The Effects of Brodalumab on the Fungal Microbiome in Patients with Psoriasis
Abstract
1. Introduction
2. Results
2.1. Demographics
2.2. Raw Data
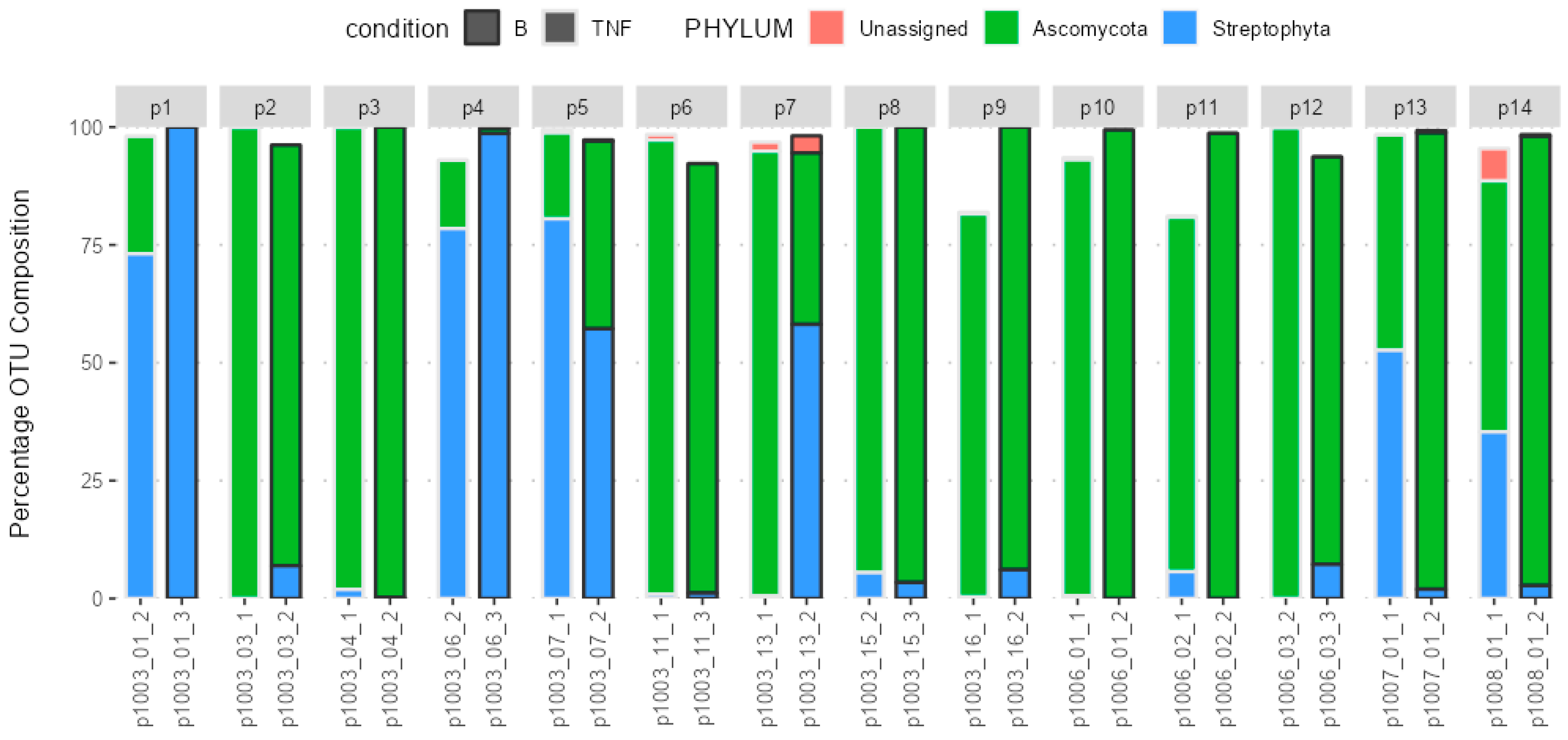
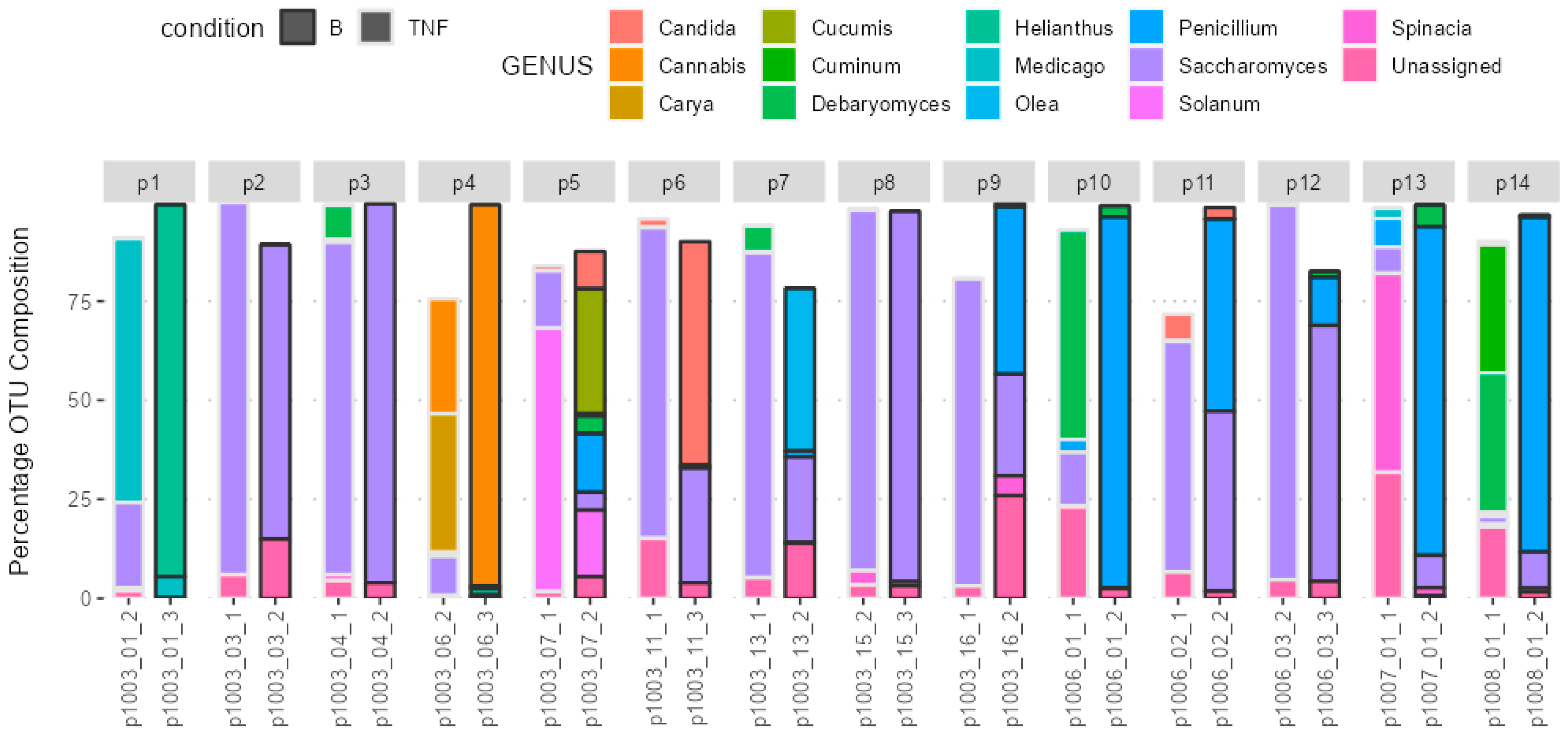
2.3. Composition of the Gut Microbiota
2.4. Difference in Groups before and after the Treatment Switch
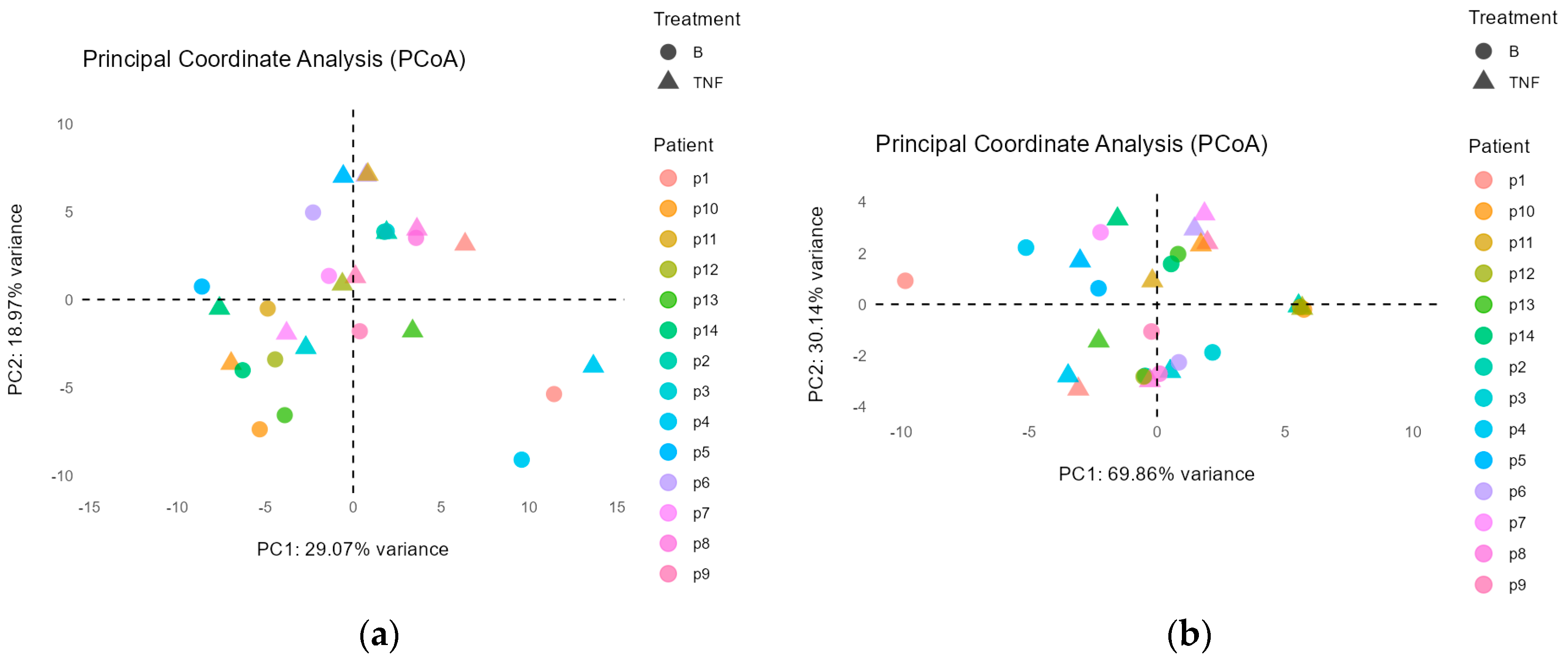
Principal Coordinate Analysis
2.5. Beta Diversity Analysis
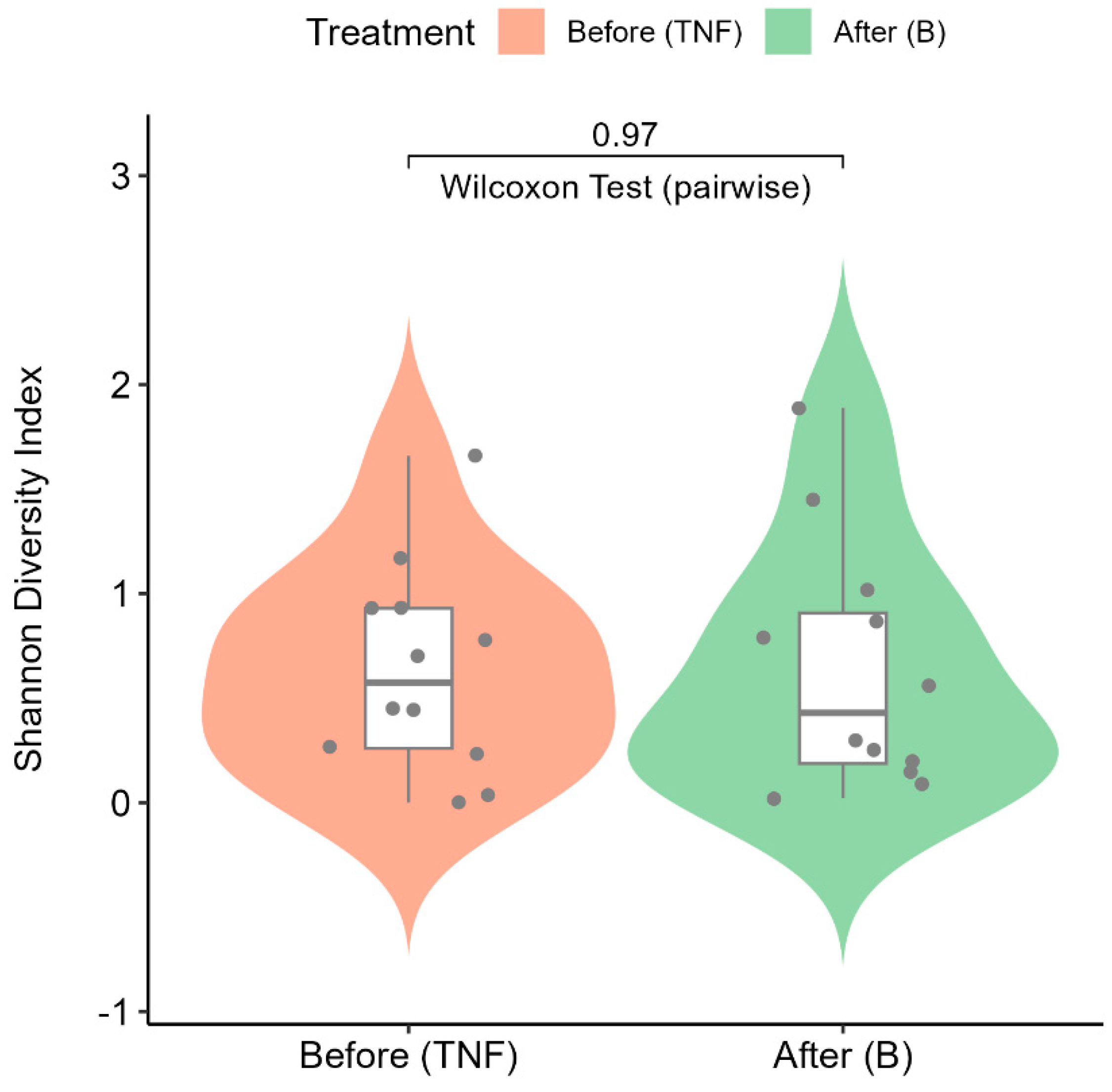
2.6. Alpha Diversity Analysis
2.7. Correlation to Clinical Variables
3. Discussion
4. Materials and Methods
4.1. Clinical Outcome Sequencing and Evaluation
4.2. Bioinformatics
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pérez, J.C. Fungi of the human gut microbiota: Roles and significance. Int. J. Med. Microbiol. 