genomic Analyses Reveal Strain-Level Differences between Microbiomes
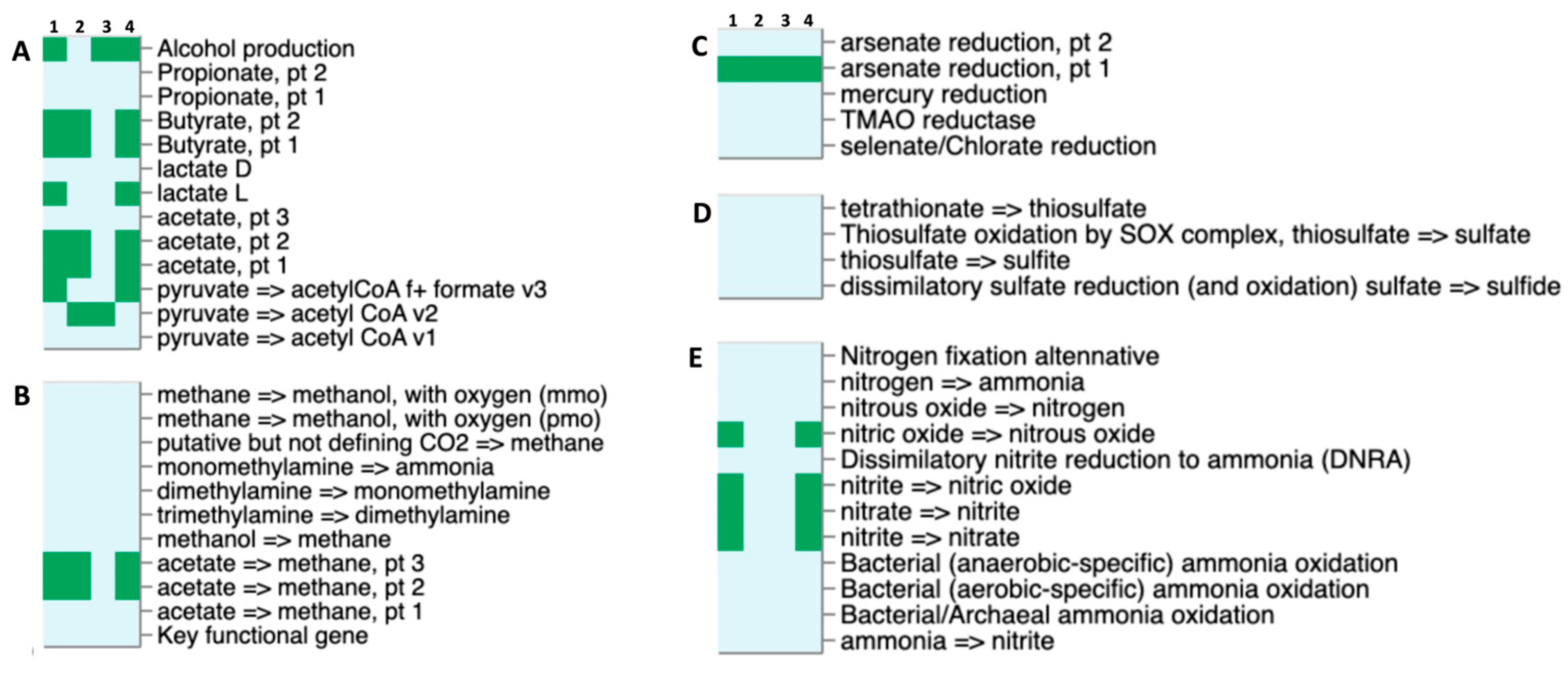
2.6. Metabolic Potential and Putative Enzymatic Activities of the Thermophilic Microbiomes
3. Discussion
4. Materials and Methods
4.1. Media and Chemicals
4.2. Microbial Growth for Enzyme Production
4.3. Azo-Xylan Assay
4.4. Reducing Sugars Assay for Enzyme Units Measurement
4.5. Electrophoretic and Zymographic Analysis
4.6. Hydrolysis of LCB-Derived Polysaccharides
4.7. HPAEC-PAD Analysis
4.8. Phylogenetic Analysis and Functional Annotation of CAZymes
4.9. Modeling of the Enzyme-Substrate Complex
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mujtaba, M.; Fraceto, L.F.; Fazeli, M.; Mukherjee, S.; Savassa, S.M.; de Medeiros, G.A.; Pereira, A.D.E.S.; Mancini, S.D.; Lipponen, J.; Vilaplana, F. Lignocellulosic biomass from agricultural waste to the circular economy: A review with focus on biofuels, biocomposites and bioplastics. J. Clean. Prod. 2023, 402, 136815. [Google Scholar] [CrossRef]
- Singh, D.P.; Prabha, R.; Renu, S.; Sahu, P.K.; Singh, V. Agrowaste bioconversion and microbial fortification have prospects for soil health, crop productivity, and eco-enterprising. Int. J. Recycl. Org. Waste Agric. 2019, 8, 457–472. [Google Scholar] [CrossRef]
- Saldarriaga-Hernández, S.; Velasco-Ayala, C.; Flores, P.L.-I.; Rostro-Alanis, M.d.J.; Parra-Saldivar, R.; Iqbal, H.M.; Carrillo-Nieves, D.; Saldarriaga-Hernández, S.; Velasco-Ayala, C.; Flores, P.L.-I.; et al. Biotransformation of lignocellulosic biomass into industrially relevant products with the aid of fungi-derived lignocellulolytic enzymes. Int. J. Biol. Macromol. 