Onco-Ontogeny of Squamous Cell Cancer of the First Pharyngeal Arch Derivatives
Abstract
1. Introduction
2. Parallelisms between the Development of the Embryo and Malignant Cells
2.1. The Evolutionary Theory of Carcinoma Evolution
2.2. Carcinoma and Development
3. Molecular Embryology of Head and Neck
3.1. Molecular Development of the Face (Pharyngeal Arches, Emphasize the First Arch)
3.2. Some Strategic Genes in Head and Neck Development
3.2.1. Wnt Signal
3.2.2. HOX Genes
3.2.3. HIPPO Pathway
4. Strategic Developmental Genes in Head and Neck Carcinoma
4.1. EMX1 and EMX2, Their Relationship with Wnt Regulation and Carcinogenesis
4.2. Wnt Expression Is Related to Carcinoma
4.3. Wnt in Head and Neck Carcinoma
4.4. OTX1 and OTX2 in Head and Neck Carcinoma Oncogenesis
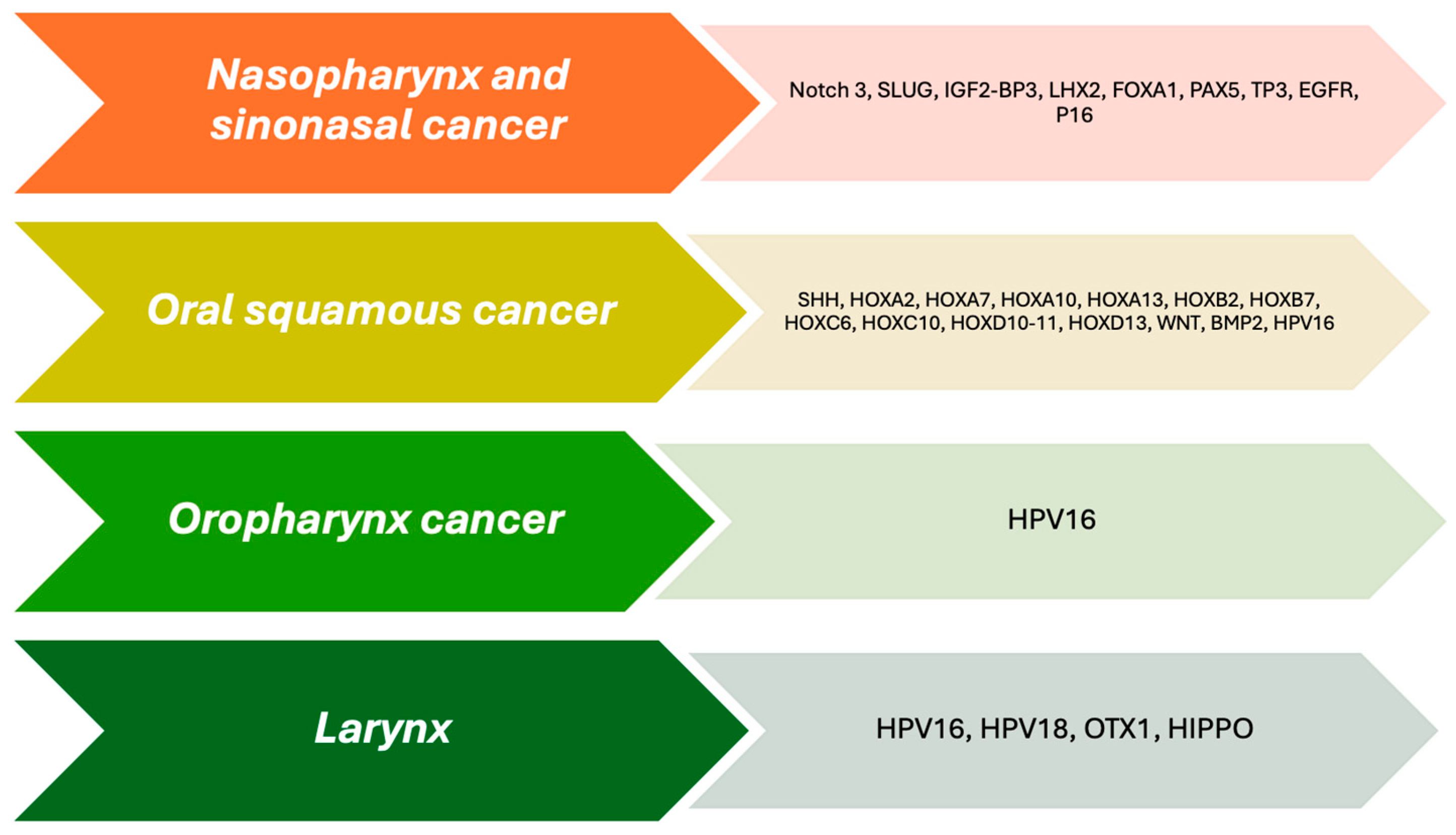
| Carcinoma | Gene | Mechanism | Observations | Reference * |
|---|---|---|---|---|
| Nasopharynx | Notch 3 | Enhanced expression | Inhibition is related to high response to cisplatin in presence of EBV | [85] |
| SLUG | Enhanced expression | Radio resistance | ||
| IGF2-BP3 | Associated to M6A | Constitutive activation Notch 3 | [85] | |
| LHX2 | Enhanced expression | Poor prognosis | [86] | |
| FOXA1 | Enhanced expression | Good prognosis associated to EBV | [87] | |
| PAX5 | Diminished expression | Advanced clinical stage at diagnosis and poor prognosis | [88] | |
| Sinonasal | TP53 | Mutated | Poor prognosis | [89] |
| EGFR | Amplification, increased number of copies, enhanced expression, activator mutation in exons 19, and 20 | Poor prognosis | [89] | |
| P16 | Mutation associated to HPV-16, 18, 31 and 33 | Improve overall survival | [89] | |
| Oral squamous carcinoma | SHH | Abnormal activation | Promoting invasion and metastasis | [89,90] |
| HOXA2 | Upregulated | Oral dysplasia | [43] | |
| HOXA7 | - | Less aggressive phenotype | [43] | |
| HOXA10 | - | Less aggressive phenotype | [43] | |
| HOXA13 | Overexpression | Good prognosis | [44] | |
| HOXB2 | - | Present in malignant lesions | [43] | |
| HOXB7 | - | Pivotal role in metastasis | [43] | |
| HOXC6 | - | Pivotal role in metastasis | [43] | |
| HOXC10 | - | Pivotal role in metastasis | [43] | |
| HOXD10-11 | Upregulated | Malignant phenotype | [43] | |
| HOXD13 | Overexpression | Poor prognosis | [44] | |
| WNT | Abnormal activation | Drug resistance | [91,92,93,94] | |
| BMP2 | Aberrant expression | Development and progression | [95] | |
| Oropharynx | HPV-16 | - | Improve overall survival | [96] |
| Larynx | HPV-16 and 18 | - | Improve overall survival | [97,98] |
| OTX1 | Overexpression | Nodal metastasis, poor prognosis and low rates overall survival | [99] | |
| HIPPO+ | Overexpression | Carcinoma development Poor prognosis | [100,101] |
4.5. BMP and Squamous Cell Carcinoma
4.6. HOX Genes during Craniofacial Development and Carcinoma Development
4.7. RA and Carcinoma Development
5. Head and Neck Carcinoma by Anatomical Regions
5.1. Nasopharynx (For This Section, See Figure 1 and Table 2)
5.2. Sinonasal Carcinoma
5.3. Oral Carcinoma
5.4. Oropharynx and Larynx Carcinoma
6. Head and Neck Squamous Cell Carcinoma and the Role of HPV in Oncogenesis
Other Alterations
7. Stem Cells in the Embryonic Period and Carcinoma
7.1. Embryonic Stem Cells
7.2. Adult Stem Cells
7.3. Cancer Stem Cells
7.4. Metabolism in Stem Cell Niches
8. Discussion
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primer 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Barsouk, A.; Aluru, J.S.; Rawla, P.; Saginala, K.; Barsouk, A. Epidemiology, Risk Factors, and Prevention of Head and Neck Squamous Cell Carcinoma. Med. Sci. 2023, 11, 42. [Google Scholar] [CrossRef]
- Head and Neck Cancers Incidence Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/head-and-neck-cancers/incidence (accessed on 2 September 2024).
