Insights into the Role of microRNAs as Clinical Tools for Diagnosis, Prognosis, and as Therapeutic Targets in Alzheimer’s Disease
Abstract
:1. Introduction
2. miRNAs: Biogenesis and Function
3. MiRNA in AD Pathophysiology and as a Therapeutic Target
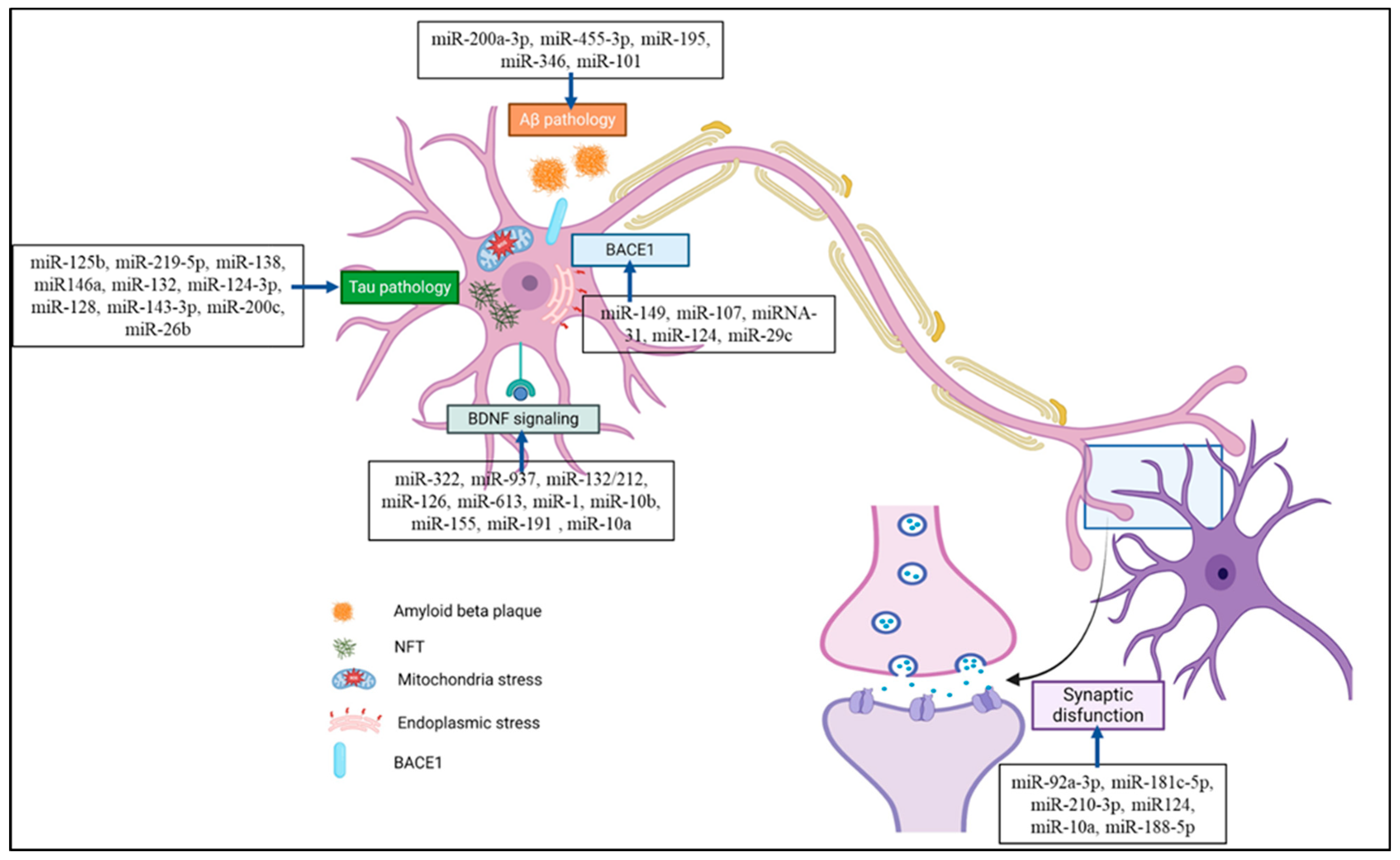
3.1. MiRNAs in Synapse Formation
3.2. MiRNAs and Brain-Derived Neurotrophic Factor (BDNF) Signaling
3.3. miRNAs and Aβ Protein
3.4. miRNAs and Tau Protein
4. miRNA as an Alzheimer’s Disease Biomarker
5. Challenges and Future Prospective
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lamptey, R.N.L.; Chaulagain, B.; Trivedi, R.; Gothwal, A.; Layek, B.; Singh, J. A Review of the Common Neurodegenerative Disorders: Current Therapeutic Approaches and the Potential Role of Nanotherapeutics. Int. J. Mol. Sci. 2022, 23, 1851. [Google Scholar] [CrossRef] [PubMed]
- Van Schependom, J.; D’haeseleer, M. Advances in Neurodegenerative Diseases. J. Clin. Med. 2023, 12, 1709. [Google Scholar] [CrossRef] [PubMed]
- Rammes, G. Molecular Mechanism of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 16837. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef]
- D’Cruz, M.; Banerjee, D. The person is not the disease–Revisiting Alzheimer’s dementia after 120 years. J. Geriatr. Ment. Health 2021, 8, 136. [Google Scholar] [CrossRef]
- Janoutová, J.; Kovalová, M.; Machaczka, O.; Ambroz, P.; Zatloukalová, A.; Němček, K.; Janout, V. Risk Factors for Alzheimer’s Disease: An Epidemiological Study. Curr. Alzheimer Res. 2021, 18, 372–379. [Google Scholar] [CrossRef]
- Twarowski, B.; Herbet, M. Inflammatory Processes in Alzheimer’s Disease—Pathomechanism, Diagnosis and Treatment: A Review. Int. J. Mol. Sci. 2023, 24, 6518. [Google Scholar] [CrossRef]
- Lee, J. Mild Cognitive Impairment in Relation to Alzheimer’s Disease: An Investigation of Principles, Classifications, Ethics, and Problems. Neuroethics 2023, 16, 16. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological Alterations in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Li, R.; Sterling, K.; Song, W. Amyloid β-based therapy for Alzheimer’s disease: Challenges, successes and future. Signal Transduct. Target. Ther. 2023, 8, 248. [Google Scholar] [CrossRef]
- Tan, J.Z.A.; Gleeson, P.A. The role of membrane trafficking in the processing of amyloid precursor protein and production of amyloid peptides in Alzheimer’s disease. Biochim. Biophys. Acta (BBA)-Biomembr. 2019, 1861, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, Y. Tau and neuroinflammation in Alzheimer’s disease: Interplay mechanisms and clinical translation. J. Neuroinflamm. 2023, 20, 165. [Google Scholar] [CrossRef] [PubMed]
- Barbier, P.; Zejneli, O.; Martinho, M.; Lasorsa, A.; Belle, V.; Smet-Nocca, C.; Tsvetkov, P.O.; Devred, F.; Landrieu, I. Role of tau as a microtubule-associated protein: Structural and functional aspects. Front. Aging Neurosci. 2019, 10, 474055. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, R.; Baglietto-Vargas, D.; Laferla, F.M. The Role of Tau in Alzheimer’s Disease and Related Disorders. CNS Neurosci. Ther. 2011, 17, 514. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.R.; Barbosa, D.J.; Remião, F.; Silva, R. Alzheimer’s disease: Insights and new prospects in disease pathophysiology, biomarkers and disease-modifying drugs. Biochem. Pharmacol. 2023, 211, 115522. [Google Scholar] [CrossRef]
- Silvestrelli, G.; Lanari, A.; Parnetti, L.; Tomassoni, D.; Amenta, F. Treatment of Alzheimer’s disease: From pharmacology to a better understanding of disease pathophysiology. Mech. Ageing Dev. 2006, 127, 148–157. [Google Scholar] [CrossRef]
- Cummings, J.; Zhou, Y.; Lee, G.; Zhong, K.; Fonseca, J.; Cheng, F. Alzheimer’s disease drug development pipeline: 2023. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2023, 9, e12385. [Google Scholar] [CrossRef]
- Vann Jones, S.A.; O’Kelly, A. Psychedelics as a Treatment for Alzheimer’s Disease Dementia. Front. Synaptic Neurosci. 2020, 12, 559980. [Google Scholar] [CrossRef]
- Satoh, J.I.; Kino, Y.; Niida, S. MicroRNA-Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer’s Disease from Public Data. Biomark Insights 2015, 10, 21. [Google Scholar] [CrossRef]
- Alawode, D.O.T.; Fox, N.C.; Zetterberg, H.; Heslegrave, A.J. Alzheimer’s Disease Biomarkers Revisited From the Amyloid Cascade Hypothesis Standpoint. Front. Neurosci. 2022, 16, 837390. [Google Scholar] [CrossRef]
- Wu, X.; Xia, P.; Yang, L.; Lu, C.; Lu, Z. The roles of long non-coding RNAs in Alzheimer’s disease diagnosis, treatment, and their involvement in Alzheimer’s disease immune responses. Non-Coding RNA Res. 2024, 9, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s Disease. Wiley Interdiscip. Rev. RNA 2018, 9, e1463. [Google Scholar] [CrossRef] [PubMed]
- Shobeiri, P.; Alilou, S.; Jaberinezhad, M.; Zare, F.; Karimi, N.; Maleki, S.; Teixeira, A.L.; Perry, G.; Rezaei, N. Circulating long non-coding RNAs as novel diagnostic biomarkers for Alzheimer’s disease (AD): A systematic review and meta-analysis. PLoS ONE 2023, 18, e0281784. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, S.; Bramanti, P.; Mazzon, E. Role of miRNAs in alzheimer’s disease and possible fields of application. Int. J. Mol. Sci. 2019, 20, 3979. [Google Scholar] [CrossRef] [PubMed]
- Tregub, P.P.; Ibrahimli, I.; Averchuk, A.S.; Salmina, A.B.; Litvitskiy, P.F.; Manasova, Z.S.; Popova, I.A. The Role of microRNAs in Epigenetic Regulation of Signaling Pathways in Neurological Pathologies. Int. J. Mol. Sci. 2023, 24, 12899. [Google Scholar] [CrossRef]
- Wang, I.F.; Ho, P.C.; Tsai, K.J. MicroRNAs in Learning and Memory and Their Impact on Alzheimer’s Disease. Biomedicines 2022, 10, 1856. [Google Scholar] [CrossRef]
- Saugstad, J.A.; Lusardi, T.A.; Van Keuren-Jensen, K.R.; Phillips, J.I.; Lind, B.; Harrington, C.A.; McFarland, T.J.; Courtright, A.L.; Reiman, R.A.; Yeri, A.S.; et al. Analysis of extracellular RNA in cerebrospinal fluid. J. Extracell. Vesicles 2017, 6, 1317577. [Google Scholar] [CrossRef]
- Lusardi, T.A.; Phillips, J.I.; Wiedrick, J.T.; Harrington, C.A.; Lind, B.; Lapidus, J.A.; Quinn, J.F.; Saugstad, J.A. MicroRNAs in Human Cerebrospinal Fluid as Biomarkers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 55, 1223. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef]
- Ho, V.; Baker, J.R.; Willison, K.R.; Barnes, P.J.; Donnelly, L.E.; Klug, D.R. Single cell quantification of microRNA from small numbers of non-invasively sampled primary human cells. Commun. Biol. 2023, 6, 458. [Google Scholar] [CrossRef]
- Dong, L.H.; Sun, L.; Zhang, W.J.; Wang, X.Y.; Li, J.M. Reduced serum miR-202 may promote the progression of Alzheimer’s disease patients via targeting amyloid precursor protein. Kaohsiung J. Med. Sci. 2021, 37, 730–738. [Google Scholar] [CrossRef]
- Aamri, M.E.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.N.; Mutharasan, R. Biosensor-based microRNA detection: Techniques, design, performance, and challenges. Analyst 2014, 139, 1576–1588. [Google Scholar] [CrossRef] [PubMed]
- Pishbin, E.; Sadri, F.; Dehghan, A.; Kiani, M.J.; Hashemi, N.; Zare, I.; Mousavi, P.; Rahi, A. Recent advances in isolation and detection of exosomal microRNAs related to Alzheimer’s disease. Environ. Res. 2023, 227, 115705. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Zhao, B.; Zhao, J.; Li, S. Potential Roles of Exosomal MicroRNAs as Diagnostic Biomarkers and Therapeutic Application in Alzheimer’s Disease. Neural Plast. 2017, 2017, 7027380. [Google Scholar] [CrossRef] [PubMed]
- Abdelmaksoud, N.M.; Sallam, A.A.M.; Abulsoud, A.I.; El-Dakroury, W.A.; Abdel Mageed, S.S.; AL-Noshokaty, T.M.; Elrebehy, M.A.; Elshaer, S.S.; Mahmoud, N.A.; Fathi, D.; et al. Unraveling the role of miRNAs in the diagnosis, progression, and therapeutic intervention of Alzheimer’s disease. Pathol. Res. Pract. 2024, 253, 155007. [Google Scholar] [CrossRef]