2020, 161, 1099–1116. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Barrett, D.M.; Delwiche, M.J.; Stroeve, P. Methods for Pretreatment of Lignocellulosic Biomass for Efficient Hydrolysis and Biofuel Production. Ind. Eng. Chem. Res. 2009, 48, 3713–3729. [Google Scholar] [CrossRef]
- Aulitto, M.; Fusco, S.; Nickel, D.B.; Bartolucci, S.; Contursi, P.; Franzén, C.J. Seed culture pre-adaptation of Bacillus coagulans MA-13 improves lactic acid production in simultaneous saccharification and fermentation. Biotechnol. Biofuels 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Matsagar, B.M.; Dhepe, P.L. Effects of cations, anions and H+ concentration of acidic ionic liquids on the valorization of polysaccharides into furfural. New J. Chem. 2017, 41, 6137–6144. [Google Scholar] [CrossRef]
- Shirkavand, E.; Baroutian, S.; Gapes, D.J.; Young, B.R. Combination of fungal and physicochemical processes for lignocellulosic biomass pretreatment—A review. Renew. Sustain. Energy Rev. 2016, 54, 217–234. [Google Scholar] [CrossRef]
- Zhu, D.; Adebisi, W.A.; Ahmad, F.; Sethupathy, S.; Danso, B.; Sun, J. Recent Development of Extremophilic Bacteria and Their Application in Biorefinery. Front. Bioeng. Biotechnol. 2020, 8, 483. [Google Scholar] [CrossRef]
- Singh, S.; Shukla, L.; Khare, S. Detection and Characterization of New Thermostable Endoglucanase from Aspergillus awamori Strain F 18. Available online: https://www.researchgate.net/publication/235622352 (accessed on 9 April 2024).