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Kartsonaki, C.; Simon, J.; Yao, P.; Guo, Y.; Lv, J.; Walters, R.G.; Chen, Y.; Fry, H.; Avery, D.; et al. Prospective Evaluation of the Relevance of Epstein-Barr Virus Antibodies for Early Detection of Nasopharyngeal Carcinoma in Chinese Adults. Int. J. Epidemiol. 2024, 53, dyae098. [Google Scholar] [CrossRef]
- Lv, J.; Xu, L.-X.; Li, Z.-X.; Lin, L.; Wu, C.-F.; Quan, T.-Q.; Zhen, Z.-C.; Li, W.-F.; Tang, L.-L.; Mao, Y.-P.; et al. Longitudinal On-Treatment Circulating Tumor DNA as a Biomarker for Real-Time Dynamic Risk Monitoring in Cancer Patients: The EP-SEASON Study. Cancer Cell 2024, 42, 1401–1414.e4. [Google Scholar] [CrossRef]
- Yu, Y.-F.; Zhou, P.; Zhou, R.; Lin, Q.; Wu, S.-G. Lobaplatin-Based Concurrent Chemoradiotherapy in Elderly Nasopharyngeal Carcinoma. Ann. Med. 2024, 56, 2383959. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Mourad, M.; Jetmore, T.; Jategaonkar, A.A.; Moubayed, S.; Moshier, E.; Urken, M.L. Epidemiological Trends of Head and Neck Cancer in the United States: A SEER Population Study. J. Oral Maxillofac. Surg. Off. J. Am. Assoc. Oral Maxillofac. Surg. 2017, 75, 2562–2572. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Cancer Prevention Research–Then and Now. Nat. Rev. Cancer 2009, 9, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.; Gupta, R.; McDowell, L.; Magarey, M.J.R.; Smith, P.N.; Schulte, K.-M.; Perriman, D.M.; Veness, M.; Porceddu, S.; Low, T.-H.H.; et al. Predictors of Distant Metastatic Recurrence in Head and Neck Cutaneous Squamous Cell Carcinoma with Lymph Node Metastases Treated with Curative Intent: A Multicenter Study. Head Neck 2024. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yao, J.; Fu, J.; Huang, Q.; Luo, Y.; You, Y.; Zhang, W.; Zhong, Q.; Xia, T.; Xia, L. ALYREF Promotes the Metastasis of Nasopharyngeal Carcinoma by Increasing the Stability of NOTCH1 mRNA. Cell Death Dis. 2024, 15, 578. [Google Scholar] [CrossRef] [PubMed]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and Neck Cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Gruber, J.; Yee, Z.; Tolwinski, N.S. Developmental Drift and the Role of Wnt Signaling in Aging. Cancers 2016, 8, 73. [Google Scholar] [CrossRef]
- Sat-Muñoz, D.; Martínez-Herrera, B.-E.; González-Rodríguez, J.-A.; Gutiérrez-Rodríguez, L.-X.; Trujillo-Hernández, B.; Quiroga-Morales, L.-A.; Alcaráz-Wong, A.-A.; Dávalos-Cobián, C.; Solórzano-Meléndez, A.; Flores-Carlos, J.-D.; et al. Phase Angle, a Cornerstone of Outcome in Head and Neck Cancer. Nutrients 2022, 14, 3030. [Google Scholar] [CrossRef]
- Martínez-Herrera, B.-E.; Gutiérrez-Rodríguez, L.-X.; Trujillo-Hernández, B.; Muñoz-García, M.-G.; Cervantes-González, L.-M.; José Ochoa, L.-L.; González-Rodríguez, J.-A.; Solórzano-Meléndez, A.; Gómez-Sánchez, E.; Carrillo-Nuñez, G.-G.; et al. Phase Angle in Head and Neck Cancer: A Sex-Differential Analysis from Biological and Clinical Behavior to Health-Related Quality of Life. Biomedicines 2023, 11, 1696. [Google Scholar] [CrossRef]
- Rauf, S.; Ullah, S.; Abid, M.A.; Ullah, A.; Khan, G.; Khan, A.U.; Ahmad, G.; Ijaz, M.; Ahmad, S.; Faisal, S. A Computational Study of Gene Expression Patterns in Head and Neck Squamous Cell Carcinoma Using TCGA Data. Future Sci. OA 2024, 10, 2380590. [Google Scholar] [CrossRef]
- Shenoy, U.S.; Morgan, R.; Hunter, K.; Kabekkodu, S.P.; Radhakrishnan, R. Integrated Computational Analysis Reveals HOX Genes Cluster as Oncogenic Drivers in Head and Neck Squamous Cell Carcinoma. Sci. Rep. 2022, 12, 7952. [Google Scholar] [CrossRef]
- Habib, I.; Anjum, F.; Mohammad, T.; Sulaimani, M.N.; Shafie, A.; Almehmadi, M.; Yadav, D.K.; Sohal, S.S.; Hassan, M.I. Differential Gene Expression and Network Analysis in Head and Neck Squamous Cell Carcinoma. Mol. Cell. Biochem. 2022, 477, 1361–1370. [Google Scholar] [CrossRef]
- Chung, J.H.; Sanford, E.; Johnson, A.; Klempner, S.J.; Schrock, A.B.; Palma, N.A.; Erlich, R.L.; Frampton, G.M.; Chalmers, Z.R.; Vergilio, J.; et al. Comprehensive Genomic Profiling of Anal Squamous Cell Carcinoma Reveals Distinct Genomically Defined Classes. Ann. Oncol. 2016, 27, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Stanger, B.Z.; Wahl, G.M. Cancer as a Disease of Development Gone Awry. Annu. Rev. Pathol. 2024, 19, 397–421. [Google Scholar] [CrossRef] [PubMed]
- Oren, O.; Smith, B.D. Eliminating Cancer Stem Cells by Targeting Embryonic Signaling Pathways. Stem Cell Rev. Rep. 2017, 13, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Solter, D. From Teratocarcinomas to Embryonic Stem Cells and beyond: A History of Embryonic Stem Cell Research. Nat. Rev. Genet. 2006, 7, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Fisher, D.E.; Garbe, C.; Gershenwald, J.E.; Grob, J.-J.; Halpern, A.; Herlyn, M.; Marchetti, M.A.; McArthur, G.; Ribas, A.; et al. Melanoma. Nat. Rev. Dis. Primer 2015, 1, 15003. [Google Scholar] [CrossRef]
- Gaggioli, C.; Sahai, E. Melanoma Invasion—Current Knowledge and Future Directions. Pigment Cell Res. 2007, 20, 161–172. [Google Scholar] [CrossRef]
- Rozhok, A.; DeGregori, J. A Generalized Theory of Age-Dependent Carcinogenesis. eLife 2019, 8, e39950. [Google Scholar] [CrossRef]
- Dobzhansky, T. Nothing in Biology Makes Sense except in the Light of Evolution. Am. Biol. Teach. 1973, 35, 125–129. [Google Scholar] [CrossRef]
- Pashov, A.; Hernandez Puente, C.V.; Ibrahim, S.M.; Monzavi-Karbassi, B.; Makhoul, I.; Kieber-Emmons, T. Thinking Cancer. Monoclon. Antibodies Immunodiagn. Immunother. 2018, 37, 117–125. [Google Scholar] [CrossRef]
- Vendramin, R.; Litchfield, K.; Swanton, C. Cancer Evolution: Darwin and Beyond. EMBO J. 2021, 40, e108389. [Google Scholar] [CrossRef]
- Panje, W.R.; Ceilley, R.I. The Influence of Embryology of the Mid-Face on the Spread of Epithelial Malignancies. Laryngoscope 1979, 89, 1914–1920. [Google Scholar] [CrossRef] [PubMed]
- Höckel, M. Morphogenetic Fields of Embryonic Development in Locoregional Cancer Spread. Lancet Oncol. 2015, 16, e148–e151. [Google Scholar] [CrossRef] [PubMed]
- Höckel, M.; Horn, L.-C.; Manthey, N.; Braumann, U.-D.; Wolf, U.; Teichmann, G.; Frauenschläger, K.; Dornhöfer, N.; Einenkel, J. Resection of the Embryologically Defined Uterovaginal (Müllerian) Compartment and Pelvic Control in Patients with Cervical Cancer: A Prospective Analysis. Lancet Oncol. 2009, 10, 683–692. [Google Scholar] [CrossRef]
- Höckel, M.; Wolf, B.; Schmidt, K.; Mende, M.; Aktas, B.; Kimmig, R.; Dornhöfer, N.; Horn, L.-C. Surgical Resection Based on Ontogenetic Cancer Field Theory for Cervical Cancer: Mature Results from a Single-Centre, Prospective, Observational, Cohort Study. Lancet Oncol. 2019, 20, 1316–1326. [Google Scholar] [CrossRef]
- Kubitschke, H.; Wolf, B.; Morawetz, E.; Horn, L.-C.; Aktas, B.; Behn, U.; Höckel, M.; Käs, J. Roadmap to Local Tumour Growth: Insights from Cervical Cancer. Sci. Rep. 2019, 9, 12768. [Google Scholar] [CrossRef]
- Höckel, M.; Behn, U. The Order of Cancer: A Theory of Malignant Progression by Inverse Morphogenesis. Front. Oncol. 2019, 9, 416. [Google Scholar] [CrossRef]
- Cofre, J.; Abdelhay, E. Cancer Is to Embryology as Mutation Is to Genetics: Hypothesis of the Cancer as Embryological Phenomenon. Sci. World J. 2017, 2017, 3578090. [Google Scholar] [CrossRef] [PubMed]
- The Embryologic Basis of Craniofacial Structure: Developmental Anatomy, Evolutionary Design, and Clinical Applications|SpringerLink. Available online: https://link.springer.com/book/10.1007/978-3-031-15636-6 (accessed on 1 March 2024).
- Carlson, B.M. Chapter 14—Head and Neck. In Human Embryology and Developmental Biology, 4th ed.; Carlson, B.M., Ed.; W.B. Saunders: Philadelphia, PL, USA, 2014; pp. 294–334. ISBN 978-1-4557-2794-0. [Google Scholar]
- Stower, M.J.; Srinivas, S. Heading Forwards: Anterior Visceral Endoderm Migration in Patterning the Mouse Embryo. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130546. [Google Scholar] [CrossRef]
- Klimova, L.; Antosova, B.; Kuzelova, A.; Strnad, H.; Kozmik, Z. Onecut1 and Onecut2 Transcription Factors Operate Downstream of Pax6 to Regulate Horizontal Cell Development. Dev. Biol. 2015, 402, 48–60. [Google Scholar] [CrossRef]
- Shioi, G.; Hoshino, H.; Abe, T.; Kiyonari, H.; Nakao, K.; Meng, W.; Furuta, Y.; Fujimori, T.; Aizawa, S. Apical Constriction in Distal Visceral Endoderm Cells Initiates Global, Collective Cell Rearrangement in Embryonic Visceral Endoderm to Form Anterior Visceral Endoderm. Dev. Biol. 2017, 429, 20–30. [Google Scholar] [CrossRef]
- Robertson, E.J. Dose-Dependent Nodal/Smad Signals Pattern the Early Mouse Embryo. Semin. Cell Dev. Biol. 2014, 32, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.L.; Bohnsack, B.L. What’s Retinoic Acid Got to Do with It? Retinoic Acid Regulation of the Neural Crest in Craniofacial and Ocular Development. Genesis 2019, 57, e23308. [Google Scholar] [CrossRef]
- Ono-Minagi, H.; Nohno, T.; Serizawa, T.; Usami, Y.; Sakai, T.; Okano, H.; Ohuchi, H. The Germinal Origin of Salivary and Lacrimal Glands and the Contributions of Neural Crest Cell-Derived Epithelium to Tissue Regeneration. Int. J. Mol. Sci. 2023, 24, 13692. [Google Scholar] [CrossRef]
- Jimenez-García, M.P.; Lucena-Cacace, A.; Otero-Albiol, D.; Carnero, A. Regulation of Sarcomagenesis by the Empty Spiracles Homeobox Genes EMX1 and EMX2. Cell Death Dis. 2021, 12, 515. [Google Scholar] [CrossRef]
- Reya, T.; Clevers, H. Wnt Signalling in Stem Cells and Cancer. Nature 2005, 434, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, A.E.; D’Souza-Schorey, C. Wnt Signaling in Cell Motility and Invasion: Drawing Parallels between Development and Cancer. Cancers 2016, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Fuerer, C.; Nusse, R.; Ten Berge, D. Wnt Signalling in Development and Disease. Max Delbrück Center for Molecular Medicine Meeting on Wnt Signaling in Development and Disease. EMBO Rep. 2008, 9, 134–138. [Google Scholar] [CrossRef]
- Clevers, H.; Loh, K.M.; Nusse, R. An Integral Program for Tissue Renewal and Regeneration: Wnt Signaling and Stem Cell Control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef]
- Schlessinger, K.; Hall, A.; Tolwinski, N. Wnt Signaling Pathways Meet Rho GTPases. Genes Dev. 2009, 23, 265–277. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/β-Catenin Signaling and Disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Willert, K.; Nusse, R. Wnt Proteins. Cold Spring Harb. Perspect. Biol. 2012, 4, a007864. [Google Scholar] [CrossRef] [PubMed]
- van Amerongen, R.; Mikels, A.; Nusse, R. Alternative Wnt Signaling Is Initiated by Distinct Receptors. Sci. Signal. 2008, 1, re9. [Google Scholar] [CrossRef] [PubMed]
- Lezzerini, M.; Budovskaya, Y. A Dual Role of the Wnt Signaling Pathway during Aging in Caenorhabditis Elegans. Aging Cell 2014, 13, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Kastury, K.; Druck, T.; Huebner, K.; Barletta, C.; Acampora, D.; Simeone, A.; Faiella, A.; Boncinelli, E. Chromosome Locations of Human EMX and OTX Genes. Genomics 1994, 22, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Simeone, A.; Puelles, E.; Acampora, D. The Otx Family. Curr. Opin. Genet. Dev. 2002, 12, 409–415. [Google Scholar] [CrossRef]
- Boncinelli, E.; Simeone, A.; Acampora, D.; Gulisano, M. Homeobox Genes in the Developing Central Nervous System. Ann. Genet. 1993, 36, 30–37. [Google Scholar] [CrossRef]
- Larsen, K.B.; Lutterodt, M.C.; Møllgård, K.; Møller, M. Expression of the Homeobox Genes OTX2 and OTX1 in the Early Developing Human Brain. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2010, 58, 669–678. [Google Scholar] [CrossRef]
- Terrinoni, A.; Micheloni, G.; Moretti, V.; Caporali, S.; Bernardini, S.; Minieri, M.; Pieri, M.; Giaroni, C.; Acquati, F.; Costantino, L.; et al. OTX Genes in Adult Tissues. Int. J. Mol. Sci. 2023, 24, 16962. [Google Scholar] [CrossRef]
- Acampora, D.; Di Giovannantonio, L.G.; Simeone, A. Otx2 Is an Intrinsic Determinant of the Embryonic Stem Cell State and Is Required for Transition to a Stable Epiblast Stem Cell Condition. Development 2013, 140, 43–55. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, T.; Shen, H.; Yan, J.; Guan, M.; Jin, X. Identification of Long Non-Coding RNA Expression Profiles and Co-Expression Genes in Thyroid Carcinoma Based on The Cancer Genome Atlas (TCGA) Database. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 9752–9769. [Google Scholar] [CrossRef]
- Yuen, H.-F.; McCrudden, C.M.; Grills, C.; Zhang, S.-D.; Huang, Y.-H.; Chan, K.-K.; Chan, Y.-P.; Wong, M.L.-Y.; Law, S.; Srivastava, G.; et al. Combinatorial Use of Bone Morphogenetic Protein 6, Noggin and SOST Significantly Predicts Cancer Progression. Cancer Sci. 2012, 103, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Spaink, H.P.; Durston, A.J. Patterning of the Vertebrate Head in Time and Space by BMP Signaling. J. Dev. Biol. 2023, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Graf, D.; Malik, Z.; Hayano, S.; Mishina, Y. Common Mechanisms in Development and Disease: BMP Signaling in Craniofacial Development. Cytokine Growth Factor Rev. 2016, 27, 129–139. [Google Scholar] [CrossRef]
- Wacker, S.A.; Jansen, H.J.; McNulty, C.L.; Houtzager, E.; Durston, A.J. Timed Interactions between the Hox Expressing Non-Organiser Mesoderm and the Spemann Organiser Generate Positional Information during Vertebrate Gastrulation. Dev. Biol. 2004, 268, 207–219. [Google Scholar] [CrossRef][Green Version]
- Durston, A.J. Time, Space and the Vertebrate Body Axis. Semin. Cell Dev. Biol. 2015, 42, 66–77. [Google Scholar] [CrossRef]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) Signaling in Development and Human Diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef]
- Yuan, J.; Deng, Y.; Zhang, Y.; Gan, X.; Gao, S.; Hu, H.; Hu, S.; Hu, J.; Liu, H.; Li, L.; et al. Bmp4 Inhibits Goose Granulosa Cell Apoptosis via PI3K/AKT/Caspase-9 Signaling Pathway. Anim. Reprod. Sci. 2019, 200, 86–95. [Google Scholar] [CrossRef]
- Padam, K.S.R.; Morgan, R.; Hunter, K.; Chakrabarty, S.; Kumar, N.A.N.; Radhakrishnan, R. Identification of HOX Signatures Contributing to Oral Cancer Phenotype. Sci. Rep. 2022, 12, 10123. [Google Scholar] [CrossRef] [PubMed]
- Aquino, G.; Franco, R.; Sabatino, R.; Mantia, E.L.; Scognamiglio, G.; Collina, F.; Longo, F.; Ionna, F.; Losito, N.S.; Liguori, G.; et al. Deregulation of Paralogous 13 HOX Genes in Oral Squamous Cell Carcinoma. Am. J. Cancer Res. 2015, 5, 3042–3055. [Google Scholar]
- Dias, A.S.; de Almeida, I.; Belmonte, J.M.; Glazier, J.A.; Stern, C.D. Somites without a Clock. Science 2014, 343, 791–795. [Google Scholar] [CrossRef]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.-L.; Luo, T.; Luo, M. The Hippo Signalling Pathway and Its Implications in Human Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Saeki, T.; Yoshimatsu, S.; Ishikawa, M.; Hon, C.-C.; Koya, I.; Shibata, S.; Hosoya, M.; Saegusa, C.; Ogawa, K.; Shin, J.W.; et al. Critical Roles of FGF, RA, and WNT Signalling in the Development of the Human Otic Placode and Subsequent Lineages in a Dish. Regen. Ther. 2022, 20, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-García, M.P.; Lucena-Cacace, A.; Otero-Albiol, D.; Carnero, A. Empty Spiracles Homeobox Genes EMX1 and EMX2 Regulate WNT Pathway Activation in Sarcomagenesis. J. Exp. Clin. Cancer Res. CR 2021, 40, 247. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Ogura, S.; Ishida, M.; Karasawa, M.; Takada, S. Gene Trap Screening as an Effective Approach for Identification of Wnt-Responsive Genes in the Mouse Embryo. Dev. Dyn. 2005, 233, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. Comparative Integromics on Non-Canonical WNT or Planar Cell Polarity Signaling Molecules: Transcriptional Mechanism of PTK7 in Colorectal Cancer and That of SEMA6A in Undifferentiated ES Cells. Int. J. Mol. Med. 2007, 20, 405–409. [Google Scholar] [CrossRef]
- de Sousa e Melo, F.; Vermeulen, L. Wnt Signaling in Cancer Stem Cell Biology. Cancers 2016, 8, 60. [Google Scholar] [CrossRef]
- Dempke, W.C.M.; Fenchel, K.; Uciechowski, P.; Chevassut, T. Targeting Developmental Pathways: The Achilles Heel of Cancer? Oncology 2017, 93, 213–223. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A Question of Life or Death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Yuan, W.; Hu, J.; Wang, M.; Li, G.; Lu, S.; Qiu, Y.; Liu, C.; Liu, Y. KDM5B Promotes Metastasis and Epithelial-Mesenchymal Transition via Wnt/β-Catenin Pathway in Squamous Cell Carcinoma of the Head and Neck. Mol. Carcinog. 2024, 63, 885–896. [Google Scholar] [CrossRef]
- Fujii, S.; Hasegawa, K.; Maehara, T.; Kurppa, K.J.; Heikinheimo, K.; Warner, K.A.; Maruyama, S.; Tajiri, Y.; Nör, J.E.; Tanuma, J.-I.; et al. Wnt/β-Catenin-C-Kit Axis May Play a Role in Adenoid Cystic Carcinoma Prognostication. Pathol. Res. Pract. 2024, 254, 155148. [Google Scholar] [CrossRef] [PubMed]
- Pirrone, C.; Chiaravalli, A.M.; Marando, A.; Conti, A.; Rainero, A.; Pistochini, A.; Lo Curto, F.; Pasquali, F.; Castelnuovo, P.; Capella, C.; et al. OTX1 and OTX2 as Possible Molecular Markers of Sinonasal Carcinomas and Olfactory Neuroblastomas. Eur. J. Histochem. EJH 2017, 61, 2730. [Google Scholar] [CrossRef][Green Version]
- Yin, J.; Jung, J.-E.; Choi, S.I.; Kim, S.S.; Oh, Y.T.; Kim, T.-H.; Choi, E.; Lee, S.J.; Kim, H.; Kim, E.O.; et al. Inhibition of BMP Signaling Overcomes Acquired Resistance to Cetuximab in Oral Squamous Cell Carcinomas. Cancer Lett. 2018, 414, 181–189. [Google Scholar] [CrossRef]
- Padam, K.S.R.; Chakrabarty, S.; Hunter, K.D.; Radhakrishnan, R. Exploring the Regulatory Interactions between Mutated Genes and Homeobox Genes in the Head and Neck Cancer Progression. Arch. Oral Biol. 2024, 159, 105872. [Google Scholar] [CrossRef]
- Li, M.; Qiu, M.; Xu, Y.; Mao, Q.; Wang, J.; Dong, G.; Xia, W.; Yin, R.; Xu, L. Differentially Expressed Protein-Coding Genes and Long Noncoding RNA in Early-Stage Lung Cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 9969–9978. [Google Scholar] [CrossRef]
- Ning, X.-H.; Qi, Y.-Y.; Wang, F.-X.; Li, S.-C.; Jia, Z.-K.; Yang, J.-J. A Three Protein-Coding Gene Prognostic Model Predicts Overall Survival in Bladder Cancer Patients. BioMed Res. Int. 2020, 2020, 7272960. [Google Scholar] [CrossRef]
- Paço, A.; de Bessa Garcia, S.A.; Freitas, R. Methylation in HOX Clusters and Its Applications in Cancer Therapy. Cells 2020, 9, 1613. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018, 2018, 3569493. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, G.d.M.; Ramão, A.; Plaça, J.R.; Simões, S.C.; Scaraboto, N.V.; Freitas-Castro, F.; Cardoso, C.; Sousa, J.d.F.; Silva, W.A. Upregulation of HOX Genes Promotes Cell Migration and Proliferation in Head and Neck Squamous Cell Carcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2021, 43, 263–278. [Google Scholar] [CrossRef]
- Lee, J.J.; Wu, X.; Hildebrandt, M.A.T.; Yang, H.; Khuri, F.R.; Kim, E.; Gu, J.; Ye, Y.; Lotan, R.; Spitz, M.R.; et al. Global Assessment of Genetic Variation Influencing Response to Retinoid Chemoprevention in Head and Neck Cancer Patients. Cancer Prev. Res. 2011, 4, 185–193. [Google Scholar] [CrossRef][Green Version]
- D’Assoro, A.B.; Leon-Ferre, R.; Braune, E.-B.; Lendahl, U. Roles of Notch Signaling in the Tumor Microenvironment. Int. J. Mol. Sci. 2022, 23, 6241. [Google Scholar] [CrossRef] [PubMed]
- Nowell, C.S.; Radtke, F. Notch as a Tumour Suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Syrjänen, S.; Syrjänen, K. HPV in Head and Neck Carcinomas: Different HPV Profiles in Oropharyngeal Carcinomas—Why? Acta Cytol. 2019, 63, 124–142. [Google Scholar] [CrossRef] [PubMed]
- Guidelines Detail. Available online: https://www.nccn.org/guidelines/guidelines-detail (accessed on 1 March 2024).
