Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage
Abstract
1. Introduction
2. Results
2.1. Cohort and Sample Description
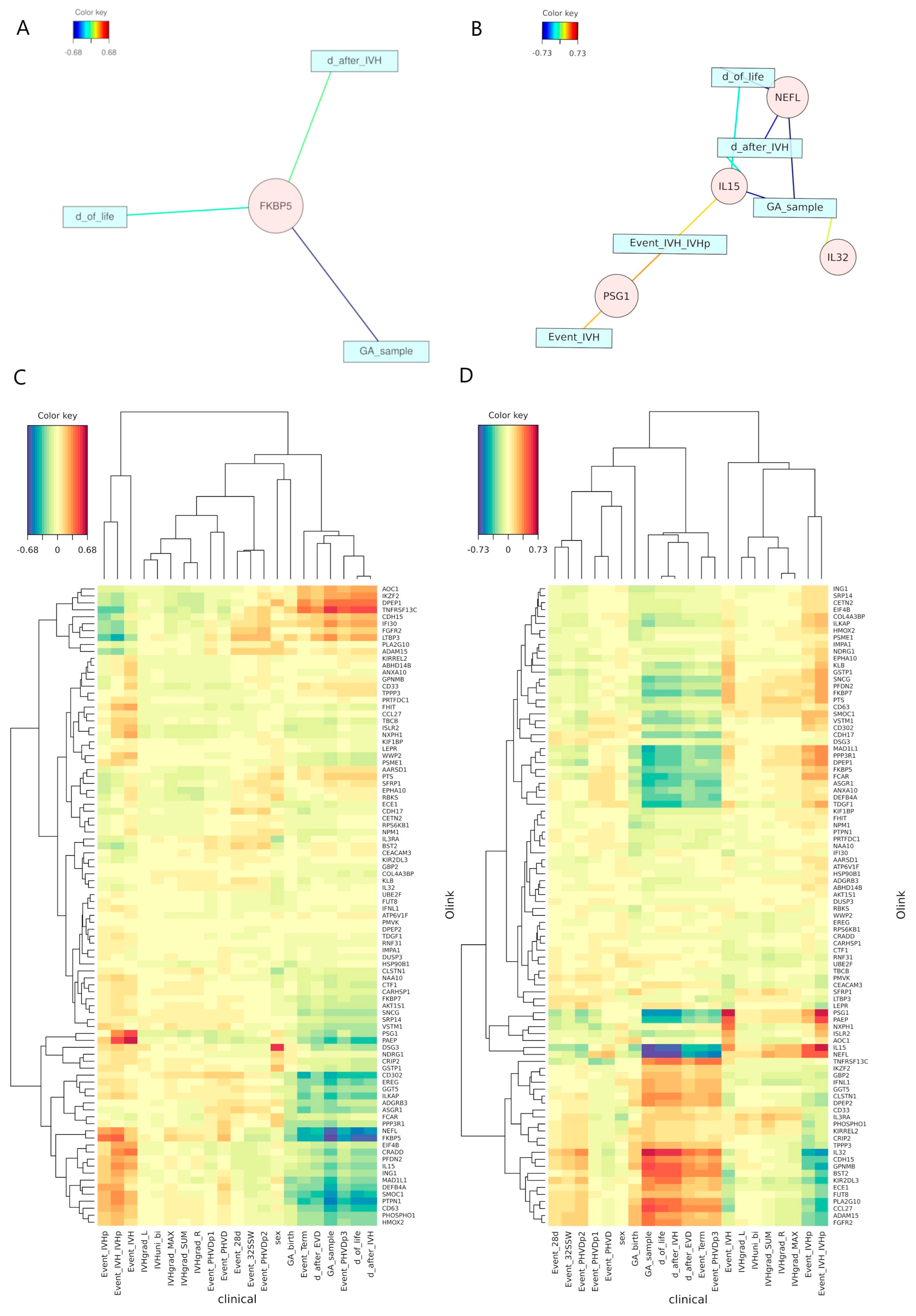
2.2. Exploratory Data Analysis
2.3. Machine Learning Reveals Potential Novel Biomarkers
2.4. Canonical Correlation Analysis Discloses Unexpected Independence
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Targeted Proteomics
4.3. Machine Learning and Biostatistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC-ROC | area under the receiver operating curve |
| BPD | bronchopulmonary dysplasia |
| CSVT | cerebral sinovenous thrombosis |
| EVD | extraventricular drainage |
| GA | gestational age |
| IVH | intraventricular hemorrhage |
| IVHgrade_L | degree of IVH in the left-brain hemisphere |
| IVHgrade_MAX | maximum degree of IVH |
| IVHgrade_R | degree of IVH in the right-brain hemisphere |
| IVHgrade_SUM | summed degree of IVH |
| IVHuni_bi | unilateral or bilateral IVH |
| ML | machine learning |
| NAIS | neonatal arterial ischemic stroke |
| NEC | necrotizing enterocolitis |
| NEFL | neurofilament light chain |
| NIH | National Institutes of Health |
| NPX | Normalized Protein eXpression |
| NSI | neurosurgical intervention |
| PCA | principal component analysis |
| PDA | persistent ductus arteriosus |
| PEA | Proximity Extension Assay |
| PHVD | posthemorrhagic ventricular dilatation |
| PHVDp | posthemorrhagic ventricular dilatation positive |
| PHVDn | posthemorrhagic ventricular dilatation negative |
| PLS-DA | partial least square discriminate analysis |
| PVCA | principal variance component analysis |
| PVL | periventricular leukomalacia |
| qPCR | quantitative polymerase chain reaction |
| RF | random forests |
| ROP | retinopathy of prematurity |
| rCCA | regularized canonical correlation analysis |
| VIP | variable importance projection |
References
- Parodi, A.; Govaert, P.; Horsch, S.; Bravo, M.C.; Ramenghi, L.A. Cranial ultrasound findings in preterm germinal matrix hemorrhage, sequelae and outcome. Pediatr. Res. 2020, 87 (Suppl. S1), 13–24. [Google Scholar] [CrossRef]
- Leijser, L.M.; de Vries, L.S. Preterm brain injury: Germinal matrix–intraventricular hemorrhage and post-hemorrhagic ventricular dilatation. Handb. Clin. Neurol. 2019, 162, 173–199. [Google Scholar] [CrossRef]
- El-Dib, M.; Limbrick, D.D., Jr.; Inder, T.; Whitelaw, A.; Kulkarni, A.V.; Warf, B.; Volpe, J.J.; de Vries, L.S. Management of Post-hemorrhagic Ventricular Dilatation in the Infant Born Preterm. J. Pediatr. 2020, 226, 16–27.e3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leijser, L.M.; Miller, S.P.; van Wezel-Meijler, G.; Brouwer, A.J.; Traubici, J.; van Haastert, I.C.; Whyte, H.E.; Groenendaal, F.; Kulkarni, A.V.; Han, K.S.; et al. Posthemorrhagic ventricular dilatation in preterm infants: When best to intervene? Neurology 2018, 90, e698–e706. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Robinson, S. Neonatal posthemorrhagic hydrocephalus from prematurity: Pathophysiology and current treatment concepts. J. Neurosurg. Pediatr. 2012, 9, 242–258. [Google Scholar] [CrossRef]
- Adams-Chapman, I.; Hansen, N.I.; Stoll, B.J.; Higgins, R. Neurodevelopmental outcome of extremely low birth weight infants with posthemorrhagic hydrocephalus requiring shunt insertion. Pediatrics 2008, 121, e1167–e1177. [Google Scholar] [CrossRef]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef]
- Aryutova, K.; Stoyanov, D.S.; Kandilarova, S.; Todeva-Radneva, A.; Kostianev, S.S. Clinical use of neurophysiological biomarkers and self-assessment scales to predict and monitor treatment response for psychotic and affective disorders. Curr. Pharm. Des. 2021, 27, 4039–4048. [Google Scholar] [CrossRef]
- Ahmad, A.; Imran, M.; Ahsan, H. Biomarkers as biomedical bioindicators: Approaches and techniques for the detection, analysis, and validation of novel biomarkers of diseases. Pharmaceutics 2023, 15, 1630. [Google Scholar] [CrossRef]
- Paik, Y.K.; Jeong, S.K.; Omenn, G.S.; Uhlen, M.; Hanash, S.; Cho, S.Y.; Lee, H.J.; Na, K.; Choi, E.Y.; Yan, F.; et al. The chromosome-centric human proteome project for cataloging proteins encoded in the genome. Nat. Biotechnol. 2012, 30, 221–223. [Google Scholar] [CrossRef]
- Baker, M.S.; Ahn, S.B.; Mohamedali, A.; Islam, M.T.; Cantor, D.; Verhaert, P.D.; Fanayan, S.; Sharma, S.; Nice, E.C.; Connor, M.; et al. Accelerating the search for the missing proteins in the human proteome. Nat. Commun. 2017, 8, 14271. [Google Scholar] [CrossRef]
- Goh, W.W.; Wong, L. Advanced bioinformatics methods for practical applications in proteomics. Brief. Bioinform. 2019, 20, 347–355. [Google Scholar] [CrossRef]
- Lundberg, M.; Eriksson, A.; Tran, B.; Assarsson, E.; Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011, 39, e102. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef]
- Carlyle, B.C.; Kitchen, R.R.; Mattingly, Z.; Celia, A.M.; Trombetta, B.A.; Das, S.; Hyman, B.T.; Kivisäkk, P.; Arnold, S.E. Technical performance evaluation of Olink proximity extension assay for blood-based biomarker discovery in longitudinal studies of Alzheimer’s disease. Front. Neurol. 2022, 13, 889647. [Google Scholar] [CrossRef]
- Dimitsaki, S.; Gavriilidis, G.I.; Dimitriadis, V.K.; Natsiavas, P. Benchmarking of machine learning classifiers on plasma proteomic for COVID-19 severity prediction through interpretable artificial intelligence. Artif. Intell. Med. 2023, 137, 102490. [Google Scholar] [CrossRef]
- Allesøe, R.L.; Lundgaard, A.T.; Hernández Medina, R.; Aguayo-Orozco, A.; Johansen, J.; Nissen, J.N.; Brorsson, C.; Mazzoni, G.; Niu, L.; Biel, J.H.; et al. Discovery of drug–omics associations in type 2 diabetes with generative deep-learning models. Nat. Biotechnol. 2023, 41, 399–408. [Google Scholar] [CrossRef]
- Pott, J.; Garcia, T.; Hauck, S.M.; Petrera, A.; Wirkner, K.; Loeffler, M.; Kirsten, H.; Peters, A.; Scholz, M. Genetically regulated gene expression and proteins revealed discordant effects. PLoS ONE 2022, 17, e0268815. [Google Scholar] [CrossRef]
- Zhang, X.; Jonassen, I.; Goksøyr, A. Machine learning approaches for biomarker discovery using gene expression data. In Bioinformatics; Exon Publications: Brisbane, Australia, 2021; pp. 53–64. [Google Scholar] [CrossRef]
- Ruiz-Perez, D.; Guan, H.; Madhivanan, P.; Mathee, K.; Narasimhan, G. So you think you can PLS-DA? BMC Bioinform. 2020, 21 (Suppl. 1), 2. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Kriegeskorte, N.; Golan, T. Neural network models and deep learning. Curr. Biol. 2019, 29, R231–R236. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Szymczak, S. A review on longitudinal data analysis with random forest. Brief Bioinform. 2023, 24, bbad002. [Google Scholar] [CrossRef] [PubMed]
- Webb, G.I. Naïve Bayes. In Encyclopedia of Machine Learning; Springer: New York, NY, USA, 2011; pp. 713–714. [Google Scholar] [CrossRef]
- van IJzendoorn, D.G.P.; Szuhai, K.; Briaire-de Bruijn, I.H.; Kostine, M.; Kuijjer, M.L.; Bovée, J.V.M.G. Machine learning analysis of gene expression data reveals novel diagnostic and prognostic biomarkers and identifies therapeutic targets for soft tissue sarcomas. PLoS Comput. Biol. 2019, 15, e1006826. [Google Scholar] [CrossRef]
- Ben Brahim, A.; Limam, M. Robust ensemble feature selection for high dimensional data sets. In Proceedings of the 2013 International Conference on High Performance Computing & Simulation (HPCS), Helsinki, Finland, 1–5 July 2013; pp. 151–157. [Google Scholar] [CrossRef]
- Seijo-Pardo, B.; Porto-Díaz, I.; Bolón-Canedo, V.; Alonso-Betanzos, A. Ensemble feature selection: Homogeneous and heterogeneous approaches. Knowl. Based Syst. 2017, 118, 124–139. [Google Scholar] [CrossRef]
- Zhang, X.; Jonassen, I. An Ensemble Feature Selection Framework Integrating Stability. In Proceedings of the 2019 International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 2792–2798. [Google Scholar] [CrossRef]
- McCauley, K.E.; Carey, E.C.; Weaver, A.L.; Mara, K.C.; Clark, R.H.; Carey, W.A.; Collura, C.A. Survival of Ventilated Extremely Premature Neonates with Severe Intraventricular Hemorrhage. Pediatrics 2021, 147, e20201584. [Google Scholar] [CrossRef]