- Böhme, B.; Moritz, B.; Wendler, J.; Hertel, T.C.; Ihling, C.; Brandt, W.; Pietzsch, M. Enzymatic activity and thermoresistance of improved microbial transglutaminase variants. Amino Acids 2020, 52, 313–326. [Google Scholar] [CrossRef]
- Zuliani, L.; Serpico, A.; De Simone, M.; Frison, N.; Fusco, S. Biorefinery Gets Hot: Thermophilic Enzymes and Microorganisms for Second-Generation Bioethanol Production. Processes 2021, 9, 1583. [Google Scholar] [CrossRef]
- Chylenski, P.; Forsberg, Z.; Ståhlberg, J.; Várnai, A.; Lersch, M.; Bengtsson, O.; Sæbø, S.; Horn, S.J.; Eijsink, V.G. Development of minimal enzyme cocktails for hydrolysis of sulfite-pulped lignocellulosic biomass. J. Biotechnol. 2017, 246, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kallioinen, A.; Puranen, T.; Siika-Aho, M. Mixtures of Thermostable Enzymes Show High Performance in Biomass Saccharification. Appl. Biochem. Biotechnol. 2014, 173, 1038–1056. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, A.V.; Yonamine, D.K.; Uyemura, S.A.; Dinamarco, T.M. A Thermostable Aspergillus fumigatus GH7 Endoglucanase Over-Expressed in Pichia pastoris Stimulates Lignocellulosic Biomass Hydrolysis. Int. J. Mol. Sci. 2019, 20, 2261. [Google Scholar] [CrossRef] [PubMed]
- Cintra, L.C.; da Costa, I.C.; de Oliveira, I.C.M.; Fernandes, A.G.; Faria, S.P.; Jesuíno, R.S.A.; Ravanal, M.C.; Eyzaguirre, J.; Ramos, L.P.; de Faria, F.P.; et al. The boosting effect of recombinant hemicellulases on the enzymatic hydrolysis of steam-treated sugarcane bagasse. Enzym. Microb. Technol. 2019, 133, 109447. [Google Scholar] [CrossRef]
- Gavande, P.V.; Goyal, A.; Fontes, C.M.G.A. Carbohydrates and Carbohydrate-Active enZymes (CAZyme): An overview. In Glycoside Hydrolases: Biochemistry, Biophysics, and Biotechnology; Elsevier: Amsterdam, The Netherlands, 2023; pp. 1–23. [Google Scholar] [CrossRef]
- Malhotra, G.; Chapadgaonkar, S.S. Production and applications of xylanases—An overview. BioTechnologia 2018, 99, 59–72. [Google Scholar] [CrossRef]