2021, 311, 151490. [Google Scholar] [CrossRef] [PubMed]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Gaitanis, G.; Velegraki, A.; Mayser, P.; Bassukas, I.D. Skin diseases associated with Malassezia yeasts: Facts and controversies. Clin. Dermatol. 2013, 31, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.L.; Lamas, B.; Liguori, G.; Hoffmann, T.W.; Sokol, H. Gut Fungal Microbiota. Inflamm. Bowel Dis. 2015, 21, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Y.; Pan, L.Y.; Zhang, X.; Zhang, Z.; Zhou, Y.Y.; Ruan, B. Altered gut bacterial–fungal interkingdom networks in patients with current depressive episode. Brain Behav. 2020, 10, e01677. [Google Scholar] [CrossRef]
- Charabati, M.; Grasmuck, C.; Ghannam, S.; Bourbonnière, L.; Fournier, A.P.; Lécuyer, M.-A.; Tastet, O.; Kebir, H.; Rébillard, R.-M.; Hoornaert, C.; et al. DICAM promotes T H 17 lymphocyte trafficking across the blood-brain barrier during autoimmune neuroinflammation. Sci. Transl. Med. 2022, 14, eabj0473. [Google Scholar] [CrossRef]
- Purzycki, C.B.; Shain, D.H. Fungal toxins and multiple sclerosis: A compelling connection. Brain Res. Bull. 2010, 82, 4–6. [Google Scholar] [CrossRef]
- Bliźniewska-Kowalska, K.; Halaris, A.; Gałecki, P.; Gałecka, M. Role of interleukin 17 (IL-17) in the inflammatory hypothesis of depression. J. Affect. Disord. Rep. 2023, 14, 100610. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Wu, L.; Xiao, S.; Ji, Y.; Tan, Y.; Jiang, C.; Zhang, G. Dysregulation of the gut-brain-skin axis and key overlapping inflammatory and immune mechanisms of psoriasis and depression. Biomed. Pharmacother. 2021, 137, 111065. [Google Scholar] [CrossRef]
- Grine, L.; Dejager, L.; Libert, C.; Vandenbroucke, R.E. An inflammatory triangle in psoriasis: TNF, type I IFNs and IL-17. Cytokine Growth Factor Rev. 2015, 26, 25–33. [Google Scholar] [CrossRef]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef] [PubMed]
- Komine, M. Recent Advances in Psoriasis Research; the Clue to Mysterious Relation to Gut Microbiome. Int. J. Mol. Sci. 2020, 21, 2582. [Google Scholar] [CrossRef] [PubMed]
- Roman, M.; Chiu, M.W. Spotlight on brodalumab in the treatment of moderate-to-severe plaque psoriasis: Design, development, and potential place in therapy. Drug Des. Dev. Ther. 2017, 11, 2065–2075. [Google Scholar] [CrossRef]
- Golbari, N.M.; Basehore, B.M.; Zito, P.M. Brodalumab. In StatPearls [Internet]; Turkderm Turkish Archives of Dermatology and Venereology; StatPearls Publishing: Treasure Island, FL, USA, 2023; Volume 56, pp. 58–60. Available online: https://www.ncbi.nlm.nih.gov/books/NBK470324/ (accessed on 13 March 2024).