- Gholampour, S.; Yamini, B.; Droessler, J.; Frim, D. A New Definition for Intracranial Compliance to Evaluate Adult Hydrocephalus After Shunting. Front. Bioeng. Biotechnol. 2022, 10, 900644. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Babendreyer, A.; Molls, L.; Simons, I.M.; Dreymueller, D.; Biller, K.; Jahr, H.; Denecke, B.; Boon, R.A.; Bette, S.; Schnakenberg, U.; et al. The metalloproteinase ADAM15 is upregulated by shear stress and promotes survival of endothelial cells. J. Mol. Cell. Cardiol. 2019, 134, 51–61. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, D.; Higham, A.; Wolosianka, S.; Gai, X.; Zhou, L.; Petersen, H.; Pinto-Plata, V.; Divo, M.; Silverman, E.K.; et al. ADAM15 expression is increased in lung CD8+ T cells, macrophages, and bronchial epithelial cells in patients with COPD and is inversely related to airflow obstruction. Respir. Res. 2020, 21, 188. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Liu, B.; Wu, C.; He, C.; Lv, X.; Jiang, Y. Association of job stress, FK506 binding protein 51 (FKBP5) gene polymorphisms and their interaction with sleep disturbance. PeerJ 2023, 11, e14794. [Google Scholar] [CrossRef] [PubMed]
- Cugliari, G. FKBP5, a Modulator of Stress Responses Involved in Malignant Mesothelioma: The Link between Stress and Cancer. Int. J. Mol. Sci. 2023, 24, 8183. [Google Scholar] [CrossRef]
- Locke, A.; Kanekar, S. Imaging of Premature Infants. Clin. Perinatol. 2022, 49, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Alazami, A.M.; Maddirevula, S.; Seidahmed, M.Z.; Albhlal, L.A.; Alkuraya, F.S. A novel ISLR2-linked autosomal recessive syndrome of congenital hydrocephalus, arthrogryposis and abdominal distension. Hum. Genet. 2019, 138, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Nice, T.J.; Diamond, M.S. Interferon-λ: Immune Functions at Barrier Surfaces and Beyond. Immunity 2015, 43, 15–28. [Google Scholar] [CrossRef]
- Cao, L.; Qian, W.; Li, W.; Ma, Z.; Xie, S. Type III interferon exerts thymic stromal lymphopoietin in mediating adaptive antiviral immune response. Front. Immunol. 2023, 14, 1250541. [Google Scholar] [CrossRef]
- Syedbasha, M.; Egli, A. Interferon Lambda: Modulating Immunity in Infectious Diseases. Front. Immunol. 2017, 8, 119. [Google Scholar] [CrossRef]
- Goeral, K.; Hauck, A.; Atkinson, A.; Wagner, M.B.; Pimpel, B.; Fuiko, R.; Klebermass-Schrehof, K.; Leppert, D.; Kuhle, J.; Berger, A.; et al. Early life serum neurofilament dynamics predict neurodevelopmental outcome of preterm infants. J. Neurol. 2021, 268, 2570–2577. [Google Scholar] [CrossRef]
