Whole-Genome and Poly(A)+Transcriptome Analysis of the Drosophila Mutant agnts3 with Cognitive Dysfunctions
Abstract
:1. Introduction
2. Results
2.1. Whole-Genome Sequencing of CS and agnts3
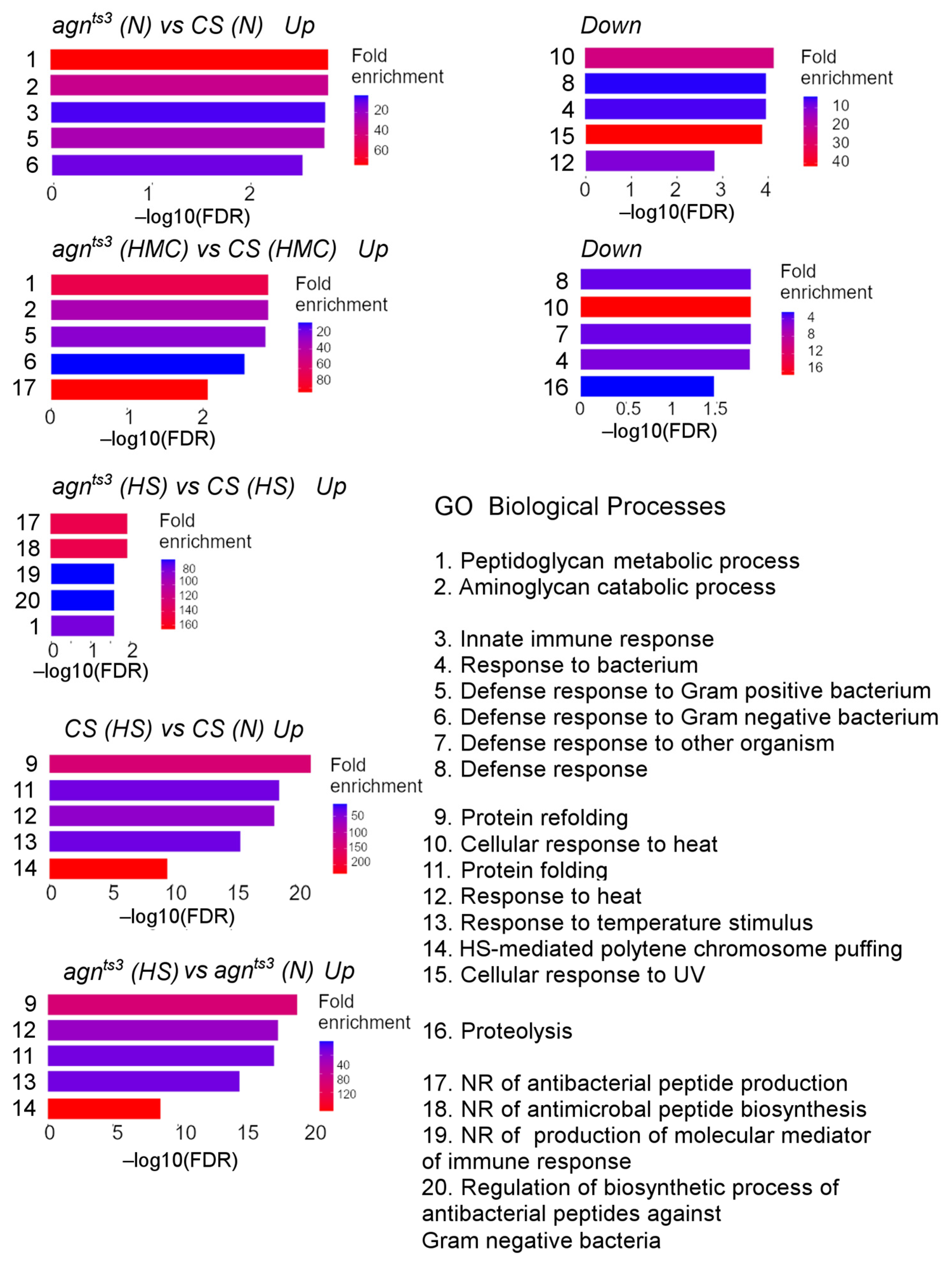
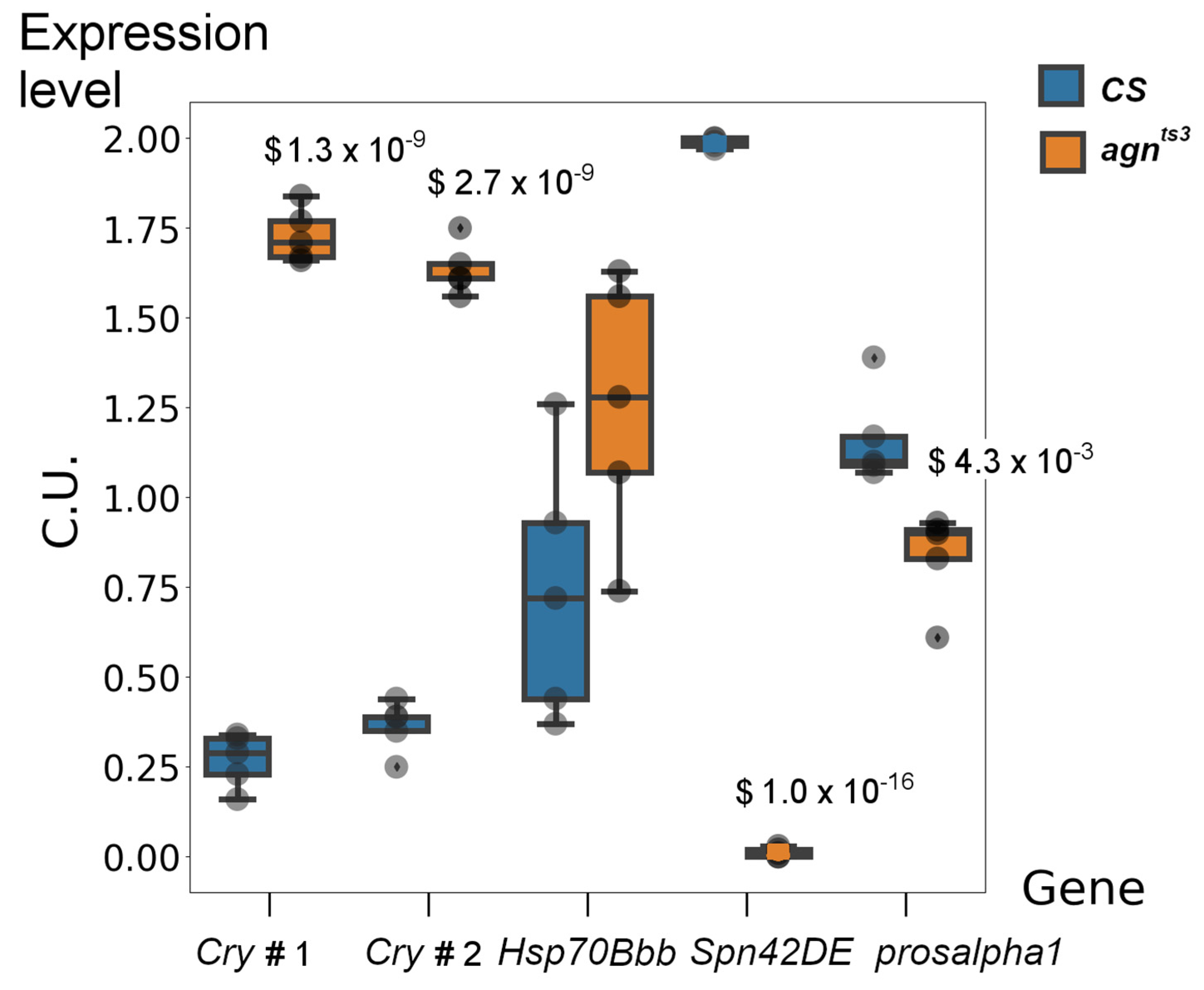
2.2. Transcription Profiles of CS and agnts3 under Normal Conditions and after Stress
3. Discussion
4. Materials and Methods
4.1. Fly Strains
4.2. Hypomagnetic Conditioning
4.3. Heat Shock Conditioning
4.4. DNA Extraction and Whole-Genome Sequencing
4.5. RNA Extraction and Transcriptome Sequencing
4.6. Genome Assembly and Annotation
4.7. Transcriptome Reads Assembly and Quantification
4.8. DEG Analysis
4.9. Reverse Transcription and Semi-Quantitative Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benzer, S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. USA 1967, 58, 1112–1119. [Google Scholar] [CrossRef]
- Quinn, W.G.; Harris, W.A.; Benzer, S. Conditioned behavior in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1974, 71, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Dudai, Y.; Jan, Y.N.; Byers, D.; Quinn, W.G.; Benzer, S. dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. USA 1976, 73, 1684–1688. [Google Scholar] [CrossRef] [PubMed]
- Byers, D.; Davis, R.L.; Kiger, J.A., Jr. Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature 1981, 289, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Tully, T. Discovery of genes involved with learning and memory: An experimental synthesis of Hirschian and Benzerian perspectives. Proc. Natl. Acad. Sci. USA 1996, 93, 13460–13467. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef]
- Kandel, E.R. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 2012, 5, 1–12. [Google Scholar] [CrossRef]
- Perkins, L.A.; Holderbaum, L.; Tao, R.; Hu, Y.; Sopko, R.; McCall, K.; Yang-Zhou, D.; Flockhart, I.; Binari, R.; Shim, H.S.; et al. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 2015, 201, 843–852. [Google Scholar] [CrossRef]
- Savvateeva, E.; Kamyshev, N. Behavioral effects of temperature sensitive mutations affecting metabolism of cAMP in D. melanogaster. Pharm. Biochem. Behav. 1981, 14, 603–611. [Google Scholar] [CrossRef]
- Medvedeva, A.; Savvateeva, E. The effects of the agnostic gene ts mutations that control calmodulin functions and learning ability on ectopic pairing of Drosophila polytene chromosomes. Dokl. Akad. Nauk SSSR 1991, 318, 733–736. [Google Scholar]