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch Signaling Pathway: Architecture, Disease, and Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Ammous-Boukhris, N.; Ayadi, W.; Derbel, M.; Allaya-Jaafar, N.; Charfi, S.; Daoud, J.; Sellami-Boudawara, T.; Mokdad-Gargouri, R. FOXA1 Expression in Nasopharyngeal Carcinoma: Association with Clinicopathological Characteristics and EMT Markers. BioMed Res. Int. 2020, 2020, 4234632. [Google Scholar] [CrossRef]
- Porcheri, C.; Mitsiadis, T.A. Notch in Head and Neck Cancer. Adv. Exp. Med. Biol. 2021, 1287, 81–103. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, L.; Huang, C.; Li, Z.; Chen, L.; Gou, L.; Chen, P.; Tong, A.; Tang, M.; Gao, F.; et al. Comparative Proteomics Approach to Screening of Potential Diagnostic and Therapeutic Targets for Oral Squamous Cell Carcinoma. Mol. Cell. Proteom. MCP 2008, 7, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.W.; Peters, A.T.; Suh, L.; Norton, J.E.; Kern, R.C.; Conley, D.B.; Chandra, R.K.; Tan, B.K.; Grammer, L.C.; Harris, K.E.; et al. A Retrospective, Cross-Sectional Study Reveals That Women with CRSwNP Have More Severe Disease than Men. Immun. Inflamm. Dis. 2015, 3, 14–22. [Google Scholar] [CrossRef]
- Mills, A.A. P63: Oncogene or Tumor Suppressor? Curr. Opin. Genet. Dev. 2006, 16, 38–44. [Google Scholar] [CrossRef]
- Candi, E.; Dinsdale, D.; Rufini, A.; Salomoni, P.; Knight, R.A.; Mueller, M.; Krammer, P.H.; Melino, G. TAp63 and DeltaNp63 in Cancer and Epidermal Development. Cell Cycle Georget. Tex 2007, 6, 274–285. [Google Scholar] [CrossRef]
- Micheloni, G.; Millefanti, G.; Conti, A.; Pirrone, C.; Marando, A.; Rainero, A.; Tararà, L.; Pistochini, A.; Lo Curto, F.; Pasquali, F.; et al. Identification of OTX1 and OTX2 As Two Possible Molecular Markers for Sinonasal Carcinomas and Olfactory Neuroblastomas. J. Vis. Exp. JoVE 2019, 144, e56880. [Google Scholar] [CrossRef]
- Khammanivong, A.; Gopalakrishnan, R.; Dickerson, E.B. SMURF1 Silencing Diminishes a CD44-High Cancer Stem Cell-like Population in Head and Neck Squamous Cell Carcinoma. Mol. Cancer 2014, 13, 260. [Google Scholar] [CrossRef]
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox Genes and Regional Patterning of the Vertebrate Body Plan. Dev. Biol. 2010, 344, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The Hox Genes and Their Roles in Oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.-T.; Jia, W.-D.; Yao, Q.-Y.; Sun, Q.-K.; Ren, W.-H.; Huang, M.; Ma, J.; Li, J.-S.; Ma, J.-L.; Yu, J.-H.; et al. Overexpression of HOXA13 as a Potential Marker for Diagnosis and Poor Prognosis of Hepatocellular Carcinoma. Tohoku J. Exp. Med. 2014, 234, 209–219. [Google Scholar] [CrossRef]
- Li, B.; Huang, Q.; Wei, G.-H. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers 2019, 11, 528. [Google Scholar] [CrossRef]
- Rauch, T.; Wang, Z.; Zhang, X.; Zhong, X.; Wu, X.; Lau, S.K.; Kernstine, K.H.; Riggs, A.D.; Pfeifer, G.P. Homeobox Gene Methylation in Lung Cancer Studied by Genome-Wide Analysis with a Microarray-Based Methylated CpG Island Recovery Assay. Proc. Natl. Acad. Sci. USA 2007, 104, 5527–5532. [Google Scholar] [CrossRef]
- Botti, G.; De Chiara, A.; Di Bonito, M.; Cerrone, M.; Malzone, M.G.; Collina, F.; Cantile, M. Noncoding RNAs within the HOX Gene Network in Tumor Pathogenesis and Progression. J. Cell. Physiol. 2018, 234, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Matsunami, M.; Sumiyama, K.; Saitou, N. Evolution of Conserved Non-Coding Sequences within the Vertebrate Hox Clusters through the Two-Round Whole Genome Duplications Revealed by Phylogenetic Footprinting Analysis. J. Mol. Evol. 2010, 71, 427–436. [Google Scholar] [CrossRef]
- Smith, J.; Zyoud, A.; Allegrucci, C. A Case of Identity: HOX Genes in Normal and Cancer Stem Cells. Cancers 2019, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Aster, J.C. Notch Signaling in Cancer: Complexity and Challenges on the Path to Clinical Translation. Semin. Cancer Biol. 2022, 85, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Huang, R.; Xia, T.; Wang, C.; Xiao, X.; Lu, S.; Chen, X.; Ouyang, Y.; Deng, X.; Miao, J.; et al. The m6A Reader IGF2BP3 Preserves NOTCH3 mRNA Stability to Sustain Notch3 Signaling and Promote Tumor Metastasis in Nasopharyngeal Carcinoma. Oncogene 2023, 42, 3564–3574. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Deng, P.; Liu, S.; Hong, J.H.; Xiao, R.; Guan, P.; Wang, Y.; Wang, P.; Gao, J.; Chen, J.; et al. Enhancer Remodeling Activates NOTCH3 Signaling to Confer Chemoresistance in Advanced Nasopharyngeal Carcinoma. Cell Death Dis. 2023, 14, 513. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Du, K.; Liu, W.; Liu, C.; Wang, B.; Tian, Y.; Li, R.; Huang, X.; Lin, J.; Jian, H.; et al. LHX2 Facilitates the Progression of Nasopharyngeal Carcinoma via Activation of the FGF1/FGFR Axis. Br. J. Cancer 2022, 127, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Huang, X.; Qin, W.; Liang, P.; Zhao, J.; Ye, Y.; Ji, H.; Peng, X.; Liang, Y.; Cai, Y. Paired Box 5 (PAX5) Gene Has Diagnostic and Prognostic Potential in Nasopharyngeal Carcinoma. Int. J. Gen. Med. 2024, 17, 487–501. [Google Scholar] [CrossRef]
- Taverna, C.; Agaimy, A.; Franchi, A. Towards a Molecular Classification of Sinonasal Carcinomas: Clinical Implications and Opportunities. Cancers 2022, 14, 1463. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Kerr, A.R. Oral Cancer Screening: Past, Present, and Future. J. Dent. Res. 2021, 100, 1313–1320. [Google Scholar] [CrossRef]
- Jaksic Karisik, M.; Lazarevic, M.; Mitic, D.; Nikolic, N.; Milosevic Markovic, M.; Jelovac, D.; Milasin, J. Osteogenic and Adipogenic Differentiation Potential of Oral Cancer Stem Cells May Offer New Treatment Modalities. Int. J. Mol. Sci. 2023, 24, 4704. [Google Scholar] [CrossRef]
- Li, P.; Liu, H.; Wang, Z.; He, F.; Wang, H.; Shi, Z.; Yang, A.; Ye, J. MicroRNAs in Laryngeal Cancer: Implications for Diagnosis, Prognosis and Therapy. Am. J. Transl. Res. 2016, 8, 1935–1944. [Google Scholar]
- Patel, H.V.; Joshi, J.S.; Shah, F.D. A Clinicopathological Exploration of Hedgehog Signaling: Implications in Oral Carcinogenesis. J. Cancer Res. Clin. Oncol. 2023, 149, 16525–16535. [Google Scholar] [CrossRef] [PubMed]