- Knoche, T.; Gaus, V.; Haffner, P.; Kowski, A. Neurofilament light chain marks severity of papilledema in idiopathic intracranial hypertension. Neurol. Sci. 2023, 44, 2131–2135. [Google Scholar] [CrossRef]
- Jacobs Sariyar, A.; van Pesch, V.; Nassogne, M.C.; Moniotte, S.; Momeni, M. Usefulness of serum neurofilament light in the assessment of neurologic outcome in the pediatric population: A systematic literature review. Eur. J. Pediatr. 2023, 182, 1941–1948. [Google Scholar] [CrossRef]
- Douglas-Escobar, M.; Weiss, M.D. Biomarkers of Brain Injury in the Premature Infant. Front. Neurol. 2013, 3, 185. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Storey, J.D. Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLoS Genet. 2007, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Goeral, K.; Kasprian, G.; Hüning, B.M.; Waldhoer, T.; Fuiko, R.; Schmidbauer, V.; Prayer, D.; Felderhoff-Müser, U.; Berger, A.; Olischar, M.; et al. A novel magnetic resonance imaging-based scoring system to predict outcome in neonates born preterm with intraventricular haemorrhage. Dev. Med. Child. Neurol. 2022, 64, 608–617. [Google Scholar] [CrossRef]
- Jansson, J.; Willing, B.; Lucio, M.; Fekete, A.; Dicksved, J.; Halfvarson, J.; Tysk, C.; Schmitt-Kopplin, P. Metabolomics Reveals Metabolic Biomarkers of Crohn’s Disease. PLoS ONE 2009, 4, e6386. [Google Scholar] [CrossRef]
- Chong, I.G.; Jun, C.H. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2005, 78, 103–112. [Google Scholar] [CrossRef]
- Broughton-Neiswanger, L.E.; Rivera-Velez, S.M.; Suarez, M.A.; Slovak, J.E.; Hwang, J.K.; Villarino, N.F. Pharmacometabolomics with a combination of PLS-DA and random forest algorithm analyses reveal meloxicam alters feline plasma metabolite profiles. J. Vet. Pharmacol. Ther. 2020, 43, 591–601. [Google Scholar] [CrossRef]
- Chen, T.; Cao, Y.; Zhang, Y.; Liu, J.; Bao, Y.; Wang, C.; Jia, W.; Zhao, A. Random Forest in Clinical Metabolomics for Phenotypic Discrimination and Biomarker Selection. Evid.-Based Complement. Altern. Med. 2013, 2013, 298183. [Google Scholar] [CrossRef]
- Rivera-Velez, S.M.; Broughton-Neiswanger, L.E.; Suarez, M.; Piñeyro, P.; Navas, J.; Chen, S.; Hwang, J.; Villarino, N.F. Repeated administration of the NSAID meloxicam alters the plasma and urine lipidome. Sci. Rep. 2019, 9, 4303. [Google Scholar] [CrossRef]
- Andersen, C.M.; Bro, R. Variable selection in regression—A tutorial. J. Chemom. 2010, 24, 728–737. [Google Scholar] [CrossRef]
- Garson, G.D. Interpreting neural-network connection weights. AI Expert. 1991, 6, 46–51. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. In R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022.
