Telomere Reprogramming and Cellular Metabolism: Is There a Link?
Abstract
1. Introduction
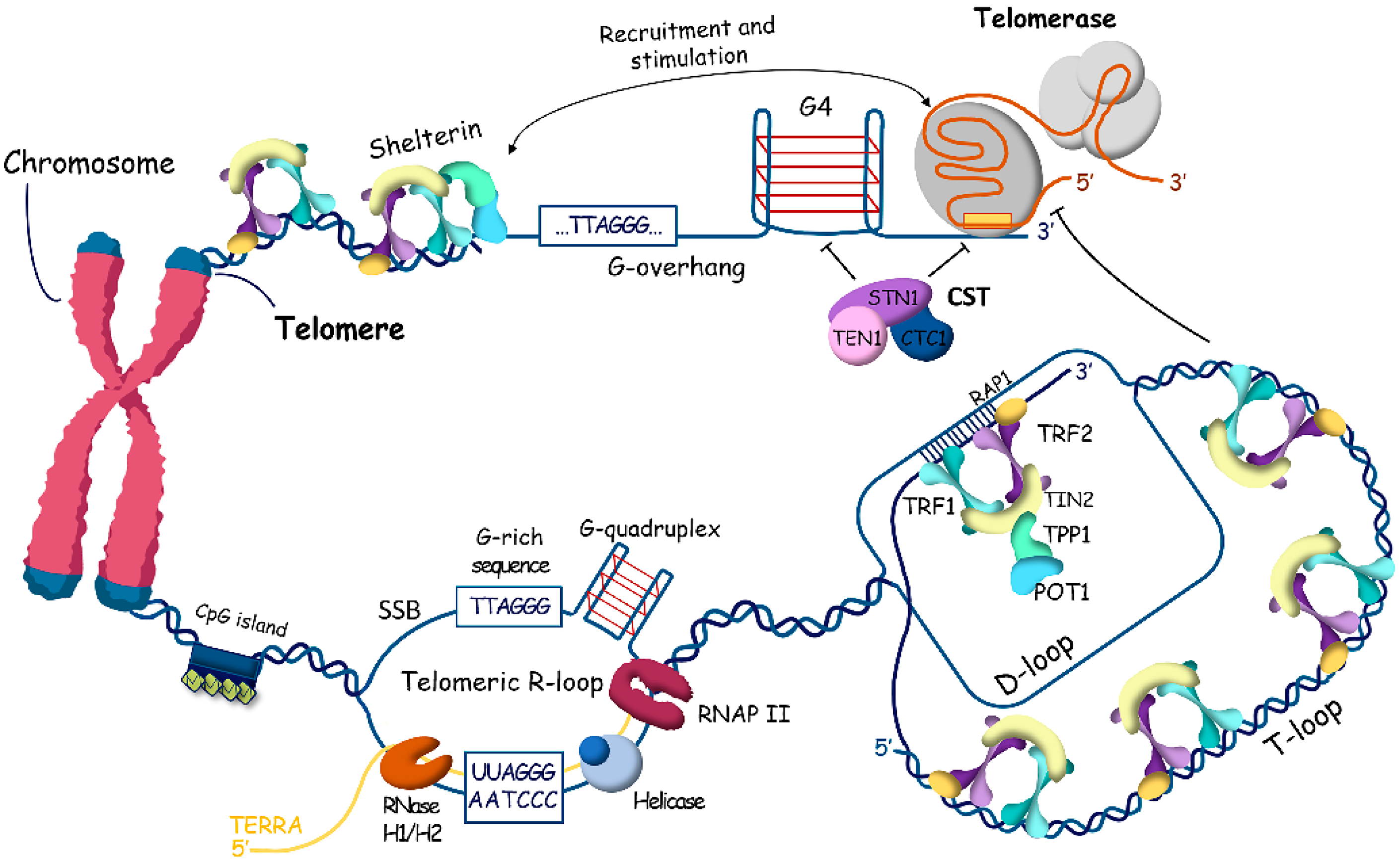
2. Telomeres: Structure and Regulation
2.1. Telomere Structure
2.2. Telomere Lengthening by Telomerase
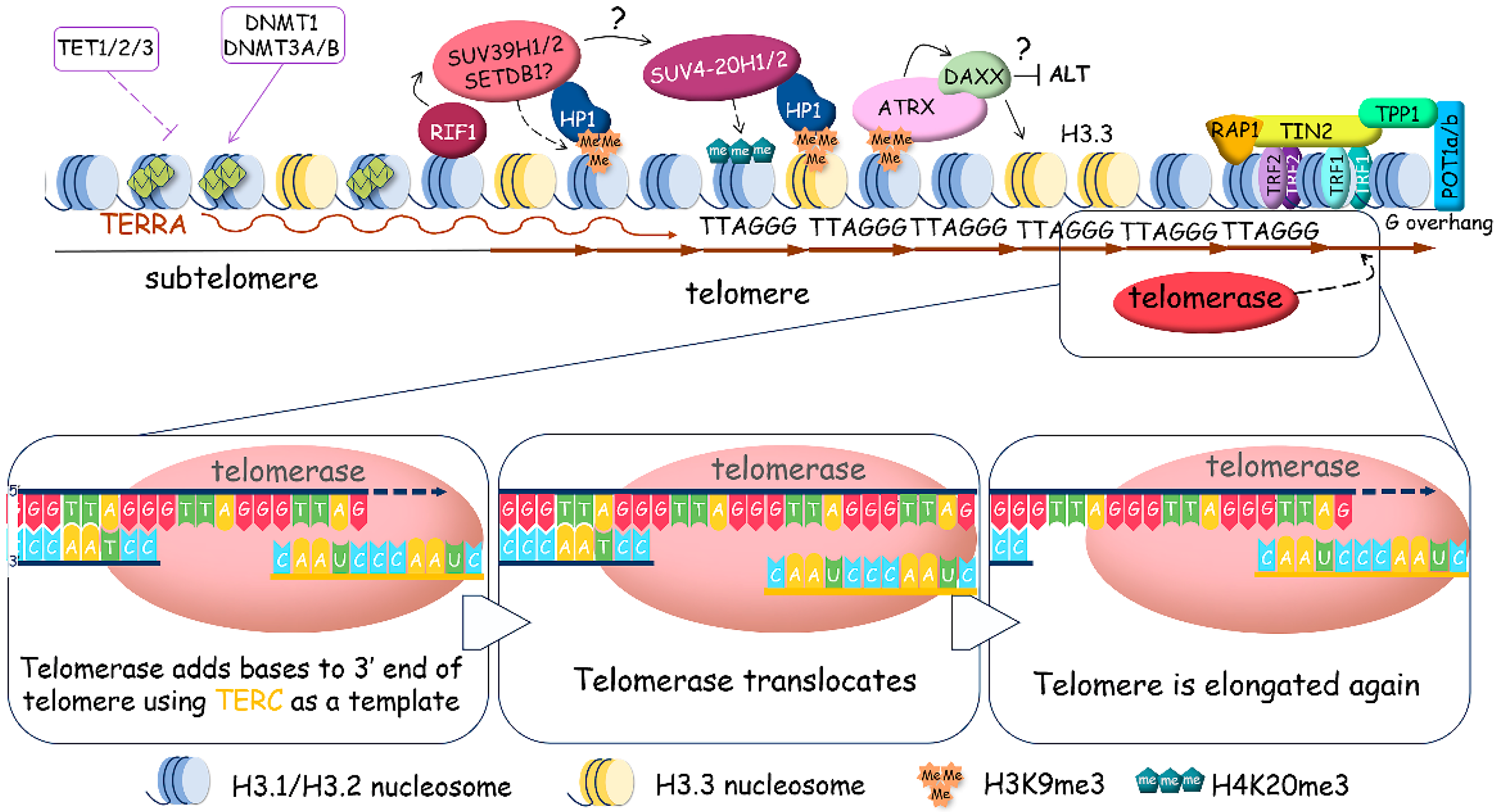
2.3. ALTernative Mechanism of Telomere Lengthening
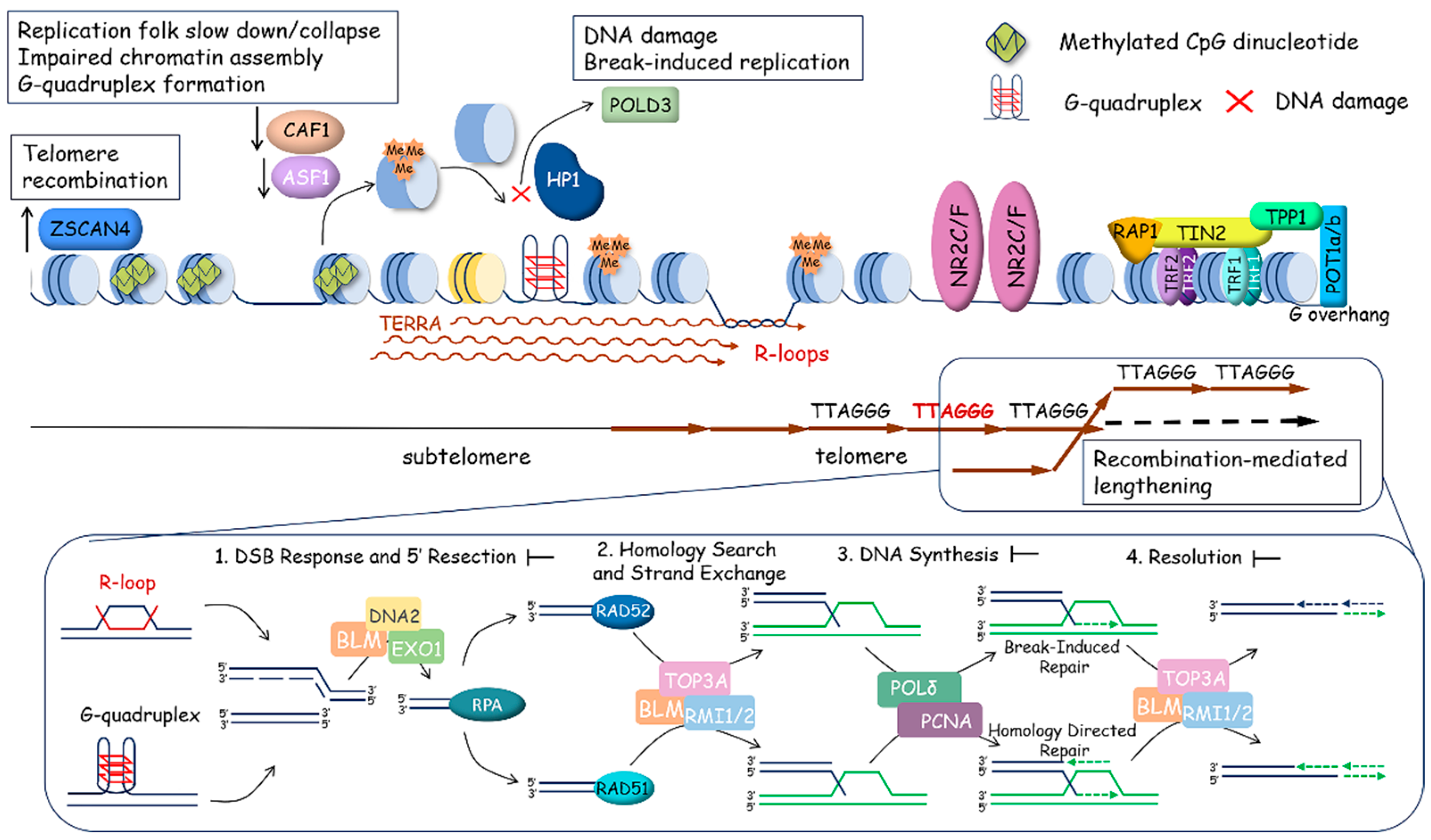
3. Non-Telomeric Roles of Telomere Components
3.1. Noncanonical Functions of Telomeric Proteins
3.2. Noncanonical Functions of Telomerase Components
3.3. Telomeric and Telomerase Components and Mitochondria
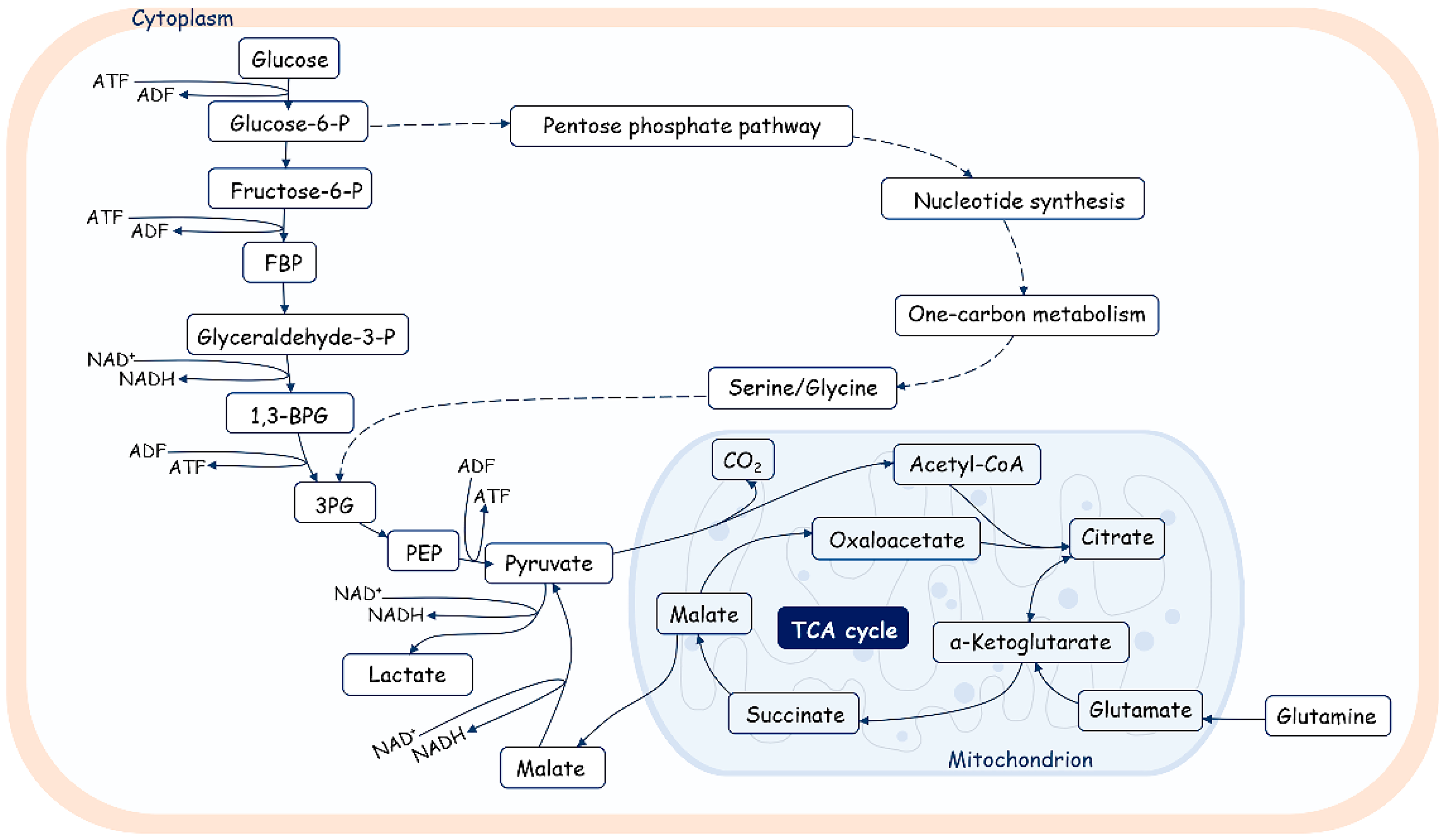
4. Metabolic Programs and Telomere Elongation
4.1. Cellular Metabolic Programs and Telomere Elongation in Cancer Cells
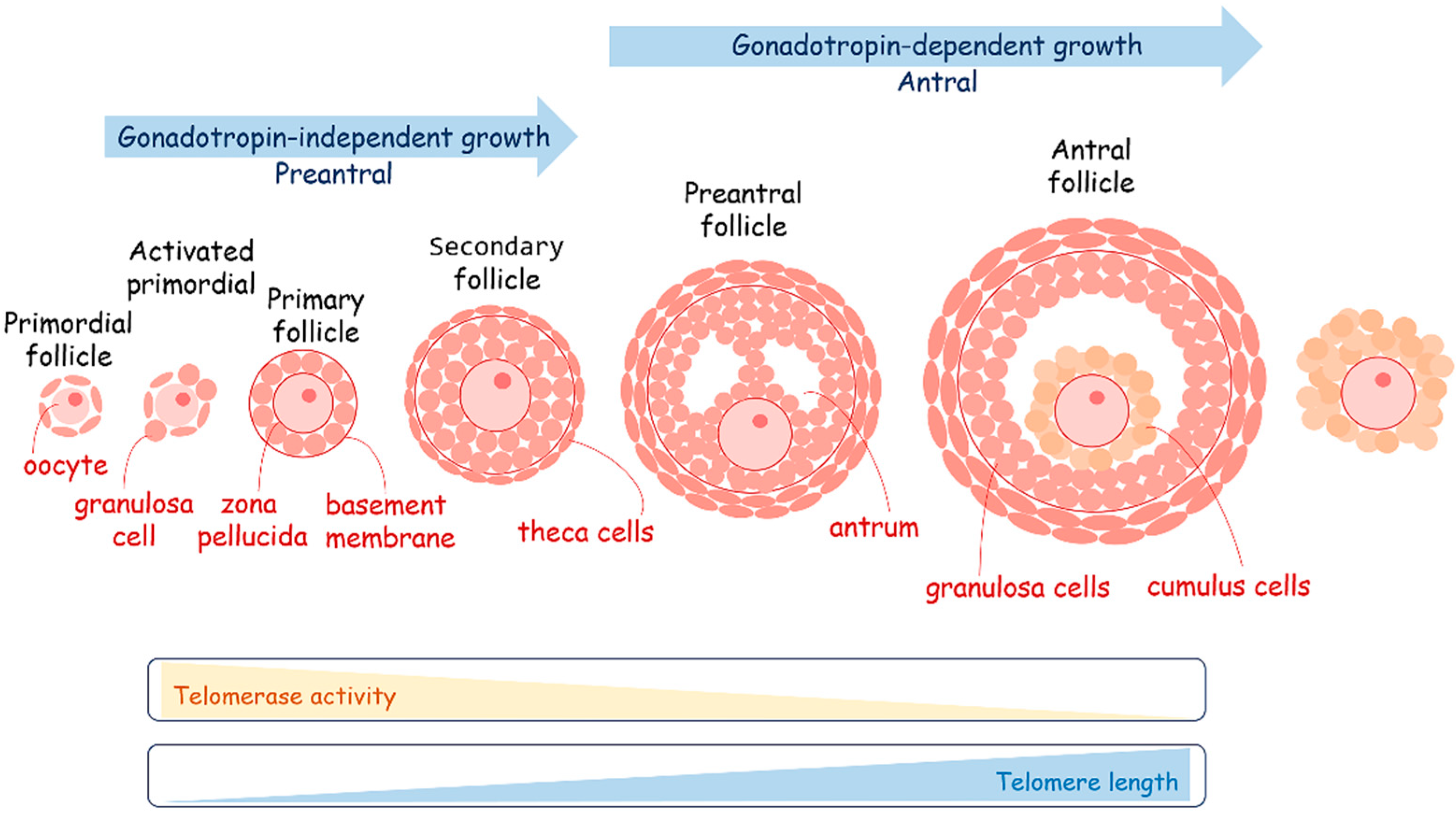
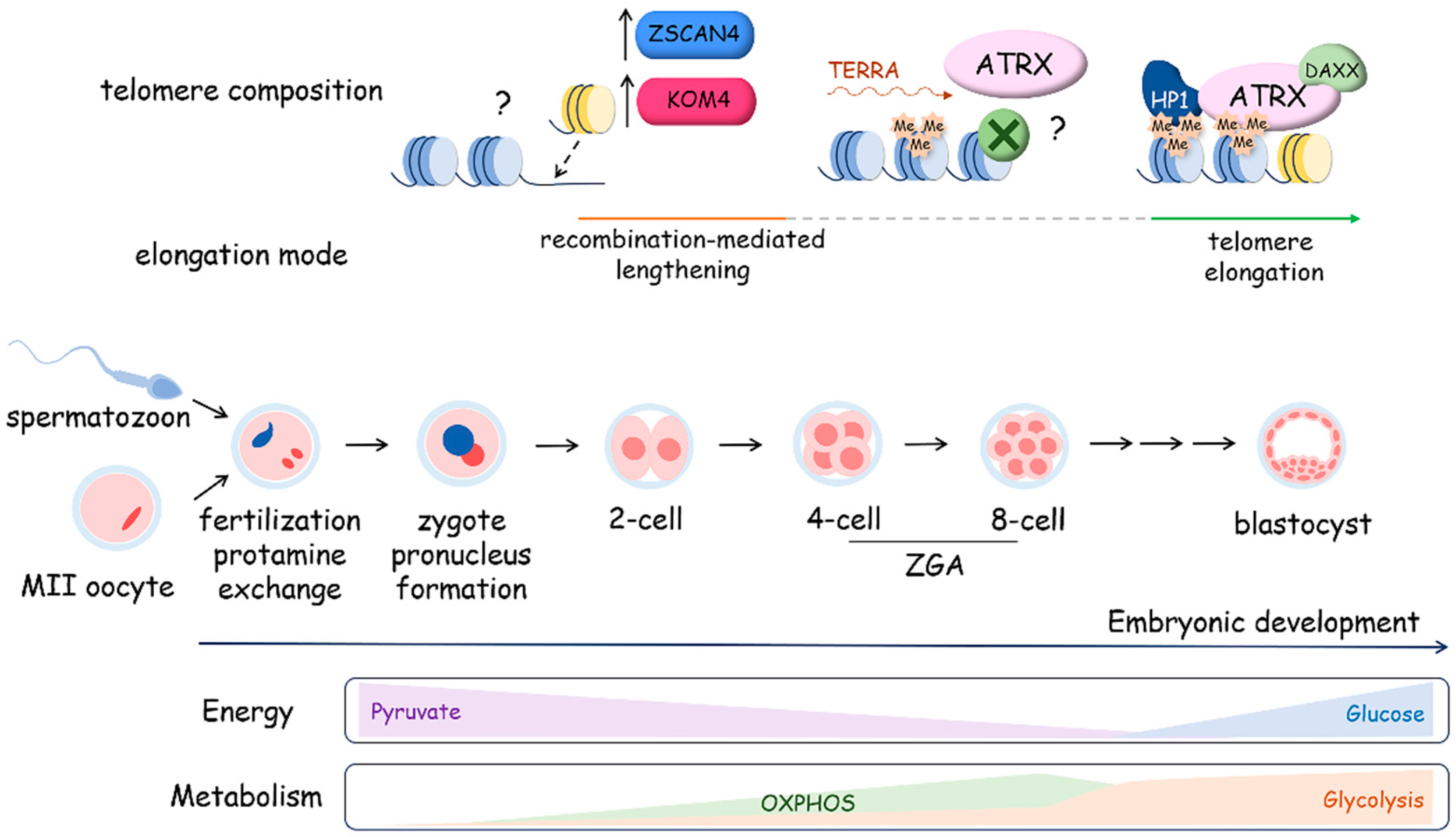
4.2. Metabolism and Telomere Lengthening during Gametogenesis and in Early Development
4.2.1. Metabolism and Telomere Lengthening during Spermatogenesis
4.2.2. Metabolism and Telomere Lengthening during Oogenesis
4.2.3. Metabolism and Telomere Lengthening during Early Development
4.3. Metabolism and Telomere Lengthening during Immune Cell Activation
5. Targets and Approaches for Telomere Reprogramming
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Olovnikov, A.M. A Theory of Marginotomy. The Incomplete Copying of Template Margin in Enzymic Synthesis of Polynucleotides and Biological Significance of the Phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Chow, T.T.; Zhao, Y.; Mak, S.S.; Shay, J.W.; Wright, W.E. Early and Late Steps in Telomere Overhang Processing in Normal Human Cells: The Position of the Final RNA Primer Drives Telomere Shortening. Genes Dev. 2012, 26, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.Z.; Allsopp, R.C.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere End-Replication Problem and Cell Aging. J. Mol. Biol. 1992, 225, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, M.; Rossiello, F.; Clerici, M.; Barozzi, S.; Cittaro, D.; Kaplunov, J.M.; Bucci, G.; Dobreva, M.; Matti, V.; Beausejour, C.M.; et al. Telomeric DNA Damage Is Irreparable and Causes Persistent DNA-Damage-Response Activation. Nat. Cell Biol. 2012, 14, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.E.; Shay, J.W. Role of Telomerase in Cellular Proliferation and Cancer. J. Cell. Physiol. 1999, 180, 10–18. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of Life-Span by Introduction of Telomerase into Normal Human Cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- Weng, N. Telomere and Adaptive Immunity. Mech. Ageing Dev. 2008, 129, 60–66. [Google Scholar] [CrossRef]
- Weng, N.P.; Levine, B.L.; June, C.H.; Hodes, R.J. Regulated Expression of Telomerase Activity in Human T Lymphocyte Development and Activation. J. Exp. Med. 1996, 183, 2471–2479. [Google Scholar] [CrossRef]
- Lu, W.-Y.; Forbes, S.J. Telomerase Activity Links to Regenerative Capacity of Hepatocytes. Transplantation 2018, 102, 1587–1588. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. A Telomeric Sequence in the RNA of Tetrahymena Telomerase Required for Telomere Repeat Synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse Transcriptase Motifs in the Catalytic Subunit of Telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; Yadav, T.; Ouyang, J.; Lan, L.; Zou, L. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 2019, 26, 955–968.e3. [Google Scholar] [CrossRef] [PubMed]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A Highly Conserved Repetitive DNA Sequence, (TTAGGG)n, Present at the Telomeres of Human Chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the Human Telomere Sequence (TTAGGG)n among Vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.L.; Hirose, Y.; Langmore, J.P. Long G Tails at Both Ends of Human Chromosomes Suggest a C Strand Degradation Mechanism for Telomere Shortening. Cell 1997, 88, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres Shorten during Ageing of Human Fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Vaziri, H.; Patterson, C.; Goldstein, S.; Younglai, E.V.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere Length Predicts Replicative Capacity of Human Fibroblasts. Proc. Natl. Acad. Sci. USA 1992, 89, 10114–10118. [Google Scholar] [CrossRef]