- Cierpikowski, P.; Lis-Nawara, A.; Bar, J. Sonic Hedgehog Is a Novel Prognostic Biomarker in Patients with Oral Squamous Cell Carcinoma. Neoplasma 2021, 68, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Xu, L.; Li, Q.; Wang, Y.; Lu, H.; Zhao, N.; Pu, Y.; Wang, L.; Guo, Y.; Guo, C. Bone Morphogenetic Protein Receptor 1α Promotes Osteolytic Lesion of Oral Squamous Cell Carcinoma by SHH-Dependent Osteoclastogenesis. Cancer Sci. 2022, 113, 1639–1651. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Li, C.; Zhou, Q.; Zhang, X.; Kou, Y.; Feng, Q.; Wang, H.; Su, R.; Hasegawa, T.; Liu, H.; et al. Notum Leads to Potential Pro-Survival of OSCC through Crosstalk between Shh and It i/β-Catenin Signaling via p-GSK3β. Int. J. Biochem. Cell Biol. 2022, 153, 106316. [Google Scholar] [CrossRef] [PubMed]
- Patni, A.P.; Harishankar, M.K.; Joseph, J.P.; Sreeshma, B.; Jayaraj, R.; Devi, A. Comprehending the Crosstalk between Notch, Wnt and Hedgehog Signaling Pathways in Oral Squamous Cell Carcinoma—Clinical Implications. Cell. Oncol. 2021, 44, 473–494. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Liu, W. Chaperonin Containing TCP1 Subunit 6A May Activate Notch and Wnt Pathways to Facilitate the Malignant Behaviors and Cancer Stemness in Oral Squamous Cell Carcinoma. Cancer Biol. Ther. 2024, 25, 2287122. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Chen, S.; Guttridge, D.C.; You, Z.; Zhang, Z.; Fribley, A.; Mayo, M.W.; Kitajewski, J.; Wang, C.-Y. WNT-1 Signaling Inhibits Apoptosis by Activating β-Catenin/T Cell Factor–Mediated Transcription. J. Cell Biol. 2001, 152, 87–96. [Google Scholar] [CrossRef]
- Liu, S.-C.; Huang, C.-S.; Huang, C.-M.; Hsieh, M.-S.; Huang, M.-S.; Fong, I.-H.; Yeh, C.-T.; Lin, C.-C. Isoorientin Inhibits Epithelial-to-Mesenchymal Properties and Cancer Stem-Cell-like Features in Oral Squamous Cell Carcinoma by Blocking Wnt/β-Catenin/STAT3 Axis. Toxicol. Appl. Pharmacol. 2021, 424, 115581. [Google Scholar] [CrossRef]
- Mohapatra, P.; Shriwas, O.; Mohanty, S.; Ghosh, A.; Smita, S.; Kaushik, S.R.; Arya, R.; Rath, R.; Das Majumdar, S.K.; Muduly, D.K.; et al. CMTM6 Drives Cisplatin Resistance by Regulating Wnt Signaling through the ENO-1/AKT/GSK3β Axis. JCI Insight 2021, 6, e143643. [Google Scholar] [CrossRef]
- Zaid, K.W.; Chantiri, M.; Bassit, G. Recombinant Human Bone Morphogenetic Protein-2 in Development and Progression of Oral Squamous Cell Carcinoma. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.X.; D’Silva, N.J. HPV–Associated Oropharyngeal Cancer: In Search of Surrogate Biomarkers for Early Lesions. Oncogene 2024, 43, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Tsinias, G.; Nikou, S.; Mastronikolis, N.; Bravou, V.; Papadaki, H. Expression and Prognostic Significance of YAP, TAZ, TEAD4 and P73 in Human Laryngeal Cancer. Histol. Histopathol. 2020, 35, 983–995. [Google Scholar] [CrossRef]
- Steuer, C.E.; El-Deiry, M.; Parks, J.R.; Higgins, K.A.; Saba, N.F. An Update on Larynx Cancer. CA. J. Clin. 2017, 67, 31–50. [Google Scholar] [CrossRef]
- Lim, Y.X.; Mierzwa, M.L.; Sartor, M.A.; D’Silva, N.J. Clinical, Morphologic and Molecular Heterogeneity of HPV–Associated Oropharyngeal Cancer. Oncogene 2023, 42, 2939–2955. [Google Scholar] [CrossRef]
- Lianou, A.D.; Ragos, V.; Fotiades, P.P.; Tsiambas, E.; Batistatou, A.; Kastanioudakis, I. Molecular Aspects in HPV–Dependent Laryngeal and Oropharyngeal Carcinoma. J. BUON Off. J. Balk. Union Oncol. 2018, 23, 19–22. [Google Scholar]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV–Associated Oropharyngeal Cancer: Epidemiology, Molecular Biology and Clinical Management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef]
- Szabolcs, M.; Sauter, J.L.; Frosina, D.; Geronimo, J.A.; Hernandez, E.; Selbs, E.; Rapkiewicz, A.V.; Rekhtman, N.; Baine, M.K.; Jäger, E.; et al. Identification of Immunohistochemical Reagents for In Situ Protein Expression Analysis of Coronavirus-Associated Changes in Human Tissues. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The Molecular Landscape of Head and Neck Cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Tu, X.-P.; Li, H.; Chen, L.-S.; Luo, X.-N.; Lu, Z.-M.; Zhang, S.-Y.; Chen, S.-H. OTX1 Exerts an Oncogenic Role and Is Negatively Regulated by miR129-5p in Laryngeal Squamous Cell Carcinoma. BMC Cancer 2020, 20, 794. [Google Scholar] [CrossRef]
- Heinrich, E.-M.; Dimmeler, S. MicroRNAs and Stem Cells: Control of Pluripotency, Reprogramming, and Lineage Commitment. Circ. Res. 2012, 110, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, J.; Bodnar, M.; Paczkowska, J.; Ustaszewski, A.; Smialek, M.J.; Szylberg, L.; Marszalek, A.; Kiwerska, K.; Grenman, R.; Szyfter, K.; et al. Loss of the MAF Transcription Factor in Laryngeal Squamous Cell Carcinoma. Biomolecules 2021, 11, 1035. [Google Scholar] [CrossRef]
- Wu, Z.; Guan, K.-L. Hippo Signaling in Embryogenesis and Development. Trends Biochem. Sci. 2021, 46, 51–63. [Google Scholar] [CrossRef]
- Puram, S.V.; Rocco, J.W. Molecular Aspects of Head and Neck Cancer Therapy. Hematol. Oncol. Clin. N. Am. 2015, 29, 971–992. [Google Scholar] [CrossRef] [PubMed]
- de Morree, A.; Rando, T.A. Regulation of Adult Stem Cell Quiescence and Its Functions in the Maintenance of Tissue Integrity. Nat. Rev. Mol. Cell Biol. 2023, 24, 334–354. [Google Scholar] [CrossRef]
- Dionne, L.K.; Driver, E.R.; Wang, X.J. Head and Neck Cancer Stem Cells. J. Dent. Res. 2015, 94, 1524–1531. [Google Scholar] [CrossRef]
- Chinn, S.B.; Darr, O.A.; Owen, J.H.; Bellile, E.; Mchugh, J.B.; Spector, M.E.; Papagerakis, S.M.; Chepeha, D.B.; Bradford, C.R.; Carey, T.E.; et al. Cancer Stem Cells Mediate Tumorigenesis and Metastasis in Head and Neck Squamous Cell Carcinoma. Head Neck 2015, 37, 317–326. [Google Scholar] [CrossRef]
- Chen, G.; Yin, S.; Zeng, H.; Li, H.; Wan, X. Regulation of Embryonic Stem Cell Self-Renewal. Life 2022, 12, 1151. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer Stem Cells: An Evolving Concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef]