- Salzano, F.; Aulitto, M.; Fiorentino, G.; Cannella, D.; Peeters, E.; Limauro, D. A novel endo-1,4-β-xylanase from Alicyclobacillus mali FL18: Biochemical characterization and its synergistic action with β-xylosidase in hemicellulose deconstruction. Int. J. Biol. Macromol. 2024, 264, 130550. [Google Scholar] [CrossRef]
- Singh, R.; Hans, M.; Kumar, S.; Yadav, Y.K. Thermophilic Anaerobic Digestion: An Advancement towards Enhanced Biogas Production from Lignocellulosic Biomass. Sustainability 2023, 15, 1859. [Google Scholar] [CrossRef]
- Ho, D.P.; Jensen, P.D.; Batstone, D.J. Methanosarcinaceae and Acetate-Oxidizing Pathways Dominate in High-Rate Thermophilic Anaerobic Digestion of Waste-Activated Sludge. Appl. Environ. Microbiol. 2013, 79, 6491–6500. [Google Scholar] [CrossRef]
- Moset, V.; Poulsen, M.; Wahid, R.; Højberg, O.; Møller, H.B. Mesophilic versus thermophilic anaerobic digestion of cattle manure: Methane productivity and microbial ecology. Microb. Biotechnol. 2015, 8, 787–800. [Google Scholar] [CrossRef]
- Suksong, W.; Kongjan, P.; Prasertsan, P.; O-Thong, S. Thermotolerant cellulolytic Clostridiaceae and Lachnospiraceae rich consortium enhanced biogas production from oil palm empty fruit bunches by solid-state anaerobic digestion. Bioresour. Technol. 2019, 291, 121851. [Google Scholar] [CrossRef]
- Jensen, M.B.; de Jonge, N.; Dolriis, M.D.; Kragelund, C.; Fischer, C.H.; Eskesen, M.R.; Noer, K.; Møller, H.B.; Ottosen, L.D.M.; Nielsen, J.L.; et al. Cellulolytic and Xylanolytic Microbial Communities Associated With Lignocellulose-Rich Wheat Straw Degradation in Anaerobic Digestion. Front. Microbiol. 2021, 12, 645174. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Cejnar, J.; Vítězová, M.; Vítěz, T.; Dordević, D.; Bomble, Y.J. Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants. Int. J. Mol. Sci. 2020, 21, 283. [Google Scholar] [CrossRef]
- Bombardi, L.; Salini, A.; Aulitto, M.; Zuliani, L.; Andreolli, M.; Bordoli, P.; Coltro, A.; Vitulo, N.; Zaccone, C.; Lampis, S.; et al. Lignocellulolytic Potential of Microbial Consortia Isolated from a Local Biogas Plant: The Case of Thermostable Xylanases Secreted by Mesophilic Bacteria. Int. J. Mol. Sci. 2024, 25, 1090. [Google Scholar] [CrossRef] [PubMed]