- Morin, G.B. The Human Telomere Terminal Transferase Enzyme Is a Ribonucleoprotein That Synthesizes TTAGGG Repeats. Cell 1989, 59, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an Alternative Mechanism for Maintaining Telomere Length in Human Tumors and Tumor-Derived Cell Lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef]
- Wright, W.E.; Piatyszek, M.A.; Rainey, W.E.; Byrd, W.; Shay, J.W. Telomerase Activity in Human Germline and Embryonic Tissues and Cells. Dev. Genet. 1996, 18, 173–179. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin: The Protein Complex That Shapes and Safeguards Human Telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; O’Connor, M.S.; Qin, J.; Songyang, Z. Telosome, a Mammalian Telomere-Associated Complex Formed by Multiple Telomeric Proteins. J. Biol. Chem. 2004, 279, 51338–51342. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; Cech, T.R. Pot1, the Putative Telomere End-Binding Protein in Fission Yeast and Humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Stansel, R.M. T-Loop Assembly in Vitro Involves Binding of TRF2 near the 3′ Telomeric Overhang. EMBO J. 2001, 20, 5532–5540. [Google Scholar] [CrossRef]
- Pisano, S.; Marchioni, E.; Galati, A.; Mechelli, R.; Savino, M.; Cacchione, S. Telomeric Nucleosomes Are Intrinsically Mobile. J. Mol. Biol. 2007, 369, 1153–1162. [Google Scholar] [CrossRef]
- Fajkus, J.; Trifonov, E.N. Columnar Packing of Telomeric Nucleosomes. Biochem. Biophys. Res. Commun. 2001, 280, 961–963. [Google Scholar] [CrossRef]
- Soman, A.; Liew, C.W.; Teo, H.L.; Berezhnoy, N.V.; Olieric, V.; Korolev, N.; Rhodes, D.; Nordenskiöld, L. The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties. Nucleic Acids Res. 2020, 48, 5383–5396. [Google Scholar] [CrossRef]
- Soman, A.; Wong, S.Y.; Korolev, N.; Surya, W.; Lattmann, S.; Vogirala, V.K.; Chen, Q.; Berezhnoy, N.V.; Van Noort, J.; Rhodes, D.; et al. Columnar Structure of Human Telomeric Chromatin. Nature 2022, 609, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Williamson, J.R.; Raghuraman, M.K.; Cech, T.R. Monovalent Cation-Induced Structure of Telomeric DNA: The G-Quartet Model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Klug, A. Telomeric DNA Dimerizes by Formation of Guanine Tetrads between Hairpin Loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef]
- Thomas, M.; White, R.L.; Davis, R.W. Hybridization of RNA to Double-Stranded DNA: Formation of R-Loops. Proc. Natl. Acad. Sci. USA 1976, 73, 2294–2298. [Google Scholar] [CrossRef] [PubMed]
- White, R.L.; Hogness, D.S. R Loop Mapping of the 18S and 28S Sequences in the Long and Short Repeating Units of Drosophila Melanogaster rDNA. Cell 1977, 10, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally Regulated Transcription of Mammalian Telomeres by DNA-Dependent RNA Polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Graf, M.; Bonetti, D.; Lockhart, A.; Serhal, K.; Kellner, V.; Maicher, A.; Jolivet, P.; Teixeira, M.T.; Luke, B. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 2017, 170, 72–85.e14. [Google Scholar] [CrossRef]
- Balk, B.; Dees, M.; Bender, K.; Luke, B. The Differential Processing of Telomeres in Response to Increased Telomeric Transcription and RNA–DNA Hybrid Accumulation. RNA Biol. 2014, 11, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Balk, B.; Maicher, A.; Dees, M.; Klermund, J.; Luke-Glaser, S.; Bender, K.; Luke, B. Telomeric RNA-DNA Hybrids Affect Telomere-Length Dynamics and Senescence. Nat. Struct. Mol. Biol. 2013, 20, 1199–1205. [Google Scholar] [CrossRef]
- Nandakumar, J.; Bell, C.F.; Weidenfeld, I.; Zaug, A.J.; Leinwand, L.A.; Cech, T.R. The TEL Patch of Telomere Protein TPP1 Mediates Telomerase Recruitment and Processivity. Nature 2012, 492, 285–289. [Google Scholar] [CrossRef]
- Zhong, F.L.; Batista, L.F.Z.; Freund, A.; Pech, M.F.; Venteicher, A.S.; Artandi, S.E. TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends. Cell 2012, 150, 481–494. [Google Scholar] [CrossRef]
- Sexton, A.N.; Youmans, D.T.; Collins, K. Specificity Requirements for Human Telomere Protein Interaction with Telomerase Holoenzyme. J. Biol. Chem. 2012, 287, 34455–34464. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Zaug, A.J.; Kufer, R.; Cech, T.R. Dynamics of Human Telomerase Recruitment Depend on Template-Telomere Base Pairing. Mol. Biol. Cell 2018, 29, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.C.; Zaug, A.J.; Cech, T.R. Live Cell Imaging Reveals the Dynamics of Telomerase Recruitment to Telomeres. Cell 2016, 166, 1188–1197.e9. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.L.; Centore, R.C.; O’Sullivan, R.J.; Rai, R.; Tse, A.; Songyang, Z.; Chang, S.; Karlseder, J.; Zou, L. TERRA and hnRNPA1 Orchestrate an RPA-to-POT1 Switch on Telomeric Single-Stranded DNA. Nature 2011, 471, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Safari, A.; O’Connor, M.S.; Chan, D.W.; Laegeler, A.; Qin, J.; Songyang, Z. PTOP Interacts with POT1 and Regulates Its Localization to Telomeres. Nat. Cell Biol. 2004, 6, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Zaug, A.J.; Podell, E.R.; Cech, T.R. Switching Human Telomerase on and off with hPOT1 Protein in Vitro. J. Biol. Chem. 2005, 280, 20449–20456. [Google Scholar] [CrossRef] [PubMed]
- Rubtsova, M.P.; Skvortsov, D.A.; Petruseva, I.O.; Lavrik, O.I.; Spirin, P.V.; Prasolov, V.S.; Kisseljov, F.L.; Dontsova, O.A. Replication Protein A Modulates the Activity of Human Telomerase in Vitro. Biochem. 2009, 74, 92–96. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Sfeir, A.J.; Shay, J.W.; Wright, W.E.; de Lange, T. POT1 Protects Telomeres from a Transient DNA Damage Response and Determines How Human Chromosomes End. EMBO J. 2005, 24, 2667–2678. [Google Scholar] [CrossRef]
- Denchi, E.L.; de Lange, T. Protection of Telomeres through Independent Control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The POT1-TPP1 Telomere Complex Is a Telomerase Processivity Factor. Nature 2007, 445, 506–510. [Google Scholar] [CrossRef]
- Heaphy, C.M.; de Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered Telomeres in Tumors with ATRX and DAXX Mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef]
- Law, M.J.; Lower, K.M.; Voon, H.P.J.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X Syndrome Protein Targets Tandem Repeats and Influences Allele-Specific Expression in a Size-Dependent Manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Iwase, S.; Xiang, B.; Ghosh, S.; Ren, T.; Lewis, P.W.; Cochrane, J.C.; Allis, C.D.; Picketts, D.J.; Patel, D.J.; Li, H.; et al. ATRX ADD Domain Links an Atypical Histone Methylation Recognition Mechanism to Human Mental-Retardation Syndrome. Nat. Struct. Mol. Biol. 2011, 18, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.-Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.-A.K.; Tönjes, M.; et al. Driver Mutations in Histone H3.3 and Chromatin Remodelling Genes in Paediatric Glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, J.; Johannessen, T.-C.; Ohba, S.; Chow, T.T.; Jones, L.; Pandita, A.; Pieper, R.O. Mutant IDH1 Cooperates with ATRX Loss to Drive the Alternative Lengthening of Telomere Phenotype in Glioma. Cancer Res. 2018, 78, 2966–2977. [Google Scholar] [CrossRef]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537.e5. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.-A.; Jones, D.T.W.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef]
- Chou, F.-J.; Liu, Y.; Lang, F.; Yang, C. D-2-Hydroxyglutarate in Glioma Biology. Cells 2021, 10, 2345. [Google Scholar] [CrossRef] [PubMed]
- Yeager, T.R.; Neumann, A.A.; Englezou, A.; Huschtscha, L.I.; Noble, J.R.; Reddel, R.R. Telomerase-Negative Immortalized Human Cells Contain a Novel Type of Promyelocytic Leukemia (PML) Body. Cancer Res. 1999, 59, 4175–4179. [Google Scholar] [PubMed]
- Henson, J.D.; Cao, Y.; Huschtscha, L.I.; Chang, A.C.; Au, A.Y.M.; Pickett, H.A.; Reddel, R.R. DNA C-Circles Are Specific and Quantifiable Markers of Alternative-Lengthening-of-Telomeres Activity. Nat. Biotechnol. 2009, 27, 1181–1185. [Google Scholar] [CrossRef]
- Wu, G.; Jiang, X.; Lee, W.-H.; Chen, P.-L. Assembly of Functional ALT-Associated Promyelocytic Leukemia Bodies Requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003, 63, 2589–2595. [Google Scholar]