- Savvateeva-Popova, E.; Peresleni, A.; Scharagina, L.; Medvedeva, A.; Korochkina, S.; Grigorieva, I.; Dyuzhikova, N.A.; Popov, A.V.; Baricheva, E.M.; Karagodin, D.; et al. Architecture of the X chromosome, expression of LIM Kinase 1, and recombination in the agnostic mutants of Drosophila: A model for human williams syndrome. Russ. J. Genet. 2004, 40, 605–624. [Google Scholar] [CrossRef]
- Hawley, R. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 1980, 94, 625–646. [Google Scholar] [CrossRef] [PubMed]
- Zhimulev, I.; Semeshin, V.; Kulichkov, V.; Belyaeva, E. Intercalary heterochromatin in Drosophila. Chromosoma 1982, 87, 197–228. [Google Scholar] [CrossRef]
- Xamena, N.; Creus, A.; Macros, R. Effect of intercalating mutagens on crossing over in Drosophila melanogaster females. Experientia 1985, 41, 1078–1081. [Google Scholar] [CrossRef]
- Belyaeva, E.; Zhimulev, I.; Volkova, E.; Alekseyenko, A.; Moshkin, Y.; Koryakov, D. Su(UR)ES: A gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 7532–7537. [Google Scholar] [CrossRef]
- Medvedeva, A.V.; Molotkov, D.A.; Nikitina, E.A.; Popov, A.V.; Karagodin, D.A.; Baricheva, E.M.; Savvateeva-Popova, E.V. Systemic regulation of genetic and cytogenetic processes by a signal cascade of actin remodeling: Locus agnostic in Drosophila. Russ. J. Genet. 2008, 44, 669–681. [Google Scholar] [CrossRef]
- Nikitina, E.; Medvedeva, A.; Zakharov, G.; Savvateeva-Popova, E. The Drosophila agnostic locus: Involvement in the formation of cognitive defects in williams syndrome. Acta Nat. 2014, 6, 53–61. [Google Scholar] [CrossRef]
- Sarimov, R.M.; Serov, D.A.; Gudkov, S.V. Hypomagnetic Conditions and Their Biological Action (Review). Biology 2023, 12, 1513. [Google Scholar] [CrossRef]
- Vasilieva, S.A.; Tokmacheva, E.V.; Medvedeva, A.V.; Ermilova, A.A.; Nikitina, E.A.; Shchegolev, B.F.; Surma, S.V.; Savvateeva-Popova, E.V. The Role of Parental Origin of Chromosomes in the Instability of the Somatic Genome in Drosophila Brain Cells and Memory Trace Formation in Norm and Stress. Cell Tiss. Biol. 2020, 14, 178–189. [Google Scholar] [CrossRef]
- Nikitina, E.A.; Zalomaeva, E.S.; Medvedeva, A.V.; Zhuravlev, A.V.; Savvateeva-Popova, E.V. Role of LIM-kinase 1 in memory process. Usp. Fiziol. Nauk. 2023, 54, 36–56. (In Russian) [Google Scholar] [CrossRef]