- BRODALUMAB Injection Food and Drug Administration Dermatologic and Ophthalmic Drugs Advisory Committee (DODAC). 2016. Available online: https://www.fda.gov/files/advisory%20committees/published/Valeant-Briefing-Information-for-the-July-19--2016-Meeting-of-the-Dermatologic-and-Ophthalmic-Drugs-Advisory-Committee.pdf (accessed on 20 September 2024).
- Papp, K.A.; Reich, K.; Paul, C.; Blauvelt, A.; Baran, W.; Bolduc, C.; Toth, D.; Langley, R.G.; Cather, J.; Gottlieb, A.B.; et al. A prospective phase III, randomized, double-blind, placebo-controlled study of brodalumab in patients with moderate-to-severe plaque psoriasis. Br. J. Dermatol. 2016, 175, 273–286. [Google Scholar] [CrossRef]
- Andersch-Björkman, Y.; Micu, E.; Seifert, O.; Lonne-Rahm, S.; Gillstedt, M.; Osmancevic, A. Effects of brodalumab on psoriasis and depressive symptoms in patients with insufficient response to TNF-α inhibitors. J. Dermatol. 2023, 50, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Mattei, P.; Corey, K.; Kimball, A. Psoriasis Area Severity Index (PASI) and the Dermatology Life Quality Index (DLQI): The correlation between disease severity and psychological burden in patients treated with biological therapies. J. Eur. Acad. Dermatol. Venereol. 2013, 28, 333–337. [Google Scholar] [CrossRef] [PubMed]
- SSDV:s Behandlingsrekommendationer för Systemisk Behandling av Psoriasis (2023-10-25). Available online: https://ssdv.se/images/SSDVs_behandlingsrekommendationer_for_systemisk_behandli_ng_av_psoriasis_2023-10-25.pdf (accessed on 20 September 2024).
- Fantino, B.; Moore, N. The self-reported Montgomery-Åsberg depression rating scale is a useful evaluative tool in major depressive disorder. BMC Psychiatry 2009, 9, 26. [Google Scholar] [CrossRef]
- Herrmann, C. International experiences with the Hospital Anxiety and Depression Scale-a review of validation data and clinical results. J. Psychosom. Res. 1997, 42, 17–41. [Google Scholar] [CrossRef]
- Fry, L.; Baker, B.S. Triggering psoriasis: The role of infections and medications. Clin. Dermatol. 2007, 25, 606–615. [Google Scholar] [CrossRef]
- Waldman, A.; Gilhar, A.; Duek, L.; Berdicevsky, I. Incidence of Candida in psoriasis—A study on the fungal flora of psoriatic patients. Mycoses 2001, 44, 77–81. [Google Scholar] [CrossRef]
- Yeung, J.; Bunce, P.E.; Lynde, C.W.; Turchin, I.; Vender, R.B. Review and practical guidance on managing fungal infections in patients with psoriasis receiving Anti-IL-17 therapies. J. Cutan. Med. Surg. 2022, 26 (Suppl. 1), 3S–23S. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 2016, 7, e01250-16. [Google Scholar] [CrossRef] [PubMed]
- Unni, R.; Pintor, K.L.; Diepold, A.; Unterweger, D. Presence and absence of type VI secretion systems in bacteria. Microbiology 2022, 168, 001151. [Google Scholar] [CrossRef]
- Ohta, A.; Nishi, K.; Hirota, K.; Matsuo, Y. Using nanopore sequencing to identify fungi from clinical samples with high phylogenetic resolution. Sci. Rep. 2023, 13, 9785. [Google Scholar] [CrossRef]
- Vižlin, A.; Björkman, Y.A.; Kumar, Y.; Göthe, M.; Gillstedt, M.; Osmančević, A. No Evidence of Gut Microbiota Alteration in Psoriasis Patients Switching to Brodalumab after Loss of TNFα Inhibition Effect. Int. J. Mol. Sci. 2024, 25, 7745. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Academic Press: Cambridge, MA, USA, 1990; Available online: https://www.researchgate.net/publication/223397588_White_T_J_T_D_Bruns_S_B_Lee_and_J_W_Taylor_Amplification_and_direct_sequencing_of_fungal_ribosomal_RNA_Genes_for_phylogenetics (accessed on 16 August 2024).