- Akunna, J.; Bizeau, C.; Moletta, R. Denitrification in anaerobic digesters: Possibilities and influence of wastewater COD/N-NOXratio. Environ. Technol. 1992, 13, 825–836. [Google Scholar] [CrossRef]
- Percheron, G.; Bernet, N.; Moletta, R. Interactions between methanogenic and nitrate reducing bacteria during the anaerobic digestion of an industrial sulfate rich wastewater. FEMS Microbiol. Ecol. 1999, 29, 341–350. [Google Scholar] [CrossRef]
- Yaoi, K.; Hiyoshi, A.; Mitsuishi, Y. Screening, Purification and Characterization of a Prokaryotic Isoprimeverose-producing Oligoxyloglucan Hydrolase from Oerskovia sp. Y1. J. Appl. Glycosci. 2007, 54, 91–94. [Google Scholar] [CrossRef]
- Bischoff, K.M.; Liu, S.; Hughes, S.R. Cloning and characterization of a recombinant family 5 endoglucanase from Bacillus licheniformis strain B-41361. Process. Biochem. 2007, 42, 1150–1154. [Google Scholar] [CrossRef]
- Liberato, M.V.; Silveira, R.L.; Prates, T.; de Araujo, E.A.; Pellegrini, V.O.A.; Camilo, C.M.; Kadowaki, M.A.; Neto, M.d.O.; Popov, A.; Skaf, M.S.; et al. Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism. Sci. Rep. 2016, 6, 23473. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Liu, Q.; Zhang, C.; Ma, Q. Molecular Cloning of Novel Cellulase Genes cel9A and cel12A from Bacillus licheniformis GXN151 and Synergism of Their Encoded Polypeptides. Curr. Microbiol. 2004, 49, 234–238. [Google Scholar] [CrossRef]
- Inácio, J.M.; Correia, I.L.; de Sá-Nogueira, I. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 2008, 154, 2719–2729. [Google Scholar] [CrossRef]
- Park, J.-M.; Jang, M.-U.; Kang, J.-H.; Kim, M.-J.; Lee, S.-W.; Song, Y.B.; Shin, C.-S.; Han, N.S.; Kim, T.-J. Detailed modes of action and biochemical characterization of endo-arabinanase from Bacillus licheniformis DSM13. J. Microbiol. 2012, 50, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.A.; Mahmud, S.; Albogami, S.; El-Shehawi, A.M.; Paul, G.K.; Islam, S.; Dutta, A.K.; Uddin, S.; Zaman, S. Biochemical and Molecular Dynamics Study of a Novel GH 43 α-l-Arabinofuranosidase/β-Xylosidase From Caldicellulosiruptor saccharolyticus DSM8903. Front. Bioeng. Biotechnol. 2022, 10, 810542. [Google Scholar] [CrossRef]