- Zakharov, G.A. Molecular Genetic Studies of the Role of the Components of the Actin Remodeling Signal Cascade in the Genesis of Behavioral Disorders in Drosophila Melanogaster. Ph.D. Thesis, Pavlov Institute of Physiology RAS, St. Petersburg, Russia, 2012. (In Russian). [Google Scholar]
- Nikitina, E.A.; Medvedeva, A.V.; Gerasimenko, M.S.; Pronikov, V.S.; Surma, S.V.; Shchegolev, B.F.; Savvateeva-Popova, E.V. A Weakened Geomagnetic Field: Effects on Genomic Transcriptiln Activity, Learning, and Memory in Drosophila Melanogaster. Neurosci. Behav. Physiol. 2018, 48, 796–803. [Google Scholar] [CrossRef]
- Savvateeva-Popova, E.V.; Zhuravlev, A.V.; Brázda, V.; Zakharov, G.A.; Kaminskaya, A.N.; Medvedeva, A.V.; Nikitina, E.A.; Tokmatcheva, E.V.; Dolgaya, J.F.; Kulikova, D.A.; et al. Drosophila model for the analysis of genesis of LIM-kinase 1-dependent Williams-Beuren syndrome cognitive phenotypes: INDELs, transposable elements of the Tc1/Mariner superfamily and MicroRNAs. Front. Genet. 2017, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Kaminker, J.S.; Bergman, C.M.; Kronmiller, B.; Carlson, J.; Svirskas, R.; Patel, S.; Frise, E.; Wheeler, D.A.; Lewis, S.E.; Rubin, G.M.; et al. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 2002, 3, RESEARCH0084. [Google Scholar] [CrossRef]
- Öztürk-Çolak, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Goutte-Gattat, D.; Jenkins, V.K.; Matthews, B.B.; Millburn, G.; Dos Santos, G.; Tabone, C.J.; et al. FlyBase: Updates to the Drosophila genes and genomes database. Genetics 2024, 227, iyad211. [Google Scholar] [CrossRef]
- Zhuravlev, A.V.; Vetrovoy, O.V.; Zalomaeva, E.S.; Egozova, E.S.; Nikitina, E.A.; Savvateeva-Popova, E.V. Overexpression of the limk1 Gene in Drosophila melanogaster Can Lead to Suppression of Courtship Memory in Males. Biochemistry 2024, 89, 393–406. [Google Scholar] [CrossRef]
- Kim, T.W.; Kwon, Y.J.; Kim, J.M.; Song, Y.H.; Kim, S.N.; Kim, Y.J. MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc. Natl. Acad. Sci. USA 2004, 101, 12153–12158. [Google Scholar] [CrossRef]
- Chu, Y.; Gómez Rosso, L.; Huang, P.; Wang, Z.; Xu, Y.; Yao, X.; Bao, M.; Yan, J.; Song, H.; Wang, G. Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity. Cell Res 2014, 24, 1250–1265. [Google Scholar] [CrossRef] [PubMed]
- Zattoni, M.; Mearelli, M.; Vanni, S.; Colini Baldeschi, A.; Tran, T.H.; Ferracin, C.; Catania, M.; Moda, F.; Di Fede, G.; Giaccone, G.; et al. Serpin Signatures in Prion and Alzheimer’s Diseases. Mol. Neurobiol. 2022, 59, 3778–3799. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Mallon, E.B.; Alghamdi, A.; Holdbrook, R.T.; Rosato, E. Immune stimulation reduces sleep and memory ability in Drosophila melanogaster. PeerJ 2014, 2, e434. [Google Scholar] [CrossRef] [PubMed]