| Variable | Sub Variable | Value |
|---|---|---|
| Age (years) | Mean (SD 1) | 46.6 (11.3) |
| Range (median) | 30–70 (50.5) | |
| Sex n 2 (%) | Female | 6 (30) |
| Male | 14 (70) | |
| Ethnicity n 2 (%) | Hispanic or Latino | 1 (5.0) |
| Not Hispanic or Latino | 19 (95) | |
| Mean BMI (SD 1) | 30 (4.3) | |
| Smokers (n 2) | 3 | |
| First psoriasis diagnosis (age) | Mean (SD 1) | 21.8 (10.1) |
| Range (median) | 11–44 (17.5) | |
| Scalp involvement [n 2 (%)] | Yes | 13 (65) |
| No | 7 (35) | |
| Nail involvement [n 2 (%)] | Yes | 8 (40) |
| No | 12 (60) | |
| Psoriatic arthritis diagnosis n 2 (%) | Yes | 3 (15) |
| No | 17 (85) | |
| Depression diagnosis n 2 (%) | 2 (10) | |
| PASI | Mean (SD 1) | 9.3 (3.5) |
| DLQI | Mean (SD 1) | 10.3 (7.2) |
| HADS | Mean (SD 1) | 4.3 (2.9) |
| MADRS-S | Mean (SD 1) | (6.0) |
| Taxa | PASI p-Value (Adjusted) | DLQI p-Value (Adjusted) | MADRS-S p-Value (Adjusted) | HADS p-Value (Adjusted) |
|---|---|---|---|---|
| Ascomycota (p) | 0.25 (1.00) | 0.89 (1.00) | 0.43 (1.00) | 0.83 (1.00) |
| Pencillium (g) | 0.19 (1.00) | 0.18 (1.00) | 0.64 (1.00) | 0.41 (1.00) |
| Candida (g) | 0.93 (1.00) | 0.49 (1.00) | 0.57 (1.00) | 0.37 (1.00) |
| Debaryomyces (g) | 0.38 (1.00) | 0.22 (1.00) | 0.98 (1.00) | 0.42 (1.00) |
| Saccharomyces (g) | 0.18 (1.00) | 0.87 (1.00) | 0.80 (1.00) | 0.90 (1.00) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vižlin, A.; Bajramović, A.; Björkman, Y.A.; Kumar, Y.; Göthe, M.; Gillstedt, M.; Osmančević, A. The Effects of Brodalumab on the Fungal Microbiome in Patients with Psoriasis. Int. J. Mol. Sci. 2024, 25, 10239. https://doi.org/10.3390/ijms251910239
Vižlin A, Bajramović A, Björkman YA, Kumar Y, Göthe M, Gillstedt M, Osmančević A. The Effects of Brodalumab on the Fungal Microbiome in Patients with Psoriasis. International Journal of Molecular Sciences. 2024; 25(19):10239. https://doi.org/10.3390/ijms251910239
Chicago/Turabian StyleVižlin, Admir, Ajša Bajramović, Ylva Andersch Björkman, Yadhu Kumar, Maria Göthe, Martin Gillstedt, and Amra Osmančević. 2024. "The Effects of Brodalumab on the Fungal Microbiome in Patients with Psoriasis" International Journal of Molecular Sciences 25, no. 19: 10239. https://doi.org/10.3390/ijms251910239
APA StyleVižlin, A., Bajramović, A., Björkman, Y. A., Kumar, Y., Göthe, M., Gillstedt, M., & Osmančević, A. (2024). The Effects of Brodalumab on the Fungal Microbiome in Patients with Psoriasis. International Journal of Molecular Sciences, 25(19), 10239. https://doi.org/10.3390/ijms251910239