- Gilad, O.; Jacobsen, S.; Stuer-Lauridsen, B.; Pedersen, M.B.; Garrigues, C.; Svensson, B. Combined transcriptome and proteome analysis of bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl. Environ. Microbiol. 2010, 76, 7285–7291. [Google Scholar] [CrossRef]
- Zanphorlin, L.M.; de Morais, M.A.B.; Diogo, J.A.; Domingues, M.N.; de Souza, F.H.M.; Ruller, R.; Murakami, M.T. Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43. Biotechnol. Bioeng. 2018, 116, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Ramírez, N.; Romero-García, E.R.; Calderón, V.C.; Avitia, C.I.; Téllez-Valencia, A.; Pedraza-Reyes, M. Expression, Characterization and Synergistic Interactions of Myxobacter Sp. AL-1 Cel9 and Cel48 Glycosyl Hydrolases. Int. J. Mol. Sci. 2008, 9, 247–257. [Google Scholar] [CrossRef]
- Yu, T.; Cui, H.; Li, J.C.; Luo, Y.; Jiang, G.; Zhao, H. Enzyme function prediction using contrastive learning. Science 2023, 379, 1358–1363. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.A.B.; Coines, J.; Domingues, M.N.; Pirolla, R.A.S.; Tonoli, C.C.C.; Santos, C.R.; Correa, J.B.L.; Gozzo, F.C.; Rovira, C.; Murakami, M.T. Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations. Nat. Commun. 2021, 12, 367. [Google Scholar] [CrossRef]
- Deshavath, N.N.; Mukherjee, G.; Goud, V.V.; Veeranki, V.D.; Sastri, C.V. Pitfalls in the 3, 5-dinitrosalicylic acid (DNS) assay for the reducing sugars: Interference of furfural and 5-hydroxymethylfurfural. Int. J. Biol. Macromol. 2020, 156, 180–185. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Anders, N.; Humann, H.; Langhans, B.; Spieß, A.C. Simultaneous determination of acid-soluble biomass-derived compounds using high performance anion exchange chromatography coupled with pulsed amperometric detection. Anal. Methods 2015, 7, 7866–7873. [Google Scholar] [CrossRef]