- Grudic, A.; Jul-Larsen, Å.; Haring, S.J.; Wold, M.S.; Lønning, P.E.; Bjerkvig, R.; Bøe, S.O. Replication Protein A Prevents Accumulation of Single-Stranded Telomeric DNA in Cells That Use Alternative Lengthening of Telomeres. Nucleic Acids Res. 2007, 35, 7267–7278. [Google Scholar] [CrossRef] [PubMed]
- Barroso-González, J.; García-Expósito, L.; Hoang, S.M.; Lynskey, M.L.; Roncaioli, J.L.; Ghosh, A.; Wallace, C.T.; Modesti, M.; Bernstein, K.A.; Sarkar, S.N.; et al. RAD51AP1 Is an Essential Mediator of Alternative Lengthening of Telomeres. Mol. Cell 2019, 76, 217. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Kaul, Z.; Gocha, A.S.; Martinez, A.R.; Harris, J.; Parvin, J.D.; Groden, J. Association of BLM and BRCA1 during Telomere Maintenance in ALT Cells. PLoS ONE 2014, 9, e103819. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Drosopoulos, W.C.; Sethi, L.; Madireddy, A.; Schildkraut, C.L.; Zhang, D. FANCM, BRCA1, and BLM Cooperatively Resolve the Replication Stress at the ALT Telomeres. Proc. Natl. Acad. Sci. USA 2017, 114, E5940–E5949. [Google Scholar] [CrossRef] [PubMed]
- Vinayagamurthy, S.; Bagri, S.; Mergny, J.-L.; Chowdhury, S. Telomeres Expand Sphere of Influence: Emerging Molecular Impact of Telomeres in Non-Telomeric Functions. Trends Genet. 2023, 39, 59–73. [Google Scholar] [CrossRef]
- Martinez, P.; Thanasoula, M.; Carlos, A.R.; Gómez-López, G.; Tejera, A.M.; Schoeftner, S.; Dominguez, O.; Pisano, D.G.; Tarsounas, M.; Blasco, M.A. Mammalian Rap1 Controls Telomere Function and Gene Expression through Binding to Telomeric and Extratelomeric Sites. Nat. Cell Biol. 2010, 12, 768–780. [Google Scholar] [CrossRef]
- Janoušková, E.; Nečasová, I.; Pavloušková, J.; Zimmermann, M.; Hluchý, M.; Marini, V.; Nováková, M.; Hofr, C. Human Rap1 Modulates TRF2 Attraction to Telomeric DNA. Nucleic Acids Res. 2015, 43, 2691–2700. [Google Scholar] [CrossRef]
- Zizza, P.; Dinami, R.; Porru, M.; Cingolani, C.; Salvati, E.; Rizzo, A.; D’Angelo, C.; Petti, E.; Amoreo, C.A.; Mottolese, M.; et al. TRF2 Positively Regulates SULF2 Expression Increasing VEGF-A Release and Activity in Tumor Microenvironment. Nucleic Acids Res. 2019, 47, 3365–3382. [Google Scholar] [CrossRef]
- Biroccio, A.; Cherfils-Vicini, J.; Augereau, A.; Pinte, S.; Bauwens, S.; Ye, J.; Simonet, T.; Horard, B.; Jamet, K.; Cervera, L.; et al. TRF2 Inhibits a Cell-Extrinsic Pathway through Which Natural Killer Cells Eliminate Cancer Cells. Nat. Cell Biol. 2013, 15, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Wu, D.; Lin, J.; Countryman, P.; Bradford, K.C.; Erie, D.A.; Riehn, R.; Opresko, P.L.; Wang, H. Enhanced Electrostatic Force Microscopy Reveals Higher-Order DNA Looping Mediated by the Telomeric Protein TRF2. Sci. Rep. 2016, 6, 20513. [Google Scholar] [CrossRef]
- Wood, A.M.; Danielsen, J.M.R.; Lucas, C.A.; Rice, E.L.; Scalzo, D.; Shimi, T.; Goldman, R.D.; Smith, E.D.; Le Beau, M.M.; Kosak, S.T. TRF2 and Lamin A/C Interact to Facilitate the Functional Organization of Chromosome Ends. Nat. Commun. 2014, 5, 5467. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.D.; Garza-Gongora, A.G.; MacQuarrie, K.L.; Kosak, S.T. Interstitial Telomeric Loops and Implications of the Interaction between TRF2 and Lamin A/C. Differentiation 2018, 102, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Roy, S.; Kar, M.; Roy, S.; Thakur, S.; Padhi, S.; Akhter, Y.; Banerjee, B. Role of Telomeric TRF2 in Orosphere Formation and CSC Phenotype Maintenance Through Efficient DNA Repair Pathway and Its Correlation with Recurrence in OSCC. Stem Cell Rev. Rep. 2018, 14, 871–887. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.P.; Garrobo, I.; Foronda, M.; Palacios, J.A.; Marión, R.M.; Flores, I.; Ortega, S.; Blasco, M.A. TRF1 Is a Stem Cell Marker and Is Essential for the Generation of Induced Pluripotent Stem Cells. Nat. Commun. 2013, 4, 1946. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, T.; Muramatsu, Y.; Yoshida, H.; Seimiya, H. TRF1 Ensures the Centromeric Function of Aurora-B and Proper Chromosome Segregation. Mol. Cell. Biol. 2014, 34, 2464–2478. [Google Scholar] [CrossRef]