- Tuzhilina, E.; Tozzi, L.; Hastie, T. Canonical correlation analysis in high dimensions with structured regularization. Stat. Model. 2023, 23, 203–227. [Google Scholar] [CrossRef]
- Gonzalez, I.; Déjean, S.; Martin, P.; Baccini, A. CCA: An R Package to Extend Canonical Correlation Analysis. J. Stat. Softw. 2008, 23, 1–14. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]

| Defined Event | Sampling Timeframe | Serum Samples | Urine Samples | Median Day of Life (IQR) |
|---|---|---|---|---|
| IVH | 0 to 2 days after IVH (bleeding Event) | 72 | 52 | 3 (2–4) |
| IVHp | 3 to 9 days after IVH and <−2 days after NSI | 108 | 101 | 6 (5–9) |
| PHVD | −2 to 0 days after NSI for PHVD positives; equivalent timeframe: 10 to 18 days after IVH for PHVD negatives | 99 | 78 | 14 (11–17) |
| PHVDp1 | 1 to 8 days after NSI for PHVD positives; equivalent timeframe: 10 to 18 days after IVH for PHVD negatives | 96 | 109 | 18 (15–22) |
| PHVDp2 | 9 to 39 days after NSI for PHVD positives; equivalent timeframe: 19 to 49 days after IVH for PHVD negatives | 108 | 83 | 38 (30–46) |
| PHVDp3 | 40+ days after NSI for PHVD positives; equivalent timeframe: 50+ days after IVH for PHVD negatives | 131 | 120 | 79 (67–96) |
| IVH_IVHp | 0 to 9 days after IVH | 172 | 152 | 5 (3–8) |
| 28 days of life | 21 to 35 days of life (28 days ± 7 days) | 82 | 83 | 27 (24–30) |
| 32 weeks | 31.0 to 33.0 GA (32 weeks ± 7 days) | 59 | 54 | 39 (22–50) |
| term-equivalent age | predicted birth timepoint/discharge from clinic, 36.0 to 41.14 GA | 93 | 84 | 83 (70–96) |
| Defined time windows: a single sample can be classified into two or more events. | ||||
| PHVDn (n = 46) | PHVDp (n = 53) | Total (n = 99) | p Value | |
|---|---|---|---|---|
| Survival | 0.045 a | |||
| Deceased n (%) | 18 (39.1) | 11 (20.8) | 29 (29.3) | |
| Survived n (%) | 28 (60.9) | 42 (79.2) | 70 (70.7) | |
| Median day at death (IQR) (days) | 17 (10–25) | |||
| Median GA at death (IQR) (weeks) | 26.57 (25.57–29.14) | |||
| GA at birth | <0.001 b | |||
| Median (IQR) (weeks) | 24.43 (23.57–25.96) | 26.29 (25.29–28.14) | 25.57 (24.14–27.14) | |
| Range | 23.00–29.71 | 23.29–33.29 | 23.00–33.29 | |
| Sex male n (%) | 30 (65.2) | 35 (66.0) | 65 (65.7) | 0.932 a |
| IVHgrade_L | ||||
| Median (IQR) | 3 (2–4) | 3 (3–3) | 3 (2–4) | 0.167 b |
| 0 n (%) | 5 (10.9) | 1 (1.9) | 6 (6.1) | 0.002 a |
| 2 n (%) | 16 (34.8) | 8 (15.1) | 24 (24.2) | |
| 3 n (%) | 11 (23.9) | 32 (62.3) | 44 (44.4) | |