- Chivian, D.; Jungbluth, S.P.; Dehal, P.S.; Wood-Charlson, E.M.; Canon, R.S.; Allen, B.H.; Clark, M.M.; Gu, T.; Land, M.L.; Price, G.A.; et al. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat. Protoc. 2022, 18, 208–238. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Lu, W.; Zhang, J.; Huang, W.; Zhang, Z.; Jia, X.; Wang, Z.; Shi, L.; Li, C.; Wolynes, P.G.; Zheng, S. DynamicBind: Predicting ligand-speciic protein-ligand complex structure with a deep equivariant generative model. Nat. Commun. 2024, 15, 1071. [Google Scholar] [CrossRef]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.-P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.-P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Orlando, M.; Fortuna, S.; Oloketuyi, S.; Bajc, G.; Goldenzweig, A.; de Marco, A. CDR1 Composition Can Affect Nanobody Recombinant Expression Yields. Biomolecules 2021, 11, 1362. [Google Scholar] [CrossRef]

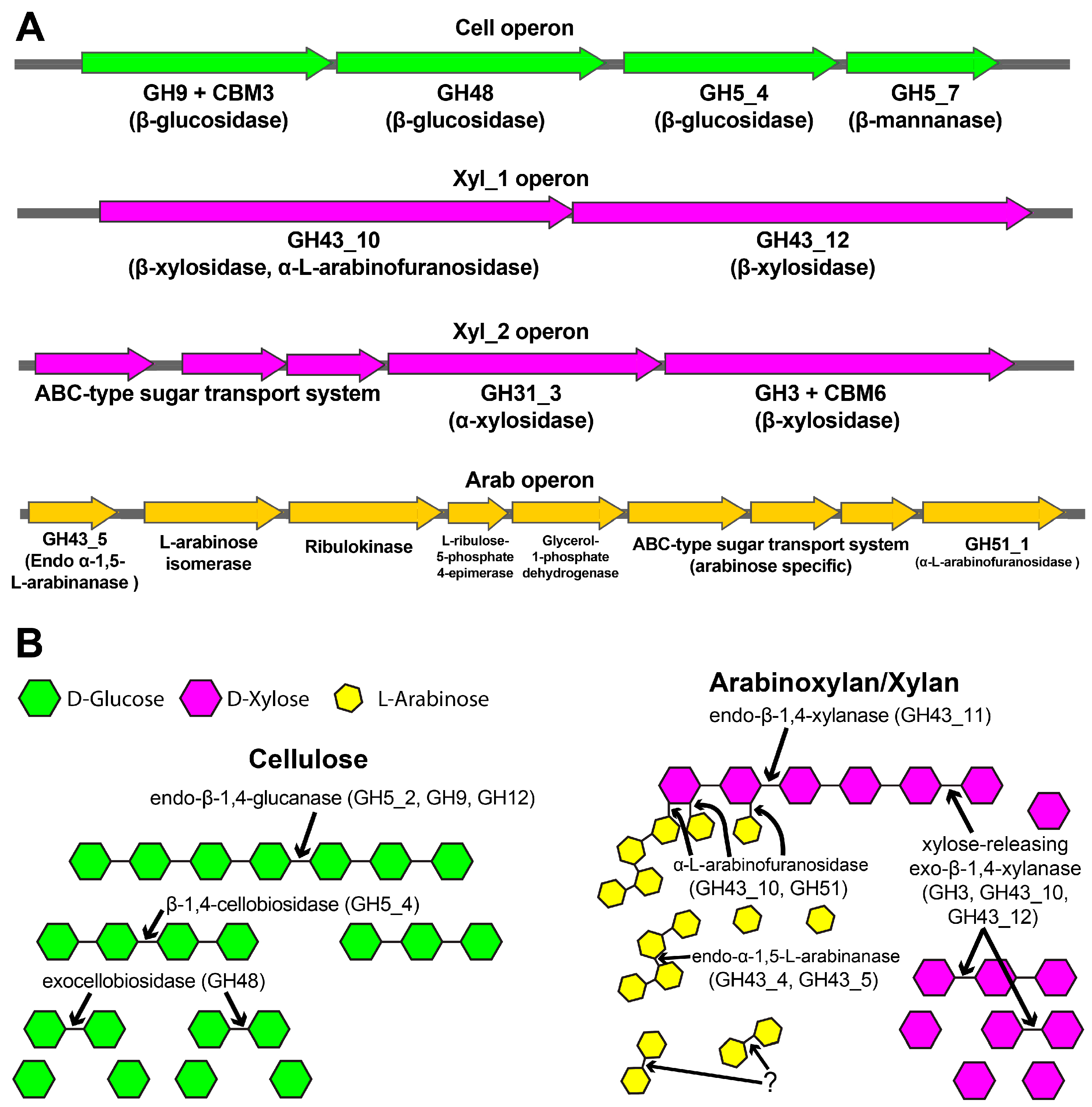
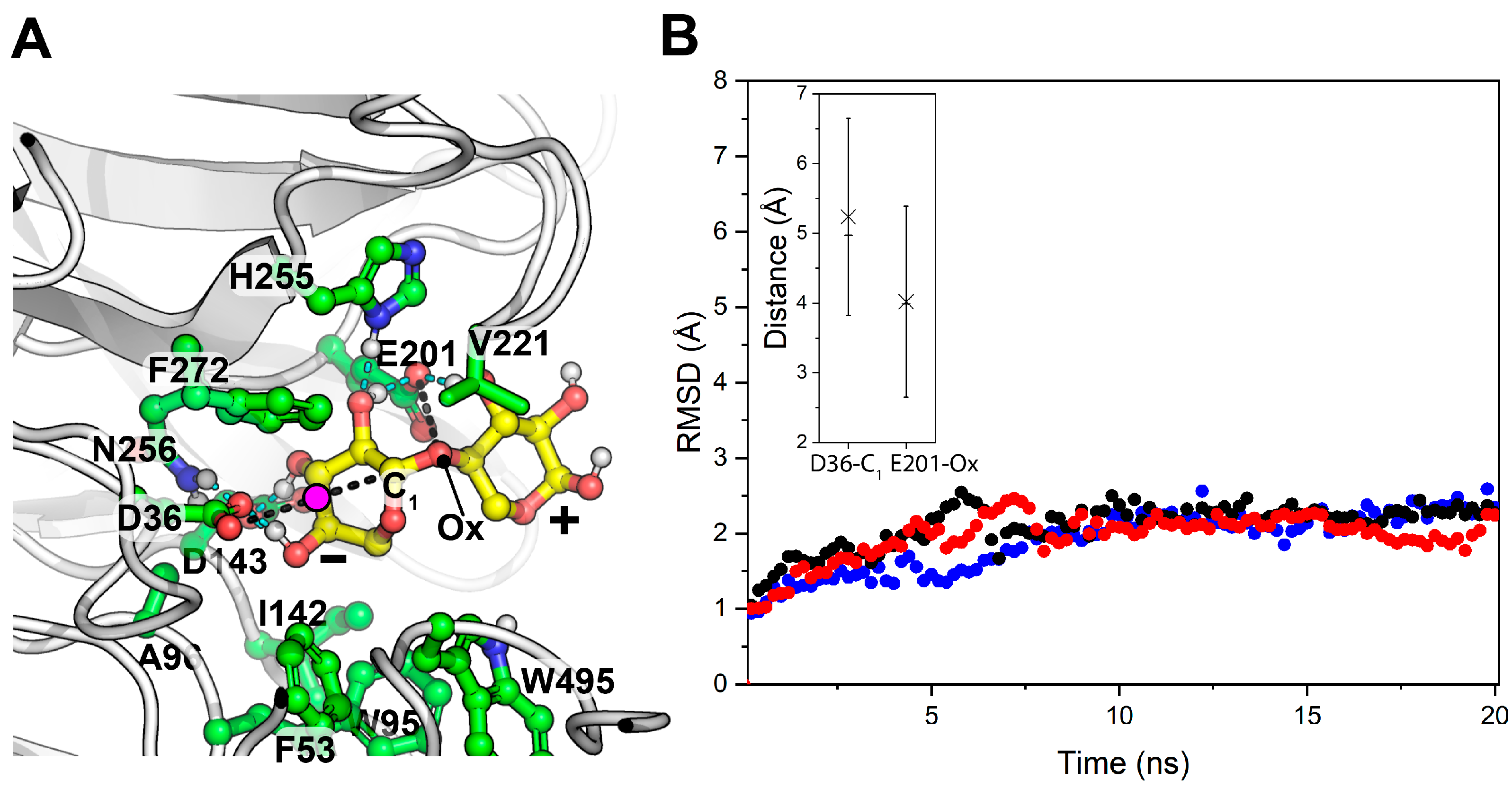
| Source | Family | Operon | Predicted E.C. Number a | Predicted Signal Peptide b | Activity MSCH (Experimental) | Uniprot c ID MSCH | Identity (%) to MSCH | Bibliography MSCH |
|---|---|---|---|---|---|---|---|---|
| XYL-50_1 | GH3 + CBM6 | Xyl_2 | 3.2.1.37 | - | Isoprimeverose-producing Oligoxyloglucan Hydrolase | I0IUK4 | 45.7 | [28] |
| CMC-50_1 | GH3 + CBM6 | Xyl_2 | 3.2.1.37 | - | Isoprimeverose-producing Oligoxyloglucan Hydrolase | I0IUK4 | 45.7 | [28] |
| XYL-50_1 | GH5_2 + CBM3 | - | 3.2.1.4 | Sec/SPI | Endo β-1,4-glucanase | Q7X3S6 | 100 | [29] |
| CMC-50_1 | GH5_2 + CBM3 | - | 3.2.1.4 | Sec/SPI | Endo β-1,4-glucanase | Q7X3S6 | 100 | [29] |
| XYL-50_1 | GH5_4 | Cell | 3.2.1.4 | Sec/SPI | β-1,4-cellobiosidase (reducing end) | AAU40777 | 100 | [30] |
| CMC-50_1 | GH5_4 | Cell | 3.2.1.4 | Sec/SPI | β-1,4-cellobiosidase (reducing end) | AAU40777 | 100 | [30] |
| XYL-50_1 | GH9 + CBM3 | Cell | 3.2.1.4 | Sec/SPI | Endo β-1,4-glucanase | Q6SYB5 | 100 | [31] |
| CMC-50_1 | GH9 + CBM3 | Cell | 3.2.1.4 | Sec/SPI | Endo β-1,4-glucanase | Q6SYB5 | 100 | [31] |
| XYL-50_1 | GH12 | - | 3.2.1.151 | Sec/SPI | Endo β-1,4-glucanase | Q7X4S4 | 100 | [31] |
| CMC-50_1 | GH12 | - | 3.2.1.151 | Sec/SPI | Endo β-1,4-glucanase | Q7X4S4 | 100 | [31] |