- Damulewicz, M.; Mazzotta, G.M. One Actor, Multiple Roles: The Performances of Cryptochrome in Drosophila. Front. Physiol. 2020, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, M.; Buchert, S.N.; Coleman, V.; McLaughlin, M.; Nguyen, A.; Sitaraman, D. Compartment specific regulation of sleep by mushroom body requires GABA and dopaminergic signaling. Sci. Rep. 2021, 11, 20067. [Google Scholar] [CrossRef] [PubMed]
- Flyer-Adams, J.G.; Rivera-Rodriguez, E.J.; Yu, J.; Mardovin, J.D.; Reed, M.L.; Griffith, L.C. Regulation of Olfactory Associative Memory by the Circadian Clock Output Signal Pigment-Dispersing Factor (PDF). J. Neurosci. 2020, 40, 9066–9077. [Google Scholar] [CrossRef]
- Ekengren, S.; Hultmark, D. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem. Biophys. Res. Commun. 2001, 284, 998–1003. [Google Scholar] [CrossRef]
- Gruntenko, N.E.; Karpova, E.K.; Babenko, V.N.; Vasiliev, G.V.; Andreenkova, O.V.; Bobrovskikh, M.A.; Menshanov, P.N.; Babenko, R.O.; Rauschenbach, I.Y. Fitness Analysis and Transcriptome Profiling Following Repeated Mild Heat Stress of Varying Frequency in Drosophila melanogaster Females. Biology 2021, 10, 1323. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Nielsen, M.M.; Kruhøffer, M.; Justesen, J.; Loeschcke, V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones 2005, 10, 312–328. [Google Scholar] [CrossRef]
- Ding, H.M.; Wang, X.; Mo, W.C.; Qin, L.L.; Wong, S.; Fu, J.P.; Tan, Y.; Liu, Y.; He, R.Q.; Hua, Q. Hypomagnetic fields cause anxiety in adult male mice. Bioelectromagnetics 2019, 40, 27–32. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, L.; Zhan, A.; Wang, M.; Tian, L.; Guo, W.; Pan, Y. Long-term exposure to a hypomagnetic field attenuates adult hippocampal neurogenesis and cognition. Nat. Commun. 2021, 12, 1174. [Google Scholar] [CrossRef]
- Mo, W.; Liu, Y.; Bartlett, P.F.; He, R. Transcriptome profile of human neuroblastoma cells in the hypomagnetic field. Sci. China Life Sci. 2014, 57, 448–461. [Google Scholar] [CrossRef]
- Yiwen, W.; Xiaohan, T.; Chunfeng, Z.; Xiaoyu, Y.; Yaodong, M.; Huanhuan, Q. Genetics of metallothioneins in Drosophila melanogaster. Chemosphere 2022, 288, 132562. [Google Scholar] [CrossRef] [PubMed]