- Lee, J.; Gollahon, L. Mitotic Perturbations Induced by Nek2 Overexpression Require Interaction with TRF1 in Breast Cancer Cells. Cell Cycle 2013, 12, 3599–3614. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ségal-Bendirdjian, E.; Geli, V. Non-Canonical Roles of Telomerase: Unraveling the Imbroglio. Front. Cell Dev. Biol. 2019, 7, 332. [Google Scholar] [CrossRef]
- Ghosh, A.; Saginc, G.; Leow, S.C.; Khattar, E.; Shin, E.M.; Yan, T.D.; Wong, M.; Zhang, Z.; Li, G.; Sung, W.-K.; et al. Telomerase Directly Regulates NF-κB-Dependent Transcription. Nat. Cell Biol. 2012, 14, 1270–1281. [Google Scholar] [CrossRef]
- Ding, D.; Xi, P.; Zhou, J.; Wang, M.; Cong, Y.-S. Human Telomerase Reverse Transcriptase Regulates MMP Expression Independently of Telomerase Activity via NF-κB-Dependent Transcription. FASEB J. 2013, 27, 4375–4383. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Hubbard, A.K.; Giardina, C. NF-κB Regulates Transcription of the Mouse Telomerase Catalytic Subunit. J. Biol. Chem. 2000, 275, 36671–36675. [Google Scholar] [CrossRef]
- Li, Y.; Tergaonkar, V. Noncanonical Functions of Telomerase: Implications in Telomerase-Targeted Cancer Therapies. Cancer Res. 2014, 74, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.A.; O’Hagan, R.C.; Deng, H.; Xiao, Q.; Hann, S.R.; Adams, R.R.; Lichtsteiner, S.; Chin, L.; Morin, G.B.; DePinho, R.A. Telomerase Reverse Transcriptase Gene Is a Direct Target of C-Myc but Is Not Functionally Equivalent in Cellular Transformation. Oncogene 1999, 18, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.J.; Grandori, C.; Amacker, M.; Simon-Vermot, N.; Polack, A.; Lingner, J.; Dalla-Favera, R. Direct Activation of TERT Transcription by C-MYC. Nat. Genet. 1999, 21, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Muller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase Regulates MYC-Driven Oncogenesis Independent of Its Reverse Transcriptase Activity. J. Clin. Investig. 2015, 125, 2109–2122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zheng, D.; Wang, M.; Cong, Y.-S. Telomerase Reverse Transcriptase Activates the Expression of Vascular Endothelial Growth Factor Independent of Telomerase Activity. Biochem. Biophys. Res. Commun. 2009, 386, 739–743. [Google Scholar] [CrossRef]
- Liu, N.; Ding, D.; Hao, W.; Yang, F.; Wu, X.; Wang, M.; Xu, X.; Ju, Z.; Liu, J.-P.; Song, Z.; et al. hTERT Promotes Tumor Angiogenesis by Activating VEGF via Interactions with the Sp1 Transcription Factor. Nucleic Acids Res. 2016, 44, 8693–8703. [Google Scholar] [CrossRef]
- Young, J.I.; Sedivy, J.M.; Smith, J.R. Telomerase Expression in Normal Human Fibroblasts Stabilizes DNA 5-Methylcytosine Transferase I. J. Biol. Chem. 2003, 278, 19904–19908. [Google Scholar] [CrossRef]
- Yu, J.; Yuan, X.; Sjöholm, L.; Liu, T.; Kong, F.; Ekström, T.J.; Björkholm, M.; Xu, D. Telomerase Reverse Transcriptase Regulates DNMT3B Expression/Aberrant DNA Methylation Phenotype and AKT Activation in Hepatocellular Carcinoma. Cancer Lett. 2018, 434, 33–41. [Google Scholar] [CrossRef]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase Modulates Wnt Signalling by Association with Target Gene Chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Ge, Y.; Liu, J.; Zhao, Y. TERC Promotes Cellular Inflammatory Response Independent of Telomerase. Nucleic Acids Res. 2019, 47, 8084–8095. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz-Pérez, F.; García-Castillo, J.; García-Moreno, D.; López-Muñoz, A.; Anchelin, M.; Angosto, D.; Zon, L.I.; Mulero, V.; Cayuela, M.L. A Non-Canonical Function of Telomerase RNA in the Regulation of Developmental Myelopoiesis in Zebrafish. Nat. Commun. 2014, 5, 3228. [Google Scholar] [CrossRef] [PubMed]
- Masutomi, K.; Possemato, R.; Wong, J.M.Y.; Currier, J.L.; Tothova, Z.; Manola, J.B.; Ganesan, S.; Lansdorp, P.M.; Collins, K.; Hahn, W.C. The Telomerase Reverse Transcriptase Regulates Chromatin State and DNA Damage Responses. Proc. Natl. Acad. Sci. USA 2005, 102, 8222–8227. [Google Scholar] [CrossRef] [PubMed]