- Pereira, S.L.; Rodrigues, A.S.; Sousa, M.I.; Correia, M.; Perestrelo, T.; Ramalho-Santos, J. From Gametogenesis and Stem Cells to Cancer: Common Metabolic Themes. Hum. Reprod. Update 2014, 20, 924–943. [Google Scholar] [CrossRef]
- Anand, G.M.; Megale, H.C.; Murphy, S.H.; Weis, T.; Lin, Z.; He, Y.; Wang, X.; Liu, J.; Ramanathan, S. Controlling Organoid Symmetry Breaking Uncovers an Excitable System Underlying Human Axial Elongation. Cell 2023, 186, 497–512.e23. [Google Scholar] [CrossRef]
- Yu, M.; Qin, K.; Fan, J.; Zhao, G.; Zhao, P.; Zeng, W.; Chen, C.; Wang, A.; Wang, Y.; Zhong, J.; et al. The Evolving Roles of Wnt Signaling in Stem Cell Proliferation and Differentiation, the Development of Human Diseases, and Therapeutic Opportunities. Genes Dis. 2024, 11, 101026. [Google Scholar] [CrossRef] [PubMed]
- Cooper, F.; Souilhol, C.; Haston, S.; Gray, S.; Boswell, K.; Gogolou, A.; Frith, T.J.R.; Stavish, D.; James, B.M.; Bose, D.; et al. Notch Signalling Influences Cell Fate Decisions and HOX Gene Induction in Axial Progenitors. Development 2024, 151, dev202098. [Google Scholar] [CrossRef] [PubMed]
- Semprich, C.I.; Davidson, L.; Amorim Torres, A.; Patel, H.; Briscoe, J.; Metzis, V.; Storey, K.G. ERK1/2 Signalling Dynamics Promote Neural Differentiation by Regulating Chromatin Accessibility and the Polycomb Repressive Complex. PLoS Biol. 2022, 20, e3000221. [Google Scholar] [CrossRef] [PubMed]
- Wind, M.; Gogolou, A.; Manipur, I.; Granata, I.; Butler, L.; Andrews, P.W.; Barbaric, I.; Ning, K.; Guarracino, M.R.; Placzek, M.; et al. Defining the Signalling Determinants of a Posterior Ventral Spinal Cord Identity in Human Neuromesodermal Progenitor Derivatives. Development 2021, 148, dev194415. [Google Scholar] [CrossRef] [PubMed]
- Gogolou, A.; Souilhol, C.; Granata, I.; Wymeersch, F.J.; Manipur, I.; Wind, M.; Frith, T.J.; Guarini, M.; Bertero, A.; Bock, C.; et al. Early Anteroposterior Regionalisation of Human Neural Crest Is Shaped by a Pro-Mesodermal Factor. eLife 2022, 11, e74263. [Google Scholar] [CrossRef]
- Fuchs, E.; Blau, H.M. Tissue Stem Cells: Architects of Their Niches. Cell Stem Cell 2020, 27, 532–556. [Google Scholar] [CrossRef]
- Läubli, H.; Borsig, L. Selectins Promote Tumor Metastasis. Semin. Cancer Biol. 2010, 20, 169–177. [Google Scholar] [CrossRef]
- Jackson, B.T.; Finley, L.W.S. Metabolic Regulation of the Hallmarks of Stem Cell Biology. Cell Stem Cell 2024, 31, 161–180. [Google Scholar] [CrossRef]
- Routray, S.; Mohanty, N. Cancer Stem Cells Accountability in Progression of Head and Neck Squamous Cell Carcinoma: The Most Recent Trends! Mol. Biol. Int. 2014, 2014, 375325. [Google Scholar] [CrossRef][Green Version]
- Chinn, S.B.; Darr, O.A.; Peters, R.D.; Prince, M.E. The Role of Head and Neck Squamous Cell Carcinoma Cancer Stem Cells in Tumorigenesis, Metastasis, and Treatment Failure. Front. Endocrinol. 2012, 3, 90. [Google Scholar] [CrossRef] [PubMed]
- Conley, S.J.; Gheordunescu, E.; Kakarala, P.; Newman, B.; Korkaya, H.; Heath, A.N.; Clouthier, S.G.; Wicha, M.S. Antiangiogenic Agents Increase Breast Cancer Stem Cells via the Generation of Tumor Hypoxia. Proc. Natl. Acad. Sci. USA 2012, 109, 2784–2789. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.S.; Cirillo, N. The Molecular Markers of Cancer Stem Cells in Head and Neck Tumors. J. Cell. Physiol. 2020, 235, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Chen, X.; Luo, H.; Meng, C.; Zhu, D. Cancer Stem Cells of Head and Neck Squamous Cell Carcinoma; Distance towards Clinical Application; a Systematic Review of Literature. Am. J. Cancer Res. 2023, 13, 4315–4345. [Google Scholar]
- Hayashi, Y.; Yokota, A.; Harada, H.; Huang, G. Hypoxia/Pseudohypoxia-mediated Activation of Hypoxia-inducible Factor-1α in Cancer. Cancer Sci. 2019, 110, 1510–1517. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg Effect: 80 Years On. Biochem. Soc. Trans. 2016, 44, 1499–1505. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Jang, H.; Yang, J.; Lee, E.; Cheong, J.-H. Metabolism in Embryonic and Cancer Stemness. Arch. Pharm. Res. 2015, 38, 381–388. [Google Scholar] [CrossRef]
- Mii, Y. Understanding and Manipulating Extracellular Behaviors of Wnt Ligands. Vitr. Cell. Dev. Biol. Anim. 2024, 60, 441–448. [Google Scholar] [CrossRef]
- Andrews, P.W. The Origins of Human Pluripotent Stem Cells: The Road from a Cancer to Regenerative Medicine. Vitr. Cell. Dev. Biol. Anim. 2024, 60, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Wang, G.; Fails, D.; Nagarajan, P.; Ge, Y. Unraveling Cancer Lineage Drivers in Squamous Cell Carcinomas. Pharmacol. Ther. 2020, 206, 107448. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Fuchs, E. Stretching the Limits: From Homeostasis to Stem Cell Plasticity in Wound Healing and Cancer. Nat. Rev. Genet. 2018, 19, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Ide, K.; Kawano, T.; Shirakawa, J.; Ntege, E.H.; Miyamoto, S.; Ikegami, T.; Sunami, H.; Suzuki, M.; Shimizu, Y.; Nakamura, H. Exploring Stage-specific Embryonic Antigen 3 Involvement in Oral Cancer Progression and as a Potential Target for Taxane-based Chemotherapy. Oncol. Rep. 2023, 50, 182. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Wolf, K. Plasticity of Cell Migration: A Multiscale Tuning Model. J. Cell Biol. 2010, 188, 11–19. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac Activation and Inactivation Control Plasticity of Tumor Cell Movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Paiboonrungruang, C.; Li, Y.; Peters, H.; Kist, R.; Xiong, Z. PAX9 in Cancer Development. Int. J. Mol. Sci. 2022, 23, 5589. [Google Scholar] [CrossRef]
- Xiong, Z.; Ren, S.; Chen, H.; Liu, Y.; Huang, C.; Zhang, Y.L.; Odera, J.O.; Chen, T.; Kist, R.; Peters, H.; et al. PAX9 Regulates Squamous Cell Differentiation and Carcinogenesis in the Oro-Esophageal Epithelium. J. Pathol. 2018, 244, 164–175. [Google Scholar] [CrossRef]
- Bhol, C.S.; Patil, S.; Sahu, B.B.; Patra, S.K.; Bhutia, S.K. The Clinical Significance and Correlative Signaling Pathways of Paired Box Gene 9 in Development and Carcinogenesis. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188561. [Google Scholar] [CrossRef]