| 4 n (%) | 14 (30.4) | 11 (20.8) | 25 (25.3) | |
| IVHgrade_R | ||||
| Median (IQR) | 3 (2–4) | 3 (3–3) | 3 (2–4) | 0.138 b |
| 0 n (%) | 5 (10.9) | 1 (1.9) | 6 (6.1) | 0.042 a |
| 1 n (%) | 3 (6.5) | 1 (1.9) | 4 (4.0) | |
| 2 n (%) | 12 (26.1) | 8 (15.1) | 20 (20.2) | |
| 3 n (%) | 14 (30.4) | 32 (60.4) | 46 (46.5) | |
| 4 n (%) | 12 (26.1) | 11 (20.8) | 23 (23.2) | |
| IVHuni_bi | 0.013 a | |||
| unilateral n (%) | 9 (19.6) | 2 (3.8) | 11 (11.1) | |
| bilateral n (%) | 37 (80.4) | 51 (96.2) | 88 (88.9) | |
| IVHgrade_MAX | ||||
| Median (IQR) | 3 (2–4) | 3 (3–4) | 3 (3–4) | 0.686 b |
| 2 n (%) | 12 (26.1) | 2 (3.8) | 14 (14.1) | 0.002 a |
| 3 n (%) | 13 (28.3) | 32 (60.4) | 45 (45.4) | |
| 4 n (%) | 21 (45.7) | 19 (35.8) | 40 (40.4) | |
| IVHgrade_SUM | ||||
| Median (IQR) | 6 (4–6) | 6 (6–6) | 6 (5–6) | 0.040 b |
| 2 n (%) | 6 (13.0) | 0 (0.0) | 6 (6.1) | 0.014 a |
| 3 n (%) | 3 (6.5) | 2 (3.8) | 5 (5.1) | |
| 4 n (%) | 8 (17.4) | 3 (5.7) | 11 (11.1) | |
| 5 n (%) | 5 (10.9) | 6 (11.3) | 11 (11.1) | |
| 6 n (%) | 13 (28.3) | 30 (56.6) | 43 (43.4) | |
| 7 n (%) | 6 (13.0) | 9 (17.0) | 15 (15.2) | |
| 8 n (%) | 5 (10.9) | 3 (5.7) | 8 (8.1) | |
| Number of NSI | <0.001 b | |||
| Median (IQR) | NA | 3 (2–5) | 1 (0–4) | |
| Range | NA | 0.00–10.00 | 0.00–10.00 | |
| Asphyxia n (%) | 10 (21.7) | 13 (24.5) | 23 (23.2) | 0.743 a |
| NAISor neonatal CSVT n (%) | 0 (0.0) | 2 (3.8) | 2 (2.0) | 0.183 a |
| Encephalitis or ventriculitis n (%) | 0 (0.0) | 11 (20.8) | 11 (11.1) | 0.001 a |
| PDA n (%) c | 6 (16.2) | 6 (12.0) | 12 (13.8) | 0.218 a |
| NEC n (%) c | 5 (10.9) | 5 (9.4) | 10 (10.1) | 0.813 a |
| BPD n (%) d | 16 (55.2) | 19 (41.3) | 35 (46.7) | 0.026 a |
| ROP n (%) d | 7 (24.1) | 8 (18.2) | 15 (20.6) | 0.084 a |
| PVL n (%) d | 2 (7.1) | 3 (6.8) | 5 (6.9) | 0.087 a |
| Model | Features Selected |
|---|---|
| Urine Models predicting PHVD | |
| Urine IVH | DEFB4A; GA at birth |
| Urine IVHp | GA at birth; TDGF1 |
| Urine PHVD | – a |
| Urine IVH_IVHp | RBKS; GA at birth; PPP3R1 |
| Urine IVH_IVHp_PHVD | RBKS; GA at birth; CD33; SNCG; PP3R1 |
| Serum Models predicting PHVD | |
| Blood IVH | PPP3R1; GA at birth |
| Blood IVHp | FUT8; GA at birth; RBKS |
| Blood PHVD | KLB; GA at birth; PAEP; PTS; AOC1; ISLR2; NXPH1; IVHgrade_MAX; VSTMT |
| Blood IVH_IVHp | GA at birth; PPP3R1; FUT8 |
| Blood IVH_IVHp_PHVD | DPEP2; GA at birth |
| Urine Models predicting survival | |
| Urine IVH | GA at birth; HSP90B1; KIRREL2 |
| Urine IVHp | – a |