- Kedde, M.; le Sage, C.; Duursma, A.; Zlotorynski, E.; van Leeuwen, B.; Nijkamp, W.; Beijersbergen, R.; Agami, R. Telomerase-Independent Regulation of ATR by Human Telomerase RNA. J. Biol. Chem. 2006, 281, 40503–40514. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, F.S.; Blackburn, E.H. An Antiapoptotic Role for Telomerase RNA in Human Immune Cells Independent of Telomere Integrity or Telomerase Enzymatic Activity. Blood 2014, 124, 3675–3684. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Jiang, S.; Zhang, L.; Yu, Z. Mitochondrial Electron Transport Chain, ROS Generation and Uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How Mitochondria Produce Reactive Oxygen Species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, S.; Oikawa, S. Mechanism of Telomere Shortening by Oxidative Stress. Ann. N. Y. Acad. Sci. 2004, 1019, 278–284. [Google Scholar] [CrossRef]
- Forsyth, N.R.; Evans, A.P.; Shay, J.W.; Wright, W.E. Developmental Differences in the Immortalization of Lung Fibroblasts by Telomerase. Aging Cell 2003, 2, 235–243. [Google Scholar] [CrossRef]
- Passos, J.F.; Saretzki, G.; Ahmed, S.; Nelson, G.; Richter, T.; Peters, H.; Wappler, I.; Birket, M.J.; Harold, G.; Schaeuble, K.; et al. Mitochondrial Dysfunction Accounts for the Stochastic Heterogeneity in Telomere-Dependent Senescence. PLoS Biol. 2007, 5, e110. [Google Scholar] [CrossRef]
- Armstrong, E.; Boonekamp, J. Does Oxidative Stress Shorten Telomeres in Vivo? A Meta-Analysis. Ageing Res. Rev. 2023, 85, 101854. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, D.M.K.; Polderman, P.E.; Den Toom, W.T.F.; Keijer, J.P.; Van Roosmalen, M.J.; Leyten, T.M.F.; Lehmann, J.; Zwakenberg, S.; De Henau, S.; Van Boxtel, R.; et al. Mitochondrial H2O2 Release Does Not Directly Cause Damage to Chromosomal DNA. Nat. Commun. 2024, 15, 2725. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Zhang, Y.; Zhang, Q.; Li, H.; Luo, Z.; Fang, H.; Kim, S.H.; Qin, L.; Yotnda, P.; Xu, J.; et al. Mitochondrial Localization of Telomeric Protein TIN2 Links Telomere Regulation to Metabolic Control. Mol. Cell 2012, 47, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.; Meyer, J.N.; Skorvaga, M.; Annab, L.A.; Van Houten, B. Mitochondrial hTERT Exacerbates Free-Radical-Mediated mtDNA Damage: Mitochondrial Telomerase, J. Hertzog Santos et Al. Aging Cell 2004, 3, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.; Meyer, J.N.; Van Houten, B. Mitochondrial Localization of Telomerase as a Determinant for Hydrogen Peroxide-Induced Mitochondrial DNA Damage and Apoptosis. Hum. Mol. Genet. 2006, 15, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Passos, J.F.; Birket, M.J.; Beckmann, T.; Brings, S.; Peters, H.; Birch-Machin, M.A.; von Zglinicki, T.; Saretzki, G. Telomerase Does Not Counteract Telomere Shortening but Protects Mitochondrial Function under Oxidative Stress. J. Cell Sci. 2008, 121, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Reyes, A.; Green, P.; Caron, M.J.; Bonini, M.G.; Gordon, D.M.; Holt, I.J.; Santos, J.H. Human Telomerase Acts as a hTR-Independent Reverse Transcriptase in Mitochondria. Nucleic Acids Res. 2012, 40, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Marinaccio, J.; Micheli, E.; Udroiu, I.; Di Nottia, M.; Carrozzo, R.; Baranzini, N.; Grimaldi, A.; Leone, S.; Moreno, S.; Muzzi, M.; et al. TERT Extra-Telomeric Roles: Antioxidant Activity and Mitochondrial Protection. Int. J. Mol. Sci. 2023, 24, 4450. [Google Scholar] [CrossRef]
- Mattiussi, M.; Tilman, G.; Lenglez, S.; Decottignies, A. Human Telomerase Represses ROS-Dependent Cellular Responses to Tumor Necrosis Factor-α without Affecting NF-κB Activation. Cell. Signal. 2012, 24, 708–717. [Google Scholar] [CrossRef]
- Green, P.; Sharma, N.; Santos, J. Telomerase Impinges on the Cellular Response to Oxidative Stress Through Mitochondrial ROS-Mediated Regulation of Autophagy. Int. J. Mol. Sci. 2019, 20, 1509. [Google Scholar] [CrossRef]