| CMC-50_1 | GH43_4 | - | 3.2.1.99 | Sec/SPII | Endo α-1,5-L-arabinanase | A0A6M4JQW9 | 75.5 | [32] |
| CMC-50_1 | GH43_5 | Arab | 3.2.1.99 | Sec/SPI | Endo α-1,5-L-arabinanase | UPI000043D9A2 | 100 | [33] |
| XYL-50_1 | GH43_5 | Arab | 3.2.1.99 | Sec/SPI | Endo α-1,5-L-arabinanase | UPI000043D9A2 | 100 | [33] |
| CMC-50_1 | GH43_5 | - | 3.2.1.99 | Sec/SPII | Endo α-1,5-L-arabinanase | UPI000043FF33 | 99.1 | [33] |
| CMC-50_1 | GH43_10 | Xyl_1 | 3.2.1.55 | - | Bifunctional β-xylosidase/α-L-arabinofuranosidase | A4XGG5 | 50.6 | [34] |
| XYL-50_1 | GH43_11 | - | 3.2.1.8 | Sec/SPII | Endo-β-xylosidase | D3R467 | 100 | [35] |
| CMC-50_1 | GH43_11 | - | 3.2.1.8 | Sec/SPII | Endo-β-xylosidase | D3R467 | 100 | [35] |
| XYL-50_1 | GH43_12 | Xyl_1 | 3.2.1.37 | - | Xylose-releasing exo-β-1,4-xylanase | Q65MB6 | 99.1 | [36] |
| CMC-50_1 | GH43_12 | Xyl_1 | 3.2.1.37 | - | Xylose-releasing exo-β-1,4-xylanase | Q65MB6 | 99.1 | [36] |
| XYL-50_1 | GH48 | Cell | 3.2.1.176 | - | exocellobiohydrolase | Q19VP0 | 95 | [37] |
| CMC-50_1 | GH48 | Cell | 3.2.1.176 | - | exocellobiohydrolase | Q19VP0 | 95 | [37] |
| XYL-50_1 | GH51 | Arab | 3.2.1.55 | - | α-L-arabinofuranosidase | A0A6M4JKK3 | 72.4 | [32] |
| CMC-50_1 | GH51 | Arab | 3.2.1.55 | - | α-L-arabinofuranosidase | A0A6M4JKK3 | 72.4 | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bombardi, L.; Orlando, M.; Aulitto, M.; Fusco, S. Thermophilic Hemicellulases Secreted by Microbial Consortia Selected from an Anaerobic Digester. Int. J. Mol. Sci. 2024, 25, 9887. https://doi.org/10.3390/ijms25189887
Bombardi L, Orlando M, Aulitto M, Fusco S. Thermophilic Hemicellulases Secreted by Microbial Consortia Selected from an Anaerobic Digester. International Journal of Molecular Sciences. 2024; 25(18):9887. https://doi.org/10.3390/ijms25189887
Chicago/Turabian StyleBombardi, Luca, Marco Orlando, Martina Aulitto, and Salvatore Fusco. 2024. "Thermophilic Hemicellulases Secreted by Microbial Consortia Selected from an Anaerobic Digester" International Journal of Molecular Sciences 25, no. 18: 9887. https://doi.org/10.3390/ijms25189887