- Günther, V.; Lindert, U.; Schaffner, W. The taste of heavy metals: Gene regulation by MTF-1. Biochim. Biophys. Acta 2012, 1823, 1416–1425. [Google Scholar] [CrossRef]
- Buraczynska, M.; Stec, A.; Filipczak, A.; Ksiazek, A. Association between functional variant of inflammatory system gene (PSMA6) and end-stage kidney disease. Int. Urol. Nephrol. 2016, 48, 2083–2087. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xu, F.; Teschendorff, A.E.; Zhao, Y.; Yao, L.; Li, J.; He, Y. Insights into the role of long non-coding RNAs in DNA methylation mediated transcriptional regulation. Front. Mol. Biosci. 2022, 9, 1067406. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Guh, C.Y.; Hsieh, Y.H.; Chu, H.P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 44. [Google Scholar] [CrossRef]
- Mérel, V.; Boulesteix, M.; Fablet, M.; Vieira, C. Transposable elements in Drosophila. Mob. DNA 2020, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Biémont, C.; Aouar, A.; Arnault, C. Genome reshuffling of the copia element in an inbred line of Drosophila melanogaster. Nature 1987, 329, 742–744. [Google Scholar] [CrossRef]
- Lipatov, M.; Lenkov, K.; Petrov, D.A.; Bergman, C.M. Paucity of chimeric gene-transposable element transcripts in the Drosophila melanogaster genome. BMC Biol. 2005, 3, 24. [Google Scholar] [CrossRef]
- Bergman, C.M.; Quesneville, H.; Anxolabéhère, D.; Ashburner, M. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol 2006, 7, R112. [Google Scholar] [CrossRef]
- Rice, N.; Ho, S.; Weng, Z.; Theurkauf, W.E. Rapid disassembly and Piwi-independent reassembly of Drosophila piRNA cluster heterochromatin following acute heat shock. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kamyshev, N.G.; Iliadi, K.G.; Bragina, J.V. Drosophila conditioned courtship: Two ways of testing memory. Learn. Mem. 1999, 6, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.; Tokmatcheva, E.; Savateeva-Popova, E. Heat shock during the development of central structures of the Drosophila brain: Memory formation in the in the l(1)ts403 mutant of Drosophila melanogaster. Russ. J. Genet. 2003, 39, 33–40. (In Russian) [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 March 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. Available online: https://figshare.com/ndownloader/files/1421612 (accessed on 25 March 2024). [CrossRef]
- Samtools. Available online: https://github.com/samtools/ (accessed on 25 March 2024).
- Glotov, A.S.; Zelenkova, I.E.; Vashukova, E.S.; Shuvalova, A.R.; Zolotareva, A.D.; Polev, D.E.; Barbitoff, Y.A.; Glotov, O.S.; Sarana, A.M.; Shcherbak, S.G.; et al. RNA Sequencing of Whole Blood Defines the Signature of High Intensity Exercise at Altitude in Elite Speed Skaters. Genes 2022, 13, 574. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [PubMed]
- Ge, X. iDEP Web Application for RNA-Seq Data Analysis. In RNA Bioinformatics. Methods in Molecular Biology; Springer Protocols; Humana Press: New York, NY, YSA, 2021; Chapter 22; Volume 2284, p. 417. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021, 060012. [Google Scholar] [CrossRef]

| Strain | High | Moderate | Low | Modifier |
|---|---|---|---|---|
| CS all | All: 785; 4: 1(Tie); 3: 8(tun, CG9698, nonA CG18538, twin) | All: 15,766; 109: 19(Muc14A) | All: 23,023; 290: 1(shot) | All: 33,986; 2025: 1(dpr6) |
| agnts3 all | All: 656; 3: 13(CG18538; MED23, Tep5, Sdk, twin) | All: 14,467; 87: 1(sls) | All: 21,984; 249: 2(sls) | All: 34,052; 1885: 1(Ptp61F) |
| CS- specific | All: 418; 4: 1(Tie); 3: 1(CG9698); 2: 18(CG10183, CG13700, CG8136, CG9500, CR44091, CR45136, Pdfr, cpo, nonA, tmod) | All: 10,193; 66: 3(Strn-Mlck) | All: 16,341; 170: 1(shot) | All: 32,791; 1254: 1(dpr6) |
| agnts3- specific | All: 289; 3: 2(MED23, Tep5); 2: 5(CG43829, Spn42De, Ssl2, mthl3, rd0, Sdk, shakB) | All: 7,613; 69: 1(Msp300) | All: 13,205; 121: 1(sls) | All: 31,626; 875: 1(bru1) |
| CS/agnts3 common | All: 379; 3: 2(CG18538, twin); 2: 12(AttC, CG14692, CG31268, CG43092, CG4580, Cp1, Cyp4p1, Cyp6a14, Ugt86D, inaD, tun) | All: 10,562; 66: 1(CG31817) | All: 18,011: 128: 1(sls) | All: 32,723; 1240: 1(sick) |
| Gene | Genome Positions | Mutation Effect | Gene Functions |
|---|---|---|---|
| CS-specific genes | |||
| Tie | 3L:4510698..4532145 | start loss/frameshift | Predicted: cell survival and migration |
| CG9698 | 3R:30514777..30516980 | stop lost | Unknown |
| CG10183 | 3R:23596561..23599034 | stop gained/splicing defect | Unknown |
| CG13700 | 3L:18298742..18301260 | stop gained/ splicing defect | Unknown |
| CG8136 | 3R:8721371..8723051 | stop gained/ splicing defect | Unknown |
| CG9500 | 2L:6357005..6358318 | frameshift/ splicing defect | Unknown |
| CR44091 | 3R:31680596..31681019 | splicing defect | Unknown |
| CR45136 | 2R:10853546..10854495 | splicing defect | Unknown |
| Pdfr | X:2552206..2578640 | frameshift | Circadian processes, development of the flight motor system, regulation of mating |
| cpo | 3R:17919832..18018892 | frameshift/ splicing defect | Synaptic transmission, climate adaptation, olfaction |
| nonA | X:16361569..16371547 | stop gained/frameshift | Visual behavior |
| tmod | 3R:30532607..30578905 | splicing defect | Notch signaling regulation, actin filament organization |
| agnts3-specific genes | |||
| MED23 | 2R:22888928..222894425 | stop gained/ splicing defect | RNA polymerase II coactivator, transcription response to heat shock. The human ortholog(s) are involved in autosomal recessive intellectual developmental disorder 18. |
| Tep5 | 2L:19558916..19563049 | frameshift | Unknown (pseudogene) |
| CG43829 | 2R:15193996..15194505 | frameshift | Unknown |
| Spn42De | 2R:6885552..6888262 | start lost/ splicing defect | Predicted: negative regulation of proteolysis. The human ortholog(s) are involved in Alzheimer’s disease and familial encephalopathy. |
| Ssl2 | 3R:29030951..29032642 | frameshift | Predicted: regulation of biosynthetic processes. |
| mthl3 | 2R:17448533..17452598 | splicing defect | Predicted: G protein-coupled receptor signaling pathway, determination of adult lifespan |
| rdo | 2L:18012380..18070246 | frameshift/ stop gained | Ocelli development |
| sdk | X:686834..749496 | stop gained | Actin branching, epithelial remodeling, synapse formation |
| shakB | X:20761071..20927050 | frameshift/ splicing defect | Structural component of the gap junctions at electrical synapses |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuravlev, A.V.; Polev, D.E.; Medvedeva, A.V.; Savvateeva-Popova, E.V. Whole-Genome and Poly(A)+Transcriptome Analysis of the Drosophila Mutant agnts3 with Cognitive Dysfunctions. Int. J. Mol. Sci. 2024, 25, 9891. https://doi.org/10.3390/ijms25189891
Zhuravlev AV, Polev DE, Medvedeva AV, Savvateeva-Popova EV. Whole-Genome and Poly(A)+Transcriptome Analysis of the Drosophila Mutant agnts3 with Cognitive Dysfunctions. International Journal of Molecular Sciences. 2024; 25(18):9891. https://doi.org/10.3390/ijms25189891
Chicago/Turabian StyleZhuravlev, Aleksandr V., Dmitrii E. Polev, Anna V. Medvedeva, and Elena V. Savvateeva-Popova. 2024. "Whole-Genome and Poly(A)+Transcriptome Analysis of the Drosophila Mutant agnts3 with Cognitive Dysfunctions" International Journal of Molecular Sciences 25, no. 18: 9891. https://doi.org/10.3390/ijms25189891