- Zhao, Z.; Szczepanski, A.P.; Tsuboyama, N.; Abdala-Valencia, H.; Goo, Y.A.; Singer, B.D.; Bartom, E.T.; Yue, F.; Wang, L. PAX9 Determines Epigenetic State Transition and Cell Fate in Cancer. Cancer Res. 2021, 81, 4696–4708. [Google Scholar] [CrossRef]
- Goker Bagca, B.; Biray Avci, C.; Sezgin, B.; Veral, A.; Gode, S.; Karci, H.B. The Role of the WNT Signaling Pathway in the Maxillary Sinus Squamous Cell Carcinoma. Med. Oncol. 2022, 39, 42. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Shen, L.; Dong, B.; Fu, H.; Kang, X.; Dai, L.; Yang, Y.; Yan, W.; Chen, K. Downregulation of HOXA13 Sensitizes Human Esophageal Squamous Cell Carcinoma to Chemotherapy. Thorac. Cancer 2018, 9, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Uehara, K.; Tanabe, Y.; Hirota, S.; Higa, S.; Toyoda, Z.; Kurima, K.; Kina, S.; Nakasone, T.; Arasaki, A.; Kinjo, T. Co-Expression of Low-Risk HPV E6/E7 and EBV LMP-1 Leads to Precancerous Lesions by DNA Damage. BMC Cancer 2021, 21, 688. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zhao, R.-X.; Chen, J.; Li, Y.; Li, X.-D.; Liu, X.-L.; Zhang, W.-M.; Quan, C.-S.; Wang, Y.-S.; Zhai, Y.-X.; et al. O-Linked GlcNAcylation Elevated by HPV E6 Mediates Viral Oncogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 9333–9338. [Google Scholar] [CrossRef]
- Sakellakis, M.; Yoon, S.M.; Reet, J.; Chalkias, A. Novel Insights into Voltage-Gated Ion Channels: Translational Breakthroughs in Medical Oncology. Channels 2024, 18, 2297605. [Google Scholar] [CrossRef]
| Transcription Factors, Signaling Molecules, and Embryonic Structures | Genes Activated | Mechanisms | Function |
|---|---|---|---|
| Neural crest cells (NCCs) (cranial ectoderm) * | HOX genes | Hindbrain segmentation (rhombomeres) | |
| NCC invade the pharyngeal pouches in three streams | 1st stream cell migration 2nd stream cell migration | 1st and 2nd rhombomeres cells invade 1st arch 4th rhombomere cells invade 6th arch | |
| 3rd stream cell migration | 6th and 7th rhombomeres cells invade rest of arches | ||
| Paraxial Mesoderm | |||
| Anterior visceral endoderm (AVE)(pharyngeal pouches endoderm) | Stimulates facial ectoderm | Specialized migratory extraembryonic epithelial cells | Signaling center for multiple pattering events |
| (Ectodermic frontonasal zone) | Induces neural crest mesenchyme precursors | To form facial bones | |
| Asymmetric expression of: LEFTY I, CER I and HEX genes | Regional organizing role of AVE | It explains that most of primary tumors in H&N originate in endoderm of pharyngeal pouches (squamous cell carcinoma) | |
| Visceral endoderm | Asymmetric expression of OTX2 and DDK1 | Covers trophectoderm-derived extraembryonic ectoderm | |
| Nodal y MAPK | In response to SMAD2 phosphorylation | Transform in AVE cells | |
| Visceral distal endoderm | Interaction with Nodal and MAPK and SMAD2 phosphorylation | Induces differentiation | as AVE |
| Forebrain and associated structures | Influenced by genes LHX1, EMX1, EMX2, OTX1, and OTX2 | Induces signals on the prechordal mesoderm or the AVE | Pouch (endodermal origin)Arch (mesenchymal origin)Grooves (ectodermal covering) |
| First pharyngeal arch | Independent of HOX genes and RA Dependent on the action OTX2 | Inductor signals to form: 1st aortic arch (maxillary artery), trigeminal (V) development | Formation of muscles of mastication, tensor tympany, veli palatini tensor digastric (anterior belly), middle ear ossicles (malleus, incus), sphenomadibular ligament, Meckel cartilage, tympanic ring |
| Pharyngeal pouches 2–4 | RA in an ascendent gradient in craniocaudal sense | Inductor signals to form: 2nd aortic arch (hyoid artery, stapedial artery), facial nerve (VII) development | Origin for muscles: facial expression, stapedial, stylohyoid, digastric (posterior belly), stapes, styloid process, stylohyoid ligament, hyoid (lesser horn, and part of body) |
| 3rd aortic arch: internal carotid artery and glossopharyngeal nerve (IX)) development | Stylopharyngeus muscle, hyoid (greater horn and part of the body) | ||
| 4th aortic arch (right subclavian and aortic arteries), vagus nerve (X) development | Pharyngeal and laryngeal muscles, laryngeal cartilages |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sat-Muñoz, D.; Balderas-Peña, L.-M.-A.; Gómez-Sánchez, E.; Martínez-Herrera, B.-E.; Trujillo-Hernández, B.; Quiroga-Morales, L.-A.; Salazar-Páramo, M.; Dávalos-Rodríguez, I.-P.; Nuño-Guzmán, C.M.; Velázquez-Flores, M.-C.; et al. Onco-Ontogeny of Squamous Cell Cancer of the First Pharyngeal Arch Derivatives. Int. J. Mol. Sci. 2024, 25, 9979. https://doi.org/10.3390/ijms25189979
Sat-Muñoz D, Balderas-Peña L-M-A, Gómez-Sánchez E, Martínez-Herrera B-E, Trujillo-Hernández B, Quiroga-Morales L-A, Salazar-Páramo M, Dávalos-Rodríguez I-P, Nuño-Guzmán CM, Velázquez-Flores M-C, et al. Onco-Ontogeny of Squamous Cell Cancer of the First Pharyngeal Arch Derivatives. International Journal of Molecular Sciences. 2024; 25(18):9979. https://doi.org/10.3390/ijms25189979
Chicago/Turabian StyleSat-Muñoz, Daniel, Luz-Ma.-Adriana Balderas-Peña, Eduardo Gómez-Sánchez, Brenda-Eugenia Martínez-Herrera, Benjamín Trujillo-Hernández, Luis-Aarón Quiroga-Morales, Mario Salazar-Páramo, Ingrid-Patricia Dávalos-Rodríguez, Carlos M. Nuño-Guzmán, Martha-Cecilia Velázquez-Flores, and et al. 2024. "Onco-Ontogeny of Squamous Cell Cancer of the First Pharyngeal Arch Derivatives" International Journal of Molecular Sciences 25, no. 18: 9979. https://doi.org/10.3390/ijms25189979
APA StyleSat-Muñoz, D., Balderas-Peña, L.-M.-A., Gómez-Sánchez, E., Martínez-Herrera, B.-E., Trujillo-Hernández, B., Quiroga-Morales, L.-A., Salazar-Páramo, M., Dávalos-Rodríguez, I.-P., Nuño-Guzmán, C. M., Velázquez-Flores, M.-C., Ochoa-Plascencia, M.-R., Muciño-Hernández, M.-I., Isiordia-Espinoza, M.-A., Mireles-Ramírez, M.-A., & Hernández-Salazar, E. (2024). Onco-Ontogeny of Squamous Cell Cancer of the First Pharyngeal Arch Derivatives. International Journal of Molecular Sciences, 25(18), 9979. https://doi.org/10.3390/ijms25189979