| Urine PHVD | FGFR2; GA at birth |
| Urine IVH_IVHp | GA at birth |
| Urine IVH_IVHp_PHVD | – a |
| Urine PHVDp1 | – a |
| Urine PHVDp2 | – a |
| Urine PHVDp3 | – a |
| Urine 28 days of life | – a |
| Urine 32 weeks | – a |
| Urine term-equivalent age | Not able to perform ML |
| Serum Models predicting survival | |
| Blood IVH | PRTFDC2; GA at birth; AKT1S1; FKBP5; SNCG; DPEP2 |
| Blood IVHp | FGFR2; GA at birth; IL15; FKBP5; DPEP2; CLSTN1; IFNL1; RBKS |
| Blood PHVD | GPNMB; DSG3; FGFR2; NEFL; IL15; CDH15; ADAM15; GA at birth; KIR2DL3; PLA2G10 |
| Blood IVH_IVHp | DPEP2; GA at birth; IL15; GSTP1; COL4A3BP; PRTFDC1; SNCG |
| Blood IVH_IVHp_PHVD | GA at birth; DSG3 |
| Blood PHVDp1 | FGFR2; ADAM15; NEFL; PLA2G10; IL15; CDH15; BST2; FCAR; GA at birth |
| Blood PHVDp2 | GA at birth; TNFRSE13C; PAEP |
| Blood PHVDp3 | IFNL1; SNCG; GA at birth; TDGF1; ADGRB3; IL32 |
| Blood 28 days of life | – a |
| Blood 32 weeks | – a |
| Blood term-equivalent age | – a |
| Applied thresholds for the models: AUC-ROC ≥ 0.7; Sensitivity ≥ 0.6 and Selectivity ≥ 0.6.Features selected from models passing thresholds had to display a relative variable importance measure ≥ 50. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vignolle, G.A.; Bauerstätter, P.; Schönthaler, S.; Nöhammer, C.; Olischar, M.; Berger, A.; Kasprian, G.; Langs, G.; Vierlinger, K.; Goeral, K. Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage. Int. J. Mol. Sci. 2024, 25, 10304. https://doi.org/10.3390/ijms251910304
Vignolle GA, Bauerstätter P, Schönthaler S, Nöhammer C, Olischar M, Berger A, Kasprian G, Langs G, Vierlinger K, Goeral K. Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage. International Journal of Molecular Sciences. 2024; 25(19):10304. https://doi.org/10.3390/ijms251910304
Chicago/Turabian StyleVignolle, Gabriel A., Priska Bauerstätter, Silvia Schönthaler, Christa Nöhammer, Monika Olischar, Angelika Berger, Gregor Kasprian, Georg Langs, Klemens Vierlinger, and Katharina Goeral. 2024. "Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage" International Journal of Molecular Sciences 25, no. 19: 10304. https://doi.org/10.3390/ijms251910304
APA StyleVignolle, G. A., Bauerstätter, P., Schönthaler, S., Nöhammer, C., Olischar, M., Berger, A., Kasprian, G., Langs, G., Vierlinger, K., & Goeral, K. (2024). Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage. International Journal of Molecular Sciences, 25(19), 10304. https://doi.org/10.3390/ijms251910304