- Martens, A.; Schmid, B.; Akintola, O.; Saretzki, G. Telomerase Does Not Improve DNA Repair in Mitochondria upon Stress but Increases MnSOD Protein under Serum-Free Conditions. Int. J. Mol. Sci. 2019, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Mohd Zain, M.Z.; Ismail, N.H.; Ahmad, N.; Sulong, S.; Karsani, S.A.; Abdul Majid, N. Telomerase Reverse Transcriptase Downregulation by RNA Interference Modulates Endoplasmic Reticulum Stress and Mitochondrial Energy Production. Mol. Biol. Rep. 2020, 47, 7735–7743. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, S.S.; Skulachev, V.P.; Starkov, A.A. High Protonic Potential Actuates a Mechanism of Production of Reactive Oxygen Species in Mitochondria. FEBS Lett. 1997, 416, 15–18. [Google Scholar] [CrossRef]
- Vyssokikh, M.Y.; Holtze, S.; Averina, O.A.; Lyamzaev, K.G.; Panteleeva, A.A.; Marey, M.V.; Zinovkin, R.A.; Severin, F.F.; Skulachev, M.V.; Fasel, N.; et al. Mild Depolarization of the Inner Mitochondrial Membrane Is a Crucial Component of an Anti-Aging Program. Proc. Natl. Acad. Sci. USA 2020, 117, 6491–6501. [Google Scholar] [CrossRef] [PubMed]
- Skulachev, V.P.; Vyssokikh, M.Y.; Chernyak, B.V.; Mulkidjanian, A.Y.; Skulachev, M.V.; Shilovsky, G.A.; Lyamzaev, K.G.; Borisov, V.B.; Severin, F.F.; Sadovnichii, V.A. Six Functions of Respiration: Isn’t It Time to Take Control over ROS Production in Mitochondria, and Aging Along with It? Int. J. Mol. Sci. 2023, 24, 12540. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.; De Cubas, L.; Bautista, E.; Moral-Blanch, M.; Medraño-Fernández, I.; Sitia, R.; Boronat, S.; Ayté, J.; Hidalgo, E. Monitoring Cytosolic H2O2 Fluctuations Arising from Altered Plasma Membrane Gradients or from Mitochondrial Activity. Nat. Commun. 2019, 10, 4526. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, S. MicroRNAs and tRNA-Derived Small Fragments: Key Messengers in Nuclear–Mitochondrial Communication. Front. Mol. Biosci. 2021, 8, 643575. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, P.; Zheng, Q.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Tong, T.; et al. Mitochondrial Trafficking and Processing of Telomerase RNA TERC. Cell Rep. 2018, 24, 2589–2595. [Google Scholar] [CrossRef]
- Zheng, Q.; Liu, P.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Di, F.; Tong, T.; et al. Mitochondrion-Processed TERC Regulates Senescence without Affecting Telomerase Activities. Protein Cell 2019, 10, 631–648. [Google Scholar] [CrossRef]
- Hoffman, H.; Grigg, G.W. An Electron Microscopic Study of Mitochondria Formation. Exp. Cell Res. 1958, 15, 118–131. [Google Scholar] [CrossRef]
- Jiang, X.; Hou, B.; Xu, Y.; Li, E.; Cao, P.; Liu, S.; Xi, Z.; Yang, H.; Huo, Y.; Che, Y. Mitochondria Fragment and Reassemble to Initiate the Formation and Development of the Nucleus. bioRxiv 2020, 2020.09.29.319723. [Google Scholar] [CrossRef]
- Shin, W.H.; Chung, K.C. Human Telomerase Reverse Transcriptase Positively Regulates Mitophagy by Inhibiting the Processing and Cytoplasmic Release of Mitochondrial PINK1. Cell Death Dis. 2020, 11, 425. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, S.; Xie, W.; Wang, Q.; Luo, Q.; Huang, M.; Gu, M.; Lan, P.; Chen, D. MCCC2 Is a Novel Mediator between Mitochondria and Telomere and Functions as an Oncogene in Colorectal Cancer. Cell Mol. Biol. Lett. 2023, 28, 80. [Google Scholar] [CrossRef] [PubMed]
- Rubtsova, M.; Naraykina, Y.; Vasilkova, D.; Meerson, M.; Zvereva, M.; Prassolov, V.; Lazarev, V.; Manuvera, V.; Kovalchuk, S.; Anikanov, N.; et al. Protein Encoded in Human Telomerase RNA Is Involved in Cell Protective Pathways. Nucleic Acids Res. 2018, 46, 8966–8977. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Hall, M.N.; Lin, S.-C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020, 31, 472–492. [Google Scholar] [CrossRef]
- Shliapina, V.L.; Yurtaeva, S.V.; Rubtsova, M.P.; Dontsova, O.A. At the Crossroads: Mechanisms of Apoptosis and Autophagy in Cell Life and Death. Acta Naturae 2021, 13, 106–115. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A Nutrient and Energy Sensor That Maintains Energy Homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of Metabolism and Mitochondrial Homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]
- Shliapina, V.; Koriagina, M.; Vasilkova, D.; Govorun, V.; Dontsova, O.; Rubtsova, M. Human Telomerase RNA Protein Encoded by Telomerase RNA Is Involved in Metabolic Responses. Front. Cell Dev. Biol. 2021, 9, 754611. [Google Scholar] [CrossRef]