- Pan, J.; Wang, R.; Shang, F.; Ma, R.; Rong, Y.; Zhang, Y. Functional Micropeptides Encoded by Long Non-Coding RNAs: A Comprehensive Review. Front. Mol. Biosci. 2022, 9, 817517. [Google Scholar] [CrossRef]
- Ayers, D.; Scerri, C. Non-coding RNA influences in dementia. Non-Coding RNA Res. 2018, 3, 188–194. [Google Scholar] [CrossRef]
- Bahlakeh, G.; Gorji, A.; Soltani, H.; Ghadiri, T. MicroRNA alterations in neuropathologic cognitive disorders with an emphasis on dementia: Lessons from animal models. J. Cell. Physiol. 2021, 236, 806–823. [Google Scholar] [CrossRef]
- Yuan, M.; Bi, X. Therapeutic and Diagnostic Potential of microRNAs in Vascular Cognitive Impairment. J. Mol. Neurosci. 2020, 70, 1619–1628. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018, 9, 388354. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Kumar Patel, S.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446. [Google Scholar] [CrossRef] [PubMed]
- Keighron, C.N.; Avazzadeh, S.; Goljanek-Whysall, K.; McDonagh, B.; Howard, L.; Ritter, T.; Quinlan, L.R. Extracellular Vesicles, Cell-Penetrating Peptides and miRNAs as Future Novel Therapeutic Interventions for Parkinson’s and Alzheimer’s Disease. Biomedicines 2023, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hefu, Z.; Li, B.; Lifang, W.; Zhijie, S.; Zhou, L.; Deng, Y.; Zhili, L.; Ding, J.; Li, T.; et al. Plasma Extracellular Vesicle MicroRNA Analysis of Alzheimer’s Disease Reveals Dysfunction of a Neural Correlation Network. Research 2023, 6, 0114. [Google Scholar] [CrossRef]
- Hutchison, E.R.; Okun, E.; Mattson, M.P. The Therapeutic Potential of microRNAs in Nervous System Damage, Degeneration and Repair. Neuromol. Med. 2009, 11, 153. [Google Scholar] [CrossRef]
- Morris, G.; O’Brien, D.; Henshall, D.C. Opportunities and challenges for microRNA-targeting therapeutics for epilepsy. Trends Pharmacol. Sci. 2021, 42, 605–616. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Hussein, M.; Magdy, R. MicroRNAs in central nervous system disorders: Current advances in pathogenesis and treatment. Egypt. J. Neurol. Psychiatry Neurosurg. 2021, 57, 36. [Google Scholar] [CrossRef]
- Wang, G.; Shen, X.; Song, X.; Wang, N.; Wo, X.; Gao, Y. Protective mechanism of gold nanoparticles on human neural stem cells injured by β-amyloid protein through miR-21–5p/SOCS6 pathway. Neurotoxicology 2023, 95, 12–22. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, W.; Liu, Y.; Yang, D.; He, G.; Wang, Z. MicroRNA-511-3p regulates Aβ1–40 induced decreased cell viability and serves as a candidate biomarker in Alzheimer’s disease. Exp. Gerontol. 2023, 178, 112195. [Google Scholar] [CrossRef]
- Deng, L.J.; Wu, D.; Yang, X.F.; Li, T. miR-146a-5p Modulates Adult Hippocampal Neurogenesis Deficits Through Klf4/p-Stat3 Signaling in APP/PS1 Mice. Neuroscience 2023, 526, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Xie, Z.; Liao, D.; Li, Y.; Li, Z.; Zhao, Y.; Li, X.; Dong, M. Inhibiting microRNA-142–5p improves learning and memory in Alzheimer’s disease rats via targeted regulation of the PTPN1-mediated Akt pathway. Brain Res. Bull. 2023, 192, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, S.; Zhou, X.; Niu, X.; Chen, L.; Yang, Z.; Peng, D. Identification of hsa-miR-365b-5p’s role in Alzheimer’s disease: A combined analysis of miRNA and mRNA microarrays. Neurosci. Lett. 2022, 790, 136892. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, K.; Zhou, H.; Jiang, L.; Xie, B.; Wang, R.; Xia, W.; Yin, Y.; Gao, Z.; Cui, D.; et al. Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer’s Disease. Aging Cell 2020, 19, e13125. [Google Scholar] [CrossRef]
- Liang, X.; Fa, W.; Wang, N.; Peng, Y.; Liu, C.; Zhu, M.; Tian, N.; Wang, Y.; Han, X.; Qiu, C.; et al. Exosomal miR-532-5p induced by long-term exercise rescues blood–brain barrier function in 5XFAD mice via downregulation of EPHA4. Aging Cell 2023, 22, e13748. [Google Scholar] [CrossRef]
- Xue, B.; Qu, Y.; Zhang, X.; Xu, X.F. miRNA-126a-3p participates in hippocampal memory via alzheimer’s disease-related proteins. Cereb. Cortex 2022, 32, 4763–4781. [Google Scholar] [CrossRef]
- Dong, Z.; Gu, H.; Guo, Q.; Liu, X.; Li, F.; Liu, H.; Sun, L.; Ma, H.; Zhao, K. Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease. Cells 2022, 11, 3830. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Poon, C.H.; Zhang, Z.; Yue, M.; Chen, R.; Zhang, Y.; Hossain, F.; Pan, Y.; Zhao, J.; Rong, L.; et al. MicroRNA-128 suppresses tau phosphorylation and reduces amyloid-beta accumulation by inhibiting the expression of GSK3β, APPBP2, and mTOR in Alzheimer’s disease. CNS Neurosci. Ther. 2023, 29, 1848–1864. [Google Scholar] [CrossRef]
- H., Z.-Q.; Q., H.-H.; Z., C.; W., M.; Z., C.; L., Z.-Y.; H., J.; Wang, Y.-W. miR-146a aggravates cognitive impairment and Alzheimer disease-like pathology by triggering oxidative stress through MAPK signaling. Neurologia 2023, 38, 486–494. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, S.H.; Park, Y.; Park, J.; Lee, J.H.; Kim, B.C.; Song, W.K. miR-16-5p is upregulated by amyloid β deposition in Alzheimer’s disease models and induces neuronal cell apoptosis through direct targeting and suppression of BCL-2. Exp. Gerontol. 2020, 136, 110954. [Google Scholar] [CrossRef]
- Zheng, K.; Hu, F.; Zhou, Y.; Zhang, J.; Zheng, J.; Lai, C.; Xiong, W.; Cui, K.; Hu, Y.-Z.; Han, Z.-T.; et al. miR-135a-5p mediates memory and synaptic impairments via the Rock2/Adducin1 signaling pathway in a mouse model of Alzheimer’s disease. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zou, T.; Zhang, M.; Fan, W.; Zhang, T.; Jiang, Y.; Cai, Y.; Chen, F.; Chen, X.; Sun, Y.; et al. MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer’s disease. Theranostics 2021, 11, 4103–4121. [Google Scholar] [CrossRef] [PubMed]
- Cha, D.J.; Mengel, D.; Mustapic, M.; Liu, W.; Selkoe, D.J.; Kapogiannis, D.; Galasko, D.; Rissman, R.A.; Bennett, D.A.; Walsh, D.M. miR-212 and miR-132 Are Downregulated in Neurally Derived Plasma Exosomes of Alzheimer’s Patients. Front. Neurosci. 2019, 13, 1208. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tan, X.; Lu, Q.; Huang, K.; Tang, X.; He, Z. MiR-29c-3p May Promote the Progression of Alzheimer’s Disease through BACE1. J. Health Eng. 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Shao, Y.; Xu, T. A study on the neuroprotective effect of miR-206-3p on Alzheimer’s disease mice by regulating brain-derived neurotrophic factor. Ann. Transl. Med. 2022, 10, 85. [Google Scholar] [CrossRef]
- Mohammadipoor-Ghasemabad, L.; Sangtarash, M.H.; Sheibani, V.; Sasan, H.A.; Esmaeili-Mahani, S. Hippocampal microRNA-191a-5p Regulates BDNF Expression and Shows Correlation with Cognitive Impairment Induced by Paradoxical Sleep Deprivation. Neuroscience 2019, 414, 49–59. [Google Scholar] [CrossRef]
- Chen, W.; Wu, L.; Hu, Y.; Jiang, L.; Liang, N.; Chen, J.; Qin, H.; Tang, N. MicroRNA-107 Ameliorates Damage in a Cell Model of Alzheimer’s Disease by Mediating the FGF7/FGFR2/PI3K/Akt Pathway. J. Mol. Neurosci. 2020, 70, 1589–1597. [Google Scholar] [CrossRef]
- Wang, G.; An, T.; Lei, C.; Zhu, X.; Yang, L.; Zhang, L.; Zhang, R. Antidepressant-like effect of ginsenoside Rb1 on potentiating synaptic plasticity via the miR-134–mediated BDNF signaling pathway in a mouse model of chronic stress-induced depression. J. Ginseng. Res. 2022, 46, 376–386. [Google Scholar] [CrossRef]
- Baby, N.; Alagappan, N.; Dheen, S.T.; Sajikumar, S. MicroRNA-134-5p inhibition rescues long-term plasticity and synaptic tagging/capture in an Aβ(1-42)-induced model of Alzheimer’s disease. Aging Cell 2020, 19, e13046. [Google Scholar] [CrossRef]
- He, B.; Chen, W.; Zeng, J.; Tong, W.; Zheng, P. MicroRNA-326 decreases tau phosphorylation and neuron apoptosis through inhibition of the JNK signaling pathway by targeting VAV1 in Alzheimer’s disease. J. Cell. Physiol. 2020, 235, 480–493. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, J.; Fang, Q.; Shao, H.; Yang, D.; Sun, J.; Gao, L. MiRNA-199a-5p targets WNT2 to regulate depression through the CREB/BDNF signaling in hippocampal neuron. Brain Behav. 2021, 11, e02107. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, W.; Yi, Y.; Tong, Q. miR-219-5p inhibits tau phosphorylation by targeting TTBK1 and GSK-3β in Alzheimer’s disease. J. Cell. Biochem. 2019, 120, 9936–9946. [Google Scholar] [CrossRef] [PubMed]
- Barros-Viegas, A.T.; Carmona, V.; Ferreiro, E.; Guedes, J.; Cardoso, A.M.; Cunha, P.; de Almeida, L.P.; de Oliveira, C.R.; de Magalhães, J.P.; Peça, J. miRNA-31 Improves Cognition and Abolishes Amyloid-β Pathology by Targeting APP and BACE1 in an Animal Model of Alzheimer’s Disease. Mol. Ther. Nucleic Acids 2020, 19, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Morton, H.; Sawant, N.; Orlov, E.; Bunquin, L.E.; Pradeepkiran, J.A.; Alvir, R.; Reddy, P.H. MicroRNA-455-3p improves synaptic, cognitive functions and extends lifespan: Relevance to Alzheimer’s disease. Redox Biol. 2021, 48, 102182. [Google Scholar] [CrossRef]
- Chopra, N.; Wang, R.; Maloney, B.; Nho, K.; Beck, J.S.; Pourshafie, N.; Niculescu, A.; Saykin, A.J.; Rinaldi, C.; Counts, S.E.; et al. MicroRNA-298 reduces levels of human amyloid-β precursor protein (APP), β-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol. Psychiatry 2021, 26, 5636–5657. [Google Scholar] [CrossRef]
- Walgrave, H.; Balusu, S.; Snoeck, S.; Vanden Eynden, E.; Craessaerts, K.; Thrupp, N.; Wolfs, L.; Horré, K.; Fourne, Y.; Ronisz, A.; et al. Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell 2021, 28, 1805–1821. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, J.; Sun, X.; Ma, G.; Luo, G.; Miao, Z.; Song, L. miR-132 improves the cognitive function of rats with Alzheimer’s disease by inhibiting the MAPK1 signal pathway. Exp. Ther. Med. 2020, 20, 159. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Y.; Zhang, L. MicroRNA-132 promotes neurons cell apoptosis and activates Tau phosphorylation by targeting GTDC-1 in Alzheimer’s disease. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8523–8532. [Google Scholar] [CrossRef]
- He, C.; Su, C.; Zhang, W.; Wan, Q. MiR-485-5p alleviates Alzheimer’s disease progression by targeting PACS1. Transl. Neurosci. 2021, 12, 335–345. [Google Scholar] [CrossRef]
- Zhou, L.T.; Zhang, J.; Tan, L.; Huang, H.Z.; Zhou, Y.; Liu, Z.Q.; Lu, Y.; Zhu, L.-Q.; Yao, C.; Liu, D. Elevated Levels of miR-144-3p Induce Cholinergic Degeneration by Impairing the Maturation of NGF in Alzheimer’s Disease. Front. Cell. Dev. Biol. 2021, 9, 667412. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.V.; Mastroeni, D.; Audrain, M.; Fanutza, T.; Kim, S.H.; Blitzer, R.D.; Gandy, S.; Dudley, J.T.; Ehrlich, M.E. miR155 regulation of behavior, neuropathology, and cortical transcriptomics in Alzheimer’s disease. Acta Neuropathol. 2020, 140, 295–315. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Liu, J.; Yao, Z.; Xiao, Y.; Zhang, X.; Zhang, Y.; Xu, J. NF-κB-Induced Upregulation of miR-146a-5p Promoted Hippocampal Neuronal Oxidative Stress and Pyroptosis via TIGAR in a Model of Alzheimer’s Disease. Front. Cell. Neurosci. 2021, 15, 653881. [Google Scholar] [CrossRef]
- Fu, Y.; Hu, X.; Zheng, C.; Sun, G.; Xu, J.; Luo, S.; Cao, P. Intrahippocampal miR-342-3p inhibition reduces β-amyloid plaques and ameliorates learning and memory in Alzheimer’s disease. Metab. Brain Dis. 2019, 34, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.Y.; Zhou, Y.; Zhu, L.S.; Wang, X.; Pang, P.; Wang, D.Q.; Liuyang, Z.; Man, H.; Lu, Y.; Liu, D. Correcting abnormalities in miR-124/PTPN1 signaling rescues tau pathology in Alzheimer’s disease. J. Neurochem. 2020, 154, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Deng, J.; Chu, X.; Zhao, Y.; Guo, Y. Role of post-transcriptional control of calpain by miR-124-3p in the development of Alzheimer’s disease. J. Alzheimer’s Dis. 2019, 67, 571–581. [Google Scholar] [CrossRef]
- Zeng, L.; Jiang, H.; Ashraf, G.M.; Liu, J.; Wang, L.; Zhao, K.; Guo, Y. Implications of miR-148a-3p/p35/PTEN signaling in tau hyperphosphorylation and autoregulatory feedforward of Akt/CREB in Alzheimer’s disease. Mol. Ther. Nucleic Acids 2022, 27, 256–275. [Google Scholar] [CrossRef] [PubMed]
- Estfanous, S.; Daily, K.P.; Eltobgy, M.; Deems, N.P.; Anne, M.N.K.; Krause, K.; Badr, A.; Hamilton, K.; Carafice, C.; Hegazi, A.; et al. Elevated Expression of MiR-17 in Microglia of Alzheimer’s Disease Patients Abrogates Autophagy-Mediated Amyloid-β Degradation. Front. Immunol. 2021, 12, 705581. [Google Scholar] [CrossRef]
- Vergallo, A.; Lista, S.; Zhao, Y.; Lemercier, P.; Teipel, S.J.; Potier, M.C.; Habert, M.-O.; Dubois, B.; Lukiw, W.J.; Hampel, H. MiRNA-15b and miRNA-125b are associated with regional Aβ-PET and FDG-PET uptake in cognitively normal individuals with subjective memory complaints. Transl. Psychiatry 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Aloi, M.S.; Prater, K.E.; Sopher, B.; Davidson, S.; Jayadev, S.; Garden, G.A. The pro-inflammatory microRNA miR-155 influences fibrillar β-Amyloid1-42 catabolism by microglia. Glia 2021, 69, 1736–1748. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Peng, W.; Jia, Y.; Tang, J.; Li, W.; Zhang, J.H.; Yang, J. MicroRNA-101a Regulates Autophagy Phenomenon via the MAPK Pathway to Modulate Alzheimer’s-Associated Pathogenesis. Cell Transpl. 2019, 28, 1076–1084. [Google Scholar] [CrossRef]
- Sarkar, S.; Engler-Chiurazzi, E.B.; Cavendish, J.Z.; Povroznik, J.M.; Russell, A.E.; Quintana, D.D.; Mathers, P.; Simpkins, J. Over-expression of miR-34a induces rapid cognitive impairment and Alzheimer’s disease-like pathology. Brain Res. 2019, 1721, 146327. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Liu, Z.; Sun, G. Diagnostic value of miR-193a-3p in Alzheimer’s disease and miR-193a-3p attenuates amyloid-β induced neurotoxicity by targeting PTEN. Exp. Gerontol. 2020, 130, 110814. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chang, Q. MicroRNA miR-212 regulates PDCD4 to attenuate Aβ25–35-induced neurotoxicity via PI3K/AKT signaling pathway in Alzheimer’s disease. Biotechnol. Lett. 2020, 42, 1789–1797. [Google Scholar] [CrossRef]
- Ansari, A.; Maffioletti, E.; Milanesi, E.; Marizzoni, M.; Frisoni, G.B.; Blin, O.; Richardson, J.C.; Bordet, R.; Forloni, G.; Gennarelli, M.; et al. miR-146a and miR-181a are involved in the progression of mild cognitive impairment to Alzheimer’s disease. Neurobiol. Aging 2019, 82, 102–109. [Google Scholar] [CrossRef]
- Sun, C.; Jia, N.; Li, R.; Zhang, Z.; Zhong, Y.; Han, K. MiR-143-3p inhibition promotes neuronal survival in an Alzheimer’s disease cell model by targeting neuregulin-1. Folia Neuropathol. 2020, 58, 10–21. [Google Scholar] [CrossRef]
- Chu, T.; Shu, Y.; Qu, Y.; Gao, S.; Zhang, L. miR-26b inhibits total neurite outgrowth, promotes cells apoptosis and downregulates neprilysin in Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2018, 11, 3383. [Google Scholar]
- Nagaraj, S.; Want, A.; Laskowska-Kaszub, K.; Fesiuk, A.; Vaz, S.; Logarinho, E.; Wojda, U. Candidate alzheimer’s disease biomarker mir-483-5p lowers tau phosphorylation by direct erk1/2 repression. Int. J. Mol. Sci. 2021, 22, 3653. [Google Scholar] [CrossRef]
- Liu, Q.; Lei, C. Neuroprotective effects of miR-331-3p through improved cell viability and inflammatory marker expression: Correlation of serum miR-331-3p levels with diagnosis and severity of Alzheimer’s disease. Exp. Gerontol. 2021, 144, 111187. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, J. Clinical significance of miR-433 in the diagnosis of Alzheimer’s disease and its effect on Aβ-induced neurotoxicity by regulating JAK2. Exp. Gerontol. 2020, 141, 111080. [Google Scholar] [CrossRef]
- Hajjari, S.N.; Sadigh-Eteghad, S.; Shanehbandi, D.; Teimourian, S.; Shahbazi, A.; Mehdizadeh, M. MicroRNA-4422-5p as a Negative Regulator of Amyloidogenic Secretases: A Potential Biomarker for Alzheimer’s Disease. Neuroscience 2021, 463, 108–115. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Wang, Q.; Jiang, H.; Zeng, L.; Li, Z.; Liu, R. MicroRNA-200a-3p Mediates Neuroprotection in Alzheimer-Related Deficits and Attenuates Amyloid-Beta Overproduction and Tau Hyperphosphorylation via Coregulating BACE1 and PRKACB. Front. Pharmacol. 2019, 10, 806. [Google Scholar] [CrossRef] [PubMed]
- Ueta, M.; Nishigaki, H.; Komai, S.; Mizushima, K.; Tamagawa-Mineoka, R.; Naito, Y.; Katoh, N.; Sotozono, C.; Kinoshita, S. Positive regulation of innate immune response by miRNA-let-7a-5p. Front. Genet. 2023, 13, 1025539. [Google Scholar] [CrossRef] [PubMed]
- Sierksma, A.; Lu, A.; Salta, E.; Vanden Eynden, E.; Callaerts-Vegh, Z.; D’Hooge, R.; Blum, D.; Buée, L.; Fiers, M.; De Strooper, B. Deregulation of neuronal miRNAs induced by amyloid-β or TAU pathology. Mol. Neurodegener. 2018, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, H.; Jia, Y.; Lu, H.; Tan, Q.; Zhou, X. miR-149-5p inhibition reduces Alzheimer’s disease β-amyloid generation in 293/APPsw cells by upregulating H4K16ac via KAT8. Exp. Ther. Med. 2020, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Reddy, P.H. The role of synaptic microRNAs in Alzheimer’s disease. Biochim. Biophys. Mol. Basis Dis. 2020, 1866, 165937. [Google Scholar] [CrossRef]
- Siegel, G.; Saba, R.; Schratt, G. microRNAs in neurons: Manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 2011, 21, 491–497. [Google Scholar] [CrossRef]
- Griffiths, J.; Grant, S.G.N. Synapse pathology in Alzheimer’s disease. Semin. Cell. Dev. Biol. 2023, 139, 13–23. [Google Scholar] [CrossRef]
- Siedlecki-Wullich, D.; Miñano-Molina, A.J.; Rodríguez-álvarez, J. Micrornas as early biomarkers of alzheimer’s disease: A synaptic perspective. Cells 2021, 10, 113. [Google Scholar] [CrossRef]
- Sharma, B.; Torres, M.M.; Rodriguez, S.; Gangwani, L.; Kumar, S. MicroRNA-502-3p regulates GABAergic synapse function in hippocampal neurons. Neural Regen. Res. 2024, 19, 2698–2707. [Google Scholar] [CrossRef]
- Rodriguez-Ortiz, C.J.; Prieto, G.A.; Martini, A.C.; Forner, S.; Trujillo-Estrada, L.; LaFerla, F.M.; Baglietto-Vargas, D.; Cotman, C.W.; Kitazawa, M. miR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer’s disease. Aging Cell 2020, 19, e13118. [Google Scholar] [CrossRef]
- Ge, J.; Xue, Z.; Shu, S.; Yu, L.; Qin, R.; Tao, W.; Liu, P.; Dong, X.; Lan, Z.; Bao, X.; et al. MiR-431 attenuates synaptic plasticity and memory deficits in APPswe/PS1dE9 mice. JCI Insight 2023, 8, e166270. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Orlov, E.; Gowda, P.; Bose, C.; Swerdlow, R.H.; Lahiri, D.K.; Reddy, P.H. Synaptosome microRNAs regulate synapse functions in Alzheimer’s disease. Npj Genom. Med. 2022, 7, 1–15. [Google Scholar] [CrossRef]
- Eyileten, C.; Sharif, L.; Wicik, Z.; Jakubik, D.; Jarosz-Popek, J.; Soplinska, A.; Postula, M.; Czlonkowska, A.; Kaplon-Cieslicka, A.; Mirowska-Guzel, D. The Relation of the Brain-Derived Neurotrophic Factor with MicroRNAs in Neurodegenerative Diseases and Ischemic Stroke. Mol. Neurobiol. 2020, 58, 329–347. [Google Scholar] [CrossRef]
- You, H.J.; Park, J.H.; Pareja-Galeano, H.; Lucia, A.; Shin, J.I. Targeting MicroRNAs Involved in the BDNF Signaling Impairment in Neurodegenerative Diseases. NeuroMolecular Med. 2016, 18, 540–550. [Google Scholar] [CrossRef]
- Sun, C.; Liu, J.; Duan, F.; Cong, L.; Qi, X. The role of the microRNA regulatory network in Alzheimer’s disease: A bioinformatics analysis. Arch. Med. Sci. 2022, 18, 206–222. [Google Scholar] [CrossRef]
- Li, W.; Li, X.; Xin, X.; Kan, P.C.; Yan, Y. MicroRNA-613 regulates the expression of brain-derived neurotrophic factor in Alzheimer’s disease. Biosci. Trends 2016, 10, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qin, L.; Tang, B. MicroRNAs in Alzheimer’s disease. Front. Genet. 2019, 10, 153. [Google Scholar] [CrossRef]
- Zhou, Z.D.; Chan, C.H.S.; Ma, Q.H.; Xu, X.H.; Xiao, Z.C.; Tan, E.K. The roles of amyloid precursor protein (APP) in neurogenesis, implications to pathogenesis and therapy of Alzheimer disease (AD). Cell Adhes. Migr. 2011, 5, 280. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.C.; Wang, L.M.; Wang, M.; Song, B.; Tan, S.; Teng, J.F.; Duan, D.-X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res. Bull. 2012, 88, 596–601. [Google Scholar] [CrossRef]
- Long, J.M.; Maloney, B.; Rogers, J.T.; Lahiri, D.K. Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: Implications in Alzheimer’s disease. Mol. Psychiatry 2019, 24, 345–363. [Google Scholar] [CrossRef]
- Long, J.M.; Lahiri, D.K. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 2011, 404, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Tang, M. MicroRNAs as Possible Biomarkers for Diagnosis and Therapy of Alzheimer’s Disease by Regulating the Abnormal Expression of Genes Related to Tau. BIO Web Conf. 2023, 60, 01001. [Google Scholar] [CrossRef]
- Wang, L.; Shui, X.; Mei, Y.; Xia, Y.; Lan, G.; Hu, L.; Zhang, M.; Gan, C.-L.; Li, R.; Tian, Y.; et al. miR-143-3p Inhibits Aberrant Tau Phosphorylation and Amyloidogenic Processing of APP by Directly Targeting DAPK1 in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 7992. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Li, S.; Chen, Z.; Mushannen, T.; Gongala, S.; Wei, Z.; Balu, D.T.; Rabinovsky, R.; Cantlon, A.; Elkhal, A.; et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 2018, 136, 537–555. [Google Scholar] [CrossRef]
- Smith, P.Y.; Delay, C.; Girard, J.; Papon MA lie Planel, E.; Sergeant, N.; Buée, L.; Hébert, S.S. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 2011, 20, 4016–4024. [Google Scholar] [CrossRef] [PubMed]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. Micro RNA -125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef]
- Wang, G.; Huang, Y.; Wang, L.L.; Zhang, Y.F.; Xu, J.; Zhou, Y.; Lourenco, G.F.; Zhang, B.; Wang, Y.; Ren, R.-J.; et al. MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci. Rep. 2016, 6, 26697. [Google Scholar] [CrossRef] [PubMed]
- Walgrave, H.; Penning, A.; Tosoni, G.; Snoeck, S.; Davie, K.; Davis, E.; Wolfs, L.; Sierksma, A.; Mars, M.; Bu, T.; et al. microRNA-132 regulates gene expression programs involved in microglial homeostasis. iScience 2023, 26, 106829. [Google Scholar] [CrossRef]
- Wu, Q.; Ye, X.; Xiong, Y.; Zhu, H.; Miao, J.; Zhang, W.; Wan, J. The Protective Role of microRNA-200c in Alzheimer’s Disease Pathologies Is Induced by Beta Amyloid-Triggered Endoplasmic Reticulum Stress. Front. Mol. Neurosci. 2016, 9, 140. [Google Scholar] [CrossRef]
- Liu, H.; Chu, W.; Gong, L.; Gao, X.; Wang, W. MicroRNA-26b is upregulated in a double transgenic mouse model of Alzheimer’s disease and promotes the expression of amyloid-β by targeting insulin-like growth factor 1. Mol. Med. Rep. 2016, 13, 2809–2814. [Google Scholar] [CrossRef]
- Lauretti, E.; Dabrowski, K.; Praticò, D. The neurobiology of non-coding RNAs and Alzheimer’s disease pathogenesis: Pathways, mechanisms and translational opportunities. Ageing Res. Rev. 2021, 71, 101425. [Google Scholar] [CrossRef] [PubMed]
- Madadi, S.; Saidijam, M.; Yavari, B.; Soleimani, M. Downregulation of serum miR-106b: A potential biomarker for Alzheimer disease. Arch. Physiol. Biochem. 2022, 128, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Zhao, Y.; Lu, Y.; Wang, P.C. ABCA1-Labeled Exosomes in Serum Contain Higher MicroRNA-193b Levels in Alzheimer’s Disease. BioMed Res. Int. 2021, 2021, 1–10. [Google Scholar] [CrossRef]
- Li, Y.; Meng, S.; Di, W.; Xia, M.; Dong, L.; Zhao, Y.; Ling, S.; He, J.; Xue, X.; Chen, X.; et al. Amyloid-β protein and MicroRNA-384 in NCAM-Labeled exosomes from peripheral blood are potential diagnostic markers for Alzheimer’s disease. CNS Neurosci. Ther. 2022, 28, 1093. [Google Scholar] [CrossRef]
- Dakterzada, F.; David Benítez, I.; Targa, A.; Lladó, A.; Torres, G.; Romero, L.; de Gonzalo-Calvo, D.; Moncusí-Moix, A.; Tort-Merino, A.; Huerto, R.; et al. Reduced Levels of miR-342-5p in Plasma Are Associated With Worse Cognitive Evolution in Patients With Mild Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 705989. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Zhang, Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer’s disease. J. Clin. Lab. Anal. 2020, 34, e23006. [Google Scholar] [CrossRef]
- Taşdelen, E.; Kizil, E.T.Ö.; Tezcan, S.; Yalap, E.; Bįngöl, A.P.; Kutlay, N.Y. Determination of miR-373 and miR-204 levels in neuronal exosomes in Alzheimer’s disease. Turk. J. Med. Sci. 2022, 52, 1458–1467. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yu, J.T.; Liu, Q.Y.; Tan, M.S.; Zhang, W.; Hu, N.; Wang, Y.-L.; Sun, L.; Jiang, T.; Tan, L. Circulating miR-125b as a biomarker of Alzheimer’s disease. J. Neurol. Sci. 2014, 336, 52–56. [Google Scholar] [CrossRef]
- Souza, V.C.; Morais, G.S.; Henriques, A.D.; Machado-Silva, W.; Perez, D.I.V.; Brito, C.J.; Camargos, E.F.; Moraes, C.F.; Nóbrega, O.T. Whole-Blood Levels of MicroRNA-9 Are Decreased in Patients With Late-Onset Alzheimer Disease. Am. J. Alzheimer’s Dis. Other Dementiasr 2020, 35, 1533317520911573. [Google Scholar] [CrossRef]
- Liu, C.G.; Song, J.; Zhang, Y.Q.; Wang, P.C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014, 10, 2395–2400. [Google Scholar] [CrossRef]
- Leidinger, P.; Backes, C.; Deutscher, S.; Schmitt, K.; Mueller, S.C.; Frese, K.; Haas, J.; Ruprecht, K.; Paul, F.; Stähler, C.; et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013, 14, R78. [Google Scholar] [CrossRef] [PubMed]
- Poursaei, E.; Abolghasemi, M.; Bornehdeli, S.; Shanehbandi, D.; Asadi, M.; Sadeghzadeh, M.; Rahmanpour, D.; Sadeh, R.N. Evaluation of hsa-let-7d-5p, hsa-let-7g-5p and hsa-miR-15b-5p plasma levels in patients with Alzheimer’s disease. Psychiatr. Genet. 2022, 32, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, Q.; Yin, Y. miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 2019, 18, 2711–2718. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Gu, H.; Guo, Q.; Liang, S.; Xue, J.; Yao, F.; Liu, X.; Li, F.; Liu, H.; Sun, L.; et al. Profiling of Serum Exosome MiRNA Reveals the Potential of a MiRNA Panel as Diagnostic Biomarker for Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 3084–3094. [Google Scholar] [CrossRef]
- De Felice, B.; Montanino, C.; Oliva, M.; Bonavita, S.; Onofrio, V.; Di Coppola, C. MicroRNA Expression Signature in Mild Cognitive Impairment Due to Alzheimer’s Disease. Mol. Neurobiol. 2020, 57, 4408–4416. [Google Scholar] [CrossRef]
- Siedlecki-Wullich, D.; Català-Solsona, J.; Fábregas, C.; Hernández, I.; Clarimon, J.; Lleó, A.; Boada, M.; Saura, C.A.; Rodríguez-Álvarez, J.; Miñano-Molina, A.J. Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease. Alzheimer’s Res. Ther. 2019, 11, 1–11. [Google Scholar] [CrossRef]
- Zhang, M.; Han, W.; Xu, Y.; Li, D.; Xue, Q. Serum mir-128 serves as a potential diagnostic biomarker for Alzheimer’s disease. Neuropsychiatr. Dis. Treat. 2021, 17, 269–275. [Google Scholar] [CrossRef]
- Chia, S.Y.; Vipin, A.; Ng, K.P.; Tu, H.; Bommakanti, A.; Wang, B.Z.; Tan, Y.J.; Zailan, F.Z.; Ng, A.S.L.; Ling, S.-C.; et al. Upregulated Blood miR-150-5p in Alzheimer’s Disease Dementia Is Associated with Cognition, Cerebrospinal Fluid Amyloid-β, and Cerebral Atrophy. J. Alzheimer’s Dis. 2022, 88, 1567–1584. [Google Scholar] [CrossRef]
- Abuelezz, N.Z.; Nasr, F.E.; Abdel Aal, W.M.; Molokhia, T.; Zaky, A. Sera miR-34a, miR-29b and miR-181c as potential novel diagnostic biomarker panel for Alzheimers in the Egyptian population. Exp. Gerontol. 2022, 169, 111961. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, S.E.; Ko, Y.; Jeong, G.H.; Lee, K.H.; Lee, J.; Solmi, M.; Jacob, L.; Smith, L.; Stickley, A.; et al. Differential expression of MicroRNAs in Alzheimer’s disease: A systematic review and meta-analysis. Mol. Psychiatry 2022, 27, 2405–2413. [Google Scholar] [CrossRef]
- Takousis, P.; Sadlon, A.; Schulz, J.; Wohlers, I.; Dobricic, V.; Middleton, L.; Lill, C.M.; Perneczky, R.; Bertram, L. Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimer’s Dement. 2019, 15, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Cosín-Tomás, M.; Antonell, A.; Lladó, A.; Alcolea, D.; Fortea, J.; Ezquerra, M.; Martí, M.J.; Pallàs, M.; Sanchez-Valle, R.; Molinuevo, J.L.; et al. Plasma miR-34a-5p and miR-545-3p as Early Biomarkers of Alzheimer’s Disease: Potential and Limitations. Mol. Neurobiol. 2017, 54, 5550–5562. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, A.P.; Yin, X.; Reddy, P.H. Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2428–2440. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Lasuncion, I.; Atienza, M.; Sanchez-Espinosa, M.P.; Cantero, J.L. Aging-Related Changes in Cognition and Cortical Integrity are Associated with Serum Expression of Candidate MicroRNAs for Alzheimer Disease. Cereb. Cortex 2019, 29, 4426–4437. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.E.; Lim, C.S.; Kim, J.I.; Seo, D.; Chun, H.; Yu, N.K.; Lee, J.; Kang, S.J.; Ko, H.-G.; Choi, J.-H.; et al. The Brain-Enriched MicroRNA miR-9-3p Regulates Synaptic Plasticity and Memory. J. Neurosci. 2016, 36, 8541–8552. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Gaetani, S.; Sorgentoni, G.; Agarbati, S.; Laggetta, M.; Matacchione, G.; Gobbi, M.; Rossi, T.; Galeazzi, R.; Piccinini, G.; et al. Circulating Inflamma-miRs as Potential Biomarkers of Cognitive Impairment in Patients Affected by Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 647015. [Google Scholar] [CrossRef] [PubMed]
- Wiedrick, J.T.; Phillips, J.I.; Lusardi, T.A.; McFarland, T.J.; Lind, B.; Sandau, U.S.; Harrington, C.A.; Lapidus, J.A.; Galasko, D.R.; Quinn, J.F.; et al. Validation of MicroRNA Biomarkers for Alzheimer’s Disease in Human Cerebrospinal Fluid. J. Alzheimer’s Dis. 2019, 67, 875–891. [Google Scholar] [CrossRef]
- Kumar, P.; Dezso, Z.; MacKenzie, C.; Oestreicher, J.; Agoulnik, S.; Byrne, M.; Bernier, F.; Yanagimachi, M.; Aoshima, K.; Oda, Y. Circulating miRNA Biomarkers for Alzheimer’s Disease. PLoS ONE 2013, 8, e69807. [Google Scholar] [CrossRef]
- Wang, W.X.; Rajeev, B.W.; Stromberg, A.J.; Ren, N.; Tang, G.; Huang, Q.; Rigoutsos, I.; Nelson, P.T. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008, 28, 1213–1223. [Google Scholar] [CrossRef]
- Du, W.; Lei, C.; Dong, Y. Microrna-149 is downregulated in alzheimer’s disease and inhibits β-amyloid accumulation and ameliorates neuronal viability through targeting bace1. Genet. Mol. Biol. 2021, 44, 1–8. [Google Scholar] [CrossRef]
- Liu, C.G.; Wang, J.L.; Li, L.; Wang, P.C. MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer’s disease. Int. J. Mol. Med. 2014, 34, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Wang, J.L.; Li, L.; Xue, L.X.; Zhang, Y.Q.; Wang, P.C. MicroRNA-135a and -200b, potential Biomarkers for Alzheimer’s disease, regulate β secretase and amyloid precursor protein. Brain Res. 2014, 1583, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fan, M.; Zheng, Q.; Hao, S.; Yang, L.; Xia, Q.; Qi, C.; Ge, J. MicroRNAs in Alzheimer’s disease: Potential diagnostic markers and therapeutic targets. Biomed. Pharmacother. 2022, 148, 112681. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Zhao, L. Mechanism and Therapeutic Prospect of miRNAs in Neurodegenerative Diseases. Behav. Neurol. 2023, 2023, 1–24. [Google Scholar] [CrossRef]
- Xia, S.; Xu, C.; Liu, F.; Chen, G. Development of microRNA-based therapeutics for central nervous system diseases. Eur. J. Pharmacol. 2023, 956, 175956. [Google Scholar] [CrossRef]
- Batistela, M.S.; Josviak, N.D.; Sulzbach, C.D.; de Souza, R.L.R. An overview of circulating cell-free microRNAs as putative biomarkers in Alzheimer’s and Parkinson’s Diseases. Int. J. Neurosci. 2017, 127, 547–558. [Google Scholar] [CrossRef]

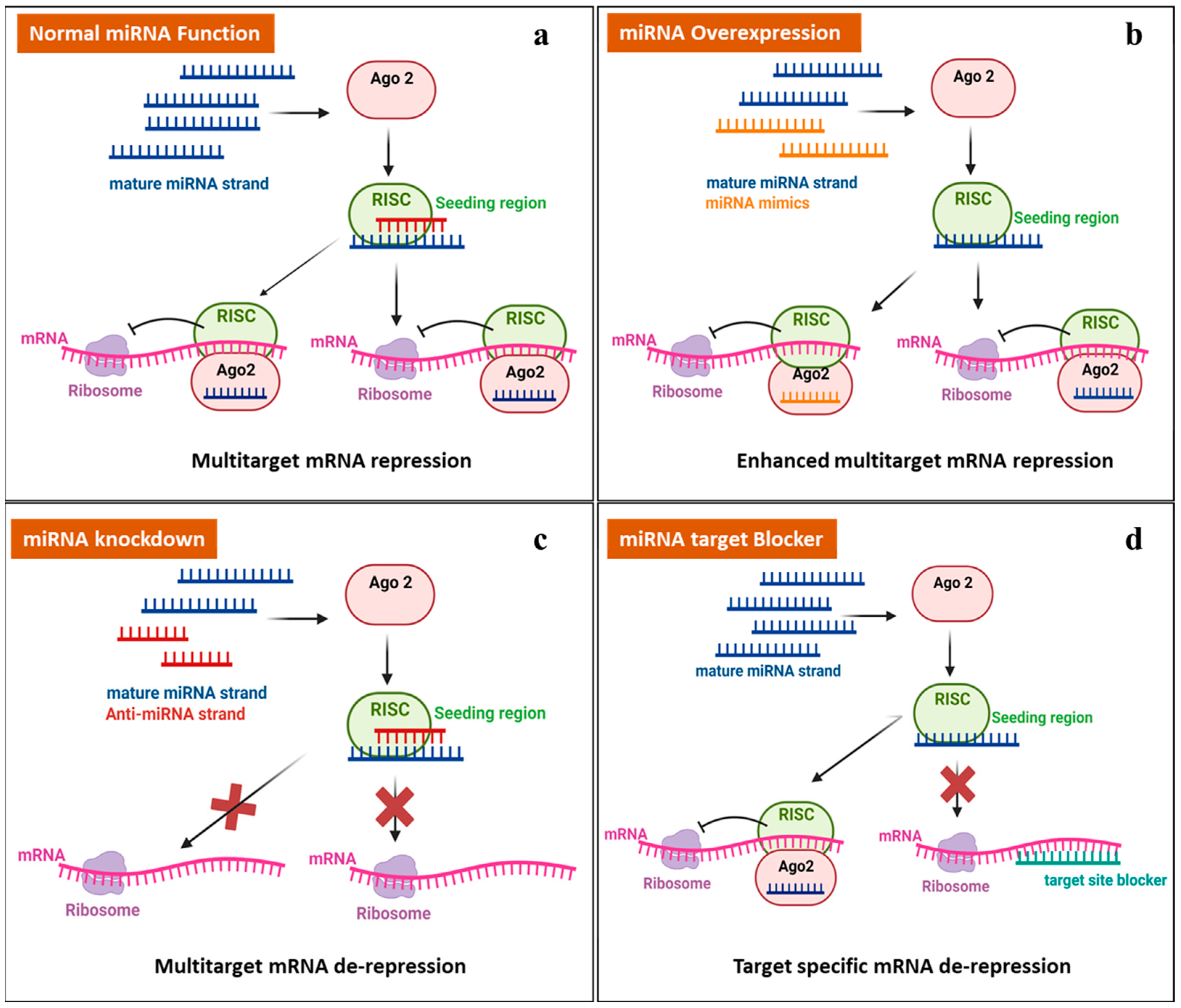
| miRNA | Experimental Model | Upregulated/ Downregulated | Outcome | Parameter Study and Technique Used | Concluding Remark | Reference |
|---|---|---|---|---|---|---|
| miR-21–5p | hNSCs | Increased | Reduced neuronal autophagy by focusing on the suppression of cytokine signaling and RAB11A | RT-qPCR and Western blot | Therapeutic target | [49] |
| miRNA-511-3p | AD patient serum samples | Downregulated | Aβ1-40-triggered cell viability and inflammation were regulated by miR-511-3p | RT-qPCR and Western blot | Candidate biomarker in AD | [50] |
| miR-146a-5p | APP/PS1 mice | Mir-146a-5p level increase in hippocampus | Modifies mature hippocampal neurogenesis abnormalities in APP/PS1 mice using Klf4/p-Stat3 signaling | RT-qPCR and Western blot | Therapeutic target | [51] |
| miRNA-142–5p | Aβ1–42 oligomer-injected rats | Overexpressed | Enhances memory and learning in rats through regulating the PTPN1-mediated Akt pathway | miRNA expression level (RT-qPCR and Western blot assessments) and behavior assay (Morris water maze (MWM) and NOR) | Therapeutic target | [52] |
| hsa-miR-365b-5p | Autopsy sample | Upregulated | Regulates neuroinflammation via Wnt and the oxidative stress pathway | Microarray bioinformatics analysis | Key therapeutic target | [53] |
| miR-34c | SAMP8 mice | Upregulated | Through the ROS-JNK-p53 pathway, increased miR-34c targets synaptotagmin 1 to mediate synaptic impairments in AD | Dual luciferase assay | Promising novel therapeutic target | [54] |
| miR-532-5p | 5XFAD mice | Downregulated in AD | Rescues BBB function via the downregulation of EPHA4 | miRNA expression level (RT-qPCR, Western blot, and in situ hybridization) and behavior assay (MWM test and eight-arm radial maze) | Potential therapeutic target | [55] |
| miRNA-126a-3p | Male APP/PS1 mice | Downregulated in AD | Participates in hippocampal memory via AD-related proteins | miRNA expression level (RT-qPCR and Western blot assessments) and behavior assay (MWM test and NOR) | Therapeutic target for AD memory disorders | [56] |
| miR-342-5p | APP mice (Exo-APP) | Upregulated/downregulated in AD | Ameliorates Aβ formation via targeting BACE1 in experimental mice | miRNA expression level (RT-qPCR, Western blot, and dual luciferase reporter assay) | Potential miRNAs in AD clinical therapy | [57] |
| miRNA-128 | 5XFAD mice | Downregulated | Decreases the accumulation of Aβ and lowers tau phosphorylation by blocking the expression of GSK3β, APPBP2, and mTOR | miRNA expression level (luciferase reporter assay) and behavior assay | Promising molecular target | [58] |
| miR-146a-5p | Aβ_1-42-injected mice; miR 146a-5p and Aβ1-42-treated mice; and miR-146a-5p inhibitor- and Aβ1-42-treated mice | NA | Exacerbates AD-like disease and cognitive impairment by activating oxidative stress via MAPK signaling | miRNA expression level (RT-qPCR and Western blot) and behavior assay (MWM test) | Molecular target | [59] |
| miR-16-5p | 5xFAD | Upregulated | miR-16-5p targets the B cell, and an anti-apoptotic factor | Microarray and RT-qPCR analysis | Therapeutic target | [60] |
| miR-135a-5p | APP/PS1 mice | Downregulated | Through the Rock2/Add1 signaling pathway, miR promotes memory impairments | Luciferase reporter assay, RT-qPCR, and behavior assay (MWM test and NOR) | Therapeutic strategy for AD | [61] |
| miR-146a | B6/JNju-Tg (APP/PS1) mice | Abnormal | Overexpression of the target gene negatively affects cognitive behaviour—learning and memory | miRNA expression level (RT-qPCR) and behavior assay (MWM test and NOR) | Therapeutic target | [62] |
| miR-212 and miR-132 | Alzheimer’s patient CSF and blood samples | Downregulated | Decreased levels of both miR-132 and miR-212 in CSF; however, miR-132 was also downregulated in blood | RT-qPCR | Potential theragnostic | [63] |
| MiR-29c-3p | Male SPF C57BL/6J mice | Upregulated | Since miR-29c-3p binds specifically to the 3′-UTR of BACE1, it adversely affects BACE1 levels, which in turn slows the advancement of AD | miRNA expression level (RT-qPCR and Western blot assessments) | Therapeutic target | [64] |
| miR-206-3p | SPF C57 mice | Upregulated | BDNF expression enriches neuronal morphology and recovers cognitive ability and memory | miRNA expression level (RT-qPCR and Western blot) and immunohistochemistry | Therapeutic target | [65] |
| miRNA-191a-5p | OVX rats | Upregulated | After miR-191 antagonization, miRNA knocks down miR-191a, BDNF expression is upregulated, and PSD-induced cognitive impairment in OVX rats is avoided | miRNA expression level (RT-qPCR) and behavior assay (MWM test) | Therapeutic target | [66] |
| miR-146a | APP/PS1 transgenic mice | NA | In transgenic mice, overexpression of the microglia-specific miR-146a prevented neuronal loss, attenuated neuroinflammation, decreased Aβ levels, and improved plaque formation | miRNA expression level (RT-qPCR), pro-inflammatory cytokines (ELISA), and behavior assay (MWM test and NOR) | Promising target | [62] |
| miR-107 | AD patients and Aβ-treated SH-SY5Y cells | Downregulated | miR-107 deactivated the FGFR2/PI3K/Akt pathway by inhibiting the expression FGF7 | RT-qPCR, luciferase reporter assay, and Western blot | Therapeutic target | [67] |
| miR-134 | Mice | NA | miR-134 could directly bind to BDNF 3′-UTR and was noticeably downregulated by Rb1 in the hippocampus of CUMS-exposed mice | miRNA expression level (RT-qPCR and Western blot) and behavior assay (OFT, TST, FST, and SPT) | Therapeutic target | [68] |
| miR-134-5p | Rats | Overexpressed | Post-transcriptional control of CREB-1 and BDNF | RT-qPCR and Western blot | Therapeutic target | [69] |
| miRNA-326 | AD mice | NA | Targeting VAV1 in AD, upregulating miR326 reduces tau phosphorylation and neuron death by targeting the JNK signaling cascade | miRNA expression level (RT-qPCR, microarray, and Western blot), quantification of Aβ1–40 and Aβ1–42 (ELISA), and behavior assay (MWM) | Therapeutic target | [70] |
| MiRNA-199a-5p | MDD patient | Upregulated | WNT2 can be targeted by miR-199a-5p to promote the onset of depression via controlling CREB/BDNF signaling | miRNA expression level (RT-qPCR and Western blot) and behavior assay (OFT, TST, FST, and SPT) | Therapeutic target | [71] |
| miR-219-5p | AD clinical sample and SH-SY5Y cells | Overexpressed in brain tissues | Inhibits tau phosphorylation by inhibiting TTBK1 and GSK-3β | miRNA expression level | Therapeutic target | [72] |
| miR-31 | 3xTg-AD animals and HT-22, HEK293, and SH-SY5Y cells | Downregulated | Enhances cognitive function and eliminates Aβ pathology by focusing on APP and BACE1 | miRNA expression level (RT-qPCR and Western blot) and behavior assay | Therapeutic target for AD | [73] |
| miR-34c | SAMP8 mice and HT-22 cells | Upregulated | Enhanced memory and synaptic impairments caused by miR-34c targeting SYT1 via the ROS-JNK-p53 pathway | miRNA expression level (RT-qPCR and Western blot) and behavior assay (MWM and NOR) | Promising novel therapeutic target | [54] |
| mir-455-3p | Transgenic mice with miR-455-3p and miR-455-3p KO mice | Downregulated | Improved neural development, lifespan, synapse, cognitive function, and mitochondrial and synaptic functioning in transgenic mice | mRNA expression level (RT-qPCR and Western blot) and behavior assay (MWM) | AD therapy | [74] |
| miR-298 | U373 and HeLa cells | NA | Soluble Aβ levels are decreased as a consequence of miR-298 negative regulation of APP and BACE1 expression | miRNA expression level (RT-qPCR and Western blot), quantification of Aβ1–40 and Aβ1–42 (ELISA), and behavior assay (MWM) | Suitable target for AD therapy | [75] |
| miR-132 | RenCell VM human NPCs, H9 cells, human hippocampal progenitor/stem cell line (HPC0A07/03C), and human hippocampi and serum | Downregulated | AD mice’s neurogenic and memory impairments are restored by overexpression in adult NSCs | miRNA level (reverse transcription and real-time PCR) and behavior assay | Therapeutic potential | [76] |
| miR-132 | Seventy SPF Sprague–Dawley rats treated with 20 μg Aβ25-35 | Upregulated | Rats with AD exhibited improved cognitive performance when miR-132 was able to suppress the development of iNOS in the hippocampus and oxidative stress by blocking MAPK1 expression | miRNA expression level (RT-qPCR and Western blot), luciferase reporter assay, and behavior assay (MWM) | Novel clinical target | [77] |
| miR-132 | Postmortem brain samples and neuron cells | Overexpressed | Control cell death and the GTDC-1/CDK-5/Tau phosphorylation signaling pathway | qRT-PCR, Western blot, and immunoprecipitation | Potential therapeutic target | [78] |
| miR-485-5p | APP/PS1 mice | Downregulated | Targets PACS1 in pericytes, promoting viability and preventing pericyte death | miRNA expression level (RT-qPCR and Western blot) and behavior assay (MWM and FCT) | Suitable target for AD therapeutic | [79] |
| miR-144-3p | APPswe/PS1dE9 mice | Upregulated | An antagonist of miR-144-3p into the hippocampi partially amended cholinergic degeneration and synaptic/memory impairments by raising tPA protein levels and modifying the proNGF/NGF ratio | miRNA expression level (RT-qPCR, Western blot) and behavior assay (Context–Place Memory Test) | Therapeutic target | [80] |
| miR155 | APP/PSEN1 mice | Downregulated | Linked to an increase in the incidence and size of Aβ plaques in mice aged 4 and 8 months | miRNA expression level (RT-qPCR, Western blot), quantification of Aβ1–40 and Aβ1–42 (ELISA), behavior assay (Barnes maze), and Field electrophysiology | Therapeutic target | [81] |
| miR-146a-5p | AD clinical sample and AD-HHNs cells | Upregulated | In hippocampus neuronal cells, NF-kB-induced overexpression of miR-146a-5p facilitated oxidative stress and pyroptosis via TIGAR | miRNA expression level (RT-qPCR and Western blot) and quantification of oxidative stress markers (ELISA) | Therapeutic target | [82] |
| miR-342-3p | 3xTg-AD mice and HT22 mouse hippocampal neuronal cells | Upregulated | Elevation of neuronal miR-342-3p in Aβ-challenged HT22 hippocampus neuronal cells is linked to an increase in c-Jun N-terminal kinase activation and, ultimately, neuronal death | miRNA expression level (RT-qPCR and Western blot), quantification of Aβ1–40 and Aβ1–42 (ELISA), and behavior assay (RAWM) | Therapeutic target | [83] |
| miR-124 | Neuroblastoma Neuro-2a cells and P301S transgenic mice | Overexpressed | Aberrant miR-124/PTPN1 signaling in the hippocampus resulted in AD-like tau pathology, which included several sites of hyperphosphorylation, insolubility, and somadendritic aggregation in addition to learning/memory deficits | miRNA expression level (RT-qPCR and Western blot) and behavior assay (fear conditioning test, Morris water maze, and elevated plus maze) | Therapeutic strategy | [84] |
| miR-124-3p | Human neural cell line, HCN-2, and APP/PS1-AD mice | Downregulated | Considerably improved AD-mouse behavior, and considerably decreased Aβ deposition. Findings point to the critical role that miR-124-3p’s post-transcriptional regulation of calpain plays in the onset of AD | RT-qPCR, Western blot, and social recognition test (SRT) | Therapeutic target | [85] |
| miR-148a-3p | Twenty-one AD patients, APP/PS1 mice, SAMP8 mice, and senescence-accelerated resistance mice | Downregulated | The direct targeting of the p35/CDK5 and PTEN/p38 MAPK pathways by the upregulation of miR-148a-3p has been observed to protect neuronal cells against Aβ-associated tau hyperphosphorylation | RNA sequencing, qRT-PCR, and Western blot | Potential therapeutic target | [86] |
| miR-17 | 5xFAD mice | Overexpressed | Impaired microglial autophagic clearance of Aβ is caused by decreased NBR1 expression and miR-17-mediated autophagy suppression, enhancing autophagy protein expression through the inhibition of increased miR-17-enhanced Aβ breakdown in vitro in microglia | miRNA expression level (RT-qPCR and Western blot) | Therapeutic interventions | [87] |
| miRNA-15b and miRNA-125b | AD clinical serum sample | Downregulated | A mechanistic connection between miRNA-125b and Aβ-independent neurotoxic pathways, as well as a possible protective anti-Aβ activity of miRNA-15b | miRNA expression level (RT-qPCR) and PET data acquisition | Therapeutic target | [88] |
| miR-155 | C57/BL6 wild-type | Up/downregulated | miR-155 influences the capacity of microglia to catabolize Aβ1-42 | miRNA expression level (RT-qPCR) | Therapeutic target | [89] |
| miRNA-101 | AD clinical blood sample, APPswe/PS1DE9 transgenic mice, and SH-SY5Y cells | Downregulated | After miRNA-101a was overexpressed in AD model cells, MAPK1 and beclin-1 were reduced, indicating that miRNA-101a may control the development of autophagy in AD | miRNA expression level (RT-qPCR and Western blot) and dual luciferase assay | Diagnosis | [90] |
| miR-34a | miR-34a+/transgenic mice | Overexpressed | Rapid onset of AD-like neuropathology and cognitive impairment is caused by the overexpression model | Behavior assay (Y-maze and T-maze) | Therapeutic target | [91] |
| miRNA-15b and miRNA-125b | AD clinical blood samples | Possible function of miRNAs-15b and -125b as putative miRNA biomarkers of AD pathogenesis | miRNA expression level (RT-qPCR) and PET data acquisition | Biomarkers of AD pathophysiology | [88] | |
| miR-193a-3p | Rat PC12 and SHSY5Y cell lines | Downregulated | miR-193a-3p reduces Aβ-induced neurotoxicity by targeting PTEN | Luciferase reporter assay | Novel biomarker | [92] |
| miR-212 | Plasma from AD patients and Aβ25–35-treated SH-SY5Y and IMR-32 cells | Downregulated | miR-212 controlled PDCD4 in Aβ25–35-treated SH-SY5Y and IMR-32 cells to regulate cell proliferation and death through the PI3K/AKT signaling pathway | qRT-PCR and Western blot | Therapeutic target | [93] |
| miR-146a and miR-181a | Forty-five patients with MCI | Upregulated | Elevated blood levels of miR-146a and miR-181a in individuals with MCI who experience a progressive loss in their cognitive function, along with a relationship between these changes and markers of the illness and AD risk factors | qRT-PCR and ApoE genotyping | Biomarker | [94] |
| miR-342-5p | Transgenic APP mice (Exo-APP) or C57BL/6 littermates (Exo-CTL), and individuals with HC (40) and AD (40) | Downregulated | Ameliorates Aβ formation via targeting BACE 1 in AD | miRNA expression level (RT-qPCR and Western blot) and dual luciferase assay | Biomarker | [57] |
| miR-143-3p | SH-SY5Y cells | Upregulated | miR-143-3p suppression targets NRG1, which enhances neuronal survival; the miR-143-3p/NRG1 axis is a possible target for therapy | miRNA expression level (RT-qPCR and Western blot) and dual luciferase assay | Biomarker | [95] |
| miR-26b | PC12 cells | Upregulated | In the PC12 cellular AD model, miR-26b causes cell death, downregulates NEP expression, and impairs neurite outgrowth | miRNA expression level (RT-qPCR and Western blot) | Biomarker as well as therapeutic target | [96] |
| miR-483-5p | HEK293 cells, neuroblastoma SK-N-MC cells, and neonatal human dermal fibroblasts | Upregulated | By controlling ERK1 and ERK2 at the mRNA and protein levels, miR-483-5p lowers tau phosphorylation through direct ERK1/2 repression and reduces the amounts of both kinases’ phosphorylated forms | miRNA expression level (RT-qPCR) and dual luciferase assay | Biomarker | [97] |
| miR-331-3p | AD patients and Aβ1–40 treated SH-SY5Y cells | Downregulated | miR-331-3p may have a neuroprotective function via controlling the expression of pro-inflammatory cytokines and cell survival in Aβ1–40-treated SH-SY5Y cells | Aβ1–40 and Aβ1–42 (ELISA) and dual luciferase assay | Therapeutic as well as diagnostic target | [98] |
| miR-433 | AD patients and Aβ-treated SH-SY5Y and SK-N-SH cells | Downregulated | In SH-SY5Y and SK-N-SH cells, overexpression of miR-433 may be able to reverse the reduction of neuronal viability caused by Aβ; the results of the luciferase activity assay indicated that miR-433 in neuronal cells targeted the gene JAK2 | miRNA expression level (RT-qPCR) and dual luciferase assay | Diagnostic biomarker as well as potential therapeutic target | [99] |
| miR-4422-5p | HEK-293T cells, SH-SY5Y cells, and the A549 cell line | Downregulated | In HEK293T cells, miR-4422-5p can bind directly to the 30 UTR of the GSAP and BACE1 genes, hence decreasing the function of these genes | Dual luciferase assays, Western blotting, and immunocytochemistry | Potential biomarker and therapeutic target | [100] |
| miR-200a-3p | APP/PS1 mice | Downregulated | Inhibits the expression of BACE1—miR-200a-3p can either directly or indirectly decrease the overproduction of Aβ; additionally, it can reduce tau hyperphosphorylation by attenuating PKA expression | Dual luciferase assays, Western blotting | Therapeutic target | [101] |
| miRNA-let-7a-5p | Nine AD patients and in vitro THP-1 cells | Upregulated | Positively regulates TLR3, RIG-I, and MDA5 | RT-qPCR assay | Therapeutic target | [102] |
| miR-10a-5p, miR-142a-5p, miR-146a-5p, miR-155-5p, miR-211-5p, and miR-455-5p | Male APPtg (APPswe/PS1L166P) and TAUtg (THY-Tau22) mice | Deregulated | miRNAs are implicated in a central pathogenic mechanism; however, they do not impair the cognitive function of wild-type mice | miRNA expression level (RT-qPCR and Western blot) and behavior assay (MWM, NOR, CFR, and SPSN) | Prognostic marker | [103] |
| miR-149-5p | AD patients and healthy volunteers | Upregulated | miR-149–5p influenced the expression of KAT8 and H4K16ac, related to AD pathogenesis | RT-qPCR, Western blot, and dual luciferase assays | Potential drug target | [104] |
| miRNA | Patients/Healthy Control (Number of Individuals)/Cell Line/Experimental Animal Model | Sample | Technique | Upregulated/Downregulated | Parameter Study and Technique Used | Concluding Remark | Reference |
|---|---|---|---|---|---|---|---|
| miR-193a-3p | Sporadic AD patients (108) and controls (93) | Serum | qRT-PCR | Downregulated | The results showed that the diagnostic sensitivity was 89.8% and the specificity was 77.4% | Diagnosis biomarker | [92] |
| miR-106b | AD patients (106) | Serum | qRT-PCR | Downregulated | qRT-PCR results showed a specificity and sensitivity of around 62% and 94%, respectively | Diagnosis biomarker | [132] |
| miRNA-193b (exosomal) | APP/PS1 mice, SCD patients (89), MCI patients (92), and DAT patients (92) | CSF and serum | TaqMan qPCR & exosomal RNA ELISA | Upregulated | miR-193b level is higher in the CSF of MCI and DAT patients | Early diagnostic biomarker | [133] |
| miR-384 | SCD patients (45), aMCI patients (50) AD (40), and healthy individuals (30) | Exosomal plasma | RT qPCR and Western blot | Upregulated | The levels of miR-384 were considerably greater in the exosomal plasma of the SCD, aMCI, and AD groups | Diagnosis of SCD | [134] |
| miR-342-5p | AD patients (19) | Plasma | RT-qPCR | Downregulated | After two years of follow-up, patients with mild AD showed a higher rate of cognitive deterioration in correlation with lower levels of miR-342-5p in plasma | Diagnostic biomarker | [135] |
| miR-103 and miR-107 | AD patients (120), PD patients (120), and controls (120) | Peripheral blood samples | RT-qPCR | Downregulated | For miR-103, the sensitivity and specificity were 80.0% and 84.2%, respectively; for miR-107, they were 77.5% and 59.2%, respectively | Biomarker able to differentiate between disease risk and disease progression stage | [136] |
| miR-373 and miR-204 | AD patients (18), moderate AD patients (18), and cognitively healthy individuals (21) | Plasma | RT-qPCR | Downregulated | Statistically noteworthy reduction in the expression of miR-204 and miR-373 in the mild and moderate AD groups compared to the healthy group | Biomarker | [137] |
| miR-202 | AD patients (121) and controls (86) | Serum | RT-qPCR | Downregulated | Serum miR-202 level could be differentiate between ACI patients and healthy individuals | Potential diagnostic biomarker | [31] |
| miR-125b | AD patients (105) | Serum | qRT-PCR | Downregulated | The specificity and sensitivity of the assay were 68.3% and 80.8%, respectively | Noninvasive biomarker for AD | [138] |
| miR-9 | AD patients (36) and controls (38) | Serum | qRT-PCR | Downregulated | The groups’ concentrations of miR-9-5p varied, with patients with AD showing a median 3-fold drop in circulating levels when compared to controls | Clinical biomarker | [139] |
| miR-193b | APP/PS1 mice; MCI and DAT patients | Serum, plasma, and CSF derived exosomes | qRT-PCR and Western blot | Decrease | Patients with DAT and MCI had lower serum and plasma levels of exosomal miR-193b than the control groups | Potential non-invasive marker for MCI and DAT patients | [140] |
| let-7F-5p, miR-1285, miR-107, miR-103a-5p, miR26b-5p, miR-532-5p, miR-151a-3p, miR-161, let-7d-3p, miR-112, and miR-5010-3p | AD patients (106) | Blood | qRT-PCR and NGS | Six upregulated and six downregulated | An accuracy of 93%, and a specificity and sensitivity of 92% and 95%, respectively | Signature biomarker | [141] |
| has-let-7d-5p and has-let-7g-5p | AD patients (50) and controls (50) | Plasma | qRT-PCR | Upregulated | A sensitivity of 0.82 and specificity of 0.34 for let-7d-5p; has-let-7g-5p showed 0.79 sensitivity and 0.28 specificity | Diagnostic biomarker | [142] |
| miR-133b | AD patients (105) and controls (98); SH-SY5Y cells with Aβ25-35 | Serum | qRT-PCR | Downregulated | Significantly lessening of the Aβ25–35-induced decrease of cell viability was achieved by overexpressing miR-133b | Potential diagnostic biomarker | [143] |
| miR-30b-5p, miR-22-3p, and miR-378a-3p | AD patients (8) and controls (8) | Serum | qRT-PCR | Deregulated | miRs were significantly deregulated in AD | Possible biomarker | [144] |
| has-mir-567 | MCI (18) and AD patients (18) | Peripheral blood samples | qRT-PCR | Upregulated | Serum from MCI-AD patients showed a higher fold change than that from controls | Signature marker in MCI diagnosis | [145] |
| miR-92a-3p, miR-181c-5p, and miR-210-3p | HCs (38), MCI (26), patients with AD dementia (56), and FTD patients (27) | Plasma | qRT-PCR and Western blot | Upregulated | An 89.3% accuracy, 84.6% sensitivity, and 85.71% sensitivity of the assay | Potential plasma biomarkers | [146] |
| miRNA-101a | AD patients and APPswe/PS1DE9 mice | Plasma | miRNA microarray assay and qRT-PCR | Downregulated | A sensitivity of 0.91 and specificity of 0.73 | Diagnostic biomarker | [90] |
| miR-128 | AD patients | Serum samples | Real-time fluorescence quantitative PCR | Upregulated | Statistically significant high levels of miR-128 | Clinical diagnostic biomarker | [147] |
| miR-150-5p | Mild DAT and MCI patients | Postmortem AD hippocampus | qRT-PCR and MRI imaging | Upregulated | miR-150-5p levels were found upregulated in the blood of DAT and MCI patients | Clinical blood-based biomarkers for DAT | [148] |
| miR-34a, miR-29b, and miR-181c | AD patients (23) | Serum | qRT-PCR | Downregulated | AD key pathological factor—Aβ42, TNF-α, and pTau—levels significantly increased in patients with high diagnostic power and assays showed high sensitivity and specificity for target miRs | Diagnostic marker | [149] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puranik, N.; Song, M. Insights into the Role of microRNAs as Clinical Tools for Diagnosis, Prognosis, and as Therapeutic Targets in Alzheimer’s Disease. Int. J. Mol. Sci. 2024, 25, 9936. https://doi.org/10.3390/ijms25189936
Puranik N, Song M. Insights into the Role of microRNAs as Clinical Tools for Diagnosis, Prognosis, and as Therapeutic Targets in Alzheimer’s Disease. International Journal of Molecular Sciences. 2024; 25(18):9936. https://doi.org/10.3390/ijms25189936
Chicago/Turabian StylePuranik, Nidhi, and Minseok Song. 2024. "Insights into the Role of microRNAs as Clinical Tools for Diagnosis, Prognosis, and as Therapeutic Targets in Alzheimer’s Disease" International Journal of Molecular Sciences 25, no. 18: 9936. https://doi.org/10.3390/ijms25189936
APA StylePuranik, N., & Song, M. (2024). Insights into the Role of microRNAs as Clinical Tools for Diagnosis, Prognosis, and as Therapeutic Targets in Alzheimer’s Disease. International Journal of Molecular Sciences, 25(18), 9936. https://doi.org/10.3390/ijms25189936