- Rubtsova, M.; Dontsova, O. How Structural Features Define Biogenesis and Function of Human Telomerase RNA Primary Transcript. Biomedicines 2022, 10, 1650. [Google Scholar] [CrossRef]
- Bayne, S.; Liu, J.-P. Hormones and Growth Factors Regulate Telomerase Activity in Ageing and Cancer. Mol. Cell. Endocrinol. 2005, 240, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.S.; Cheng, N.-L.; Kim, J.; An, J.; Dong, F.; Yang, Q.; Zou, I.; Weng, N. Human T Cell Differentiation Negatively Regulates Telomerase Expression Resulting in Reduced Activation-Induced Proliferation and Survival. Front. Immunol. 2019, 10, 1993. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.J.; Agathocleous, M.; Morrison, S.J. Metabolic Regulation of Stem Cell Function. J. Intern. Med. 2014, 276, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.; Lochner, M.; Berod, L.; Sparwasser, T. Metabolic Pathways in T Cell Activation and Lineage Differentiation. Semin. Immunol. 2016, 28, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Metabolic Reprogramming: The Emerging Concept and Associated Therapeutic Strategies. J. Exp. Clin. Cancer Res. 2015, 34, 111. [Google Scholar] [CrossRef]
- Miyazawa, H.; Aulehla, A. Revisiting the Role of Metabolism during Development. Development 2018, 145, dev131110. [Google Scholar] [CrossRef]
- Bao, Y.; Mukai, K.; Hishiki, T.; Kubo, A.; Ohmura, M.; Sugiura, Y.; Matsuura, T.; Nagahata, Y.; Hayakawa, N.; Yamamoto, T.; et al. Energy Management by Enhanced Glycolysis in G1-Phase in Human Colon Cancer Cells In Vitro and In Vivo. Mol. Cancer Res. 2013, 11, 973–985. [Google Scholar] [CrossRef]
- Mitra, K.; Wunder, C.; Roysam, B.; Lin, G.; Lippincott-Schwartz, J. A Hyperfused Mitochondrial State Achieved at G 1 –S Regulates Cyclin E Buildup and Entry into S Phase. Proc. Natl. Acad. Sci. USA 2009, 106, 11960–11965. [Google Scholar] [CrossRef] [PubMed]
- Tudzarova, S.; Colombo, S.L.; Stoeber, K.; Carcamo, S.; Williams, G.H.; Moncada, S. Two Ubiquitin Ligases, APC/C-Cdh1 and SKP1-CUL1-F (SCF)-β-TrCP, Sequentially Regulate Glycolysis during the Cell Cycle. Proc. Natl. Acad. Sci. USA 2011, 108, 5278–5283. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, M.; Candas, D.; Zhang, T.-Q.; Qin, L.; Eldridge, A.; Wachsmann-Hogiu, S.; Ahmed, K.M.; Chromy, B.A.; Nantajit, D.; et al. Cyclin B1/Cdk1 Coordinates Mitochondrial Respiration for Cell-Cycle G2/M Progression. Dev. Cell 2014, 29, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Sutter, B.M.; Li, B.; Tu, B.P. Acetyl-CoA Induces Cell Growth and Proliferation by Promoting the Acetylation of Histones at Growth Genes. Mol. Cell 2011, 42, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Intlekofer, A.M.; Dematteo, R.G.; Venneti, S.; Finley, L.W.S.; Lu, C.; Judkins, A.R.; Rustenburg, A.S.; Grinaway, P.B.; Chodera, J.D.; Cross, J.R.; et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab. 2015, 22, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J. Energy Metabolism of Cancer: Glycolysis versus Oxidative Phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Akıncılar, S.C.; Khattar, E.; Boon, P.L.S.; Unal, B.; Fullwood, M.J.; Tergaonkar, V. Long-Range Chromatin Interactions Drive Mutant TERT Promoter Activation. Cancer Discov. 2016, 6, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Lorbeer, F.K.; Shain, A.H.; McSwiggen, D.T.; Schruf, E.; Oh, A.; Ryu, J.; Darzacq, X.; Bastian, B.C.; Hockemeyer, D. Mutations in the Promoter of the Telomerase Gene TERT Contribute to Tumorigenesis by a Two-Step Mechanism. Science 2017, 357, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Hertwig, F.; Roels, F.; Dreidax, D.; Gartlgruber, M.; Menon, R.; Krämer, A.; Roncaioli, J.L.; Sand, F.; Heuckmann, J.M.; et al. Telomerase Activation by Genomic Rearrangements in High-Risk Neuroblastoma. Nature 2015, 526, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, R.; Ozturk, M.B.; Low, J.-L.; Akincilar, S.C.; Chua, J.Y.H.; Thangavelu, M.T.; Periyasamy, G.; DasGupta, R.; Tergaonkar, V. Genome-Wide Screens Identify Specific Drivers of Mutant hTERT Promoters. Proc. Natl. Acad. Sci. USA 2022, 119, e2105171119. [Google Scholar] [CrossRef]
- Ozturk, M.B.; Li, Y.; Tergaonkar, V. Current Insights to Regulation and Role of Telomerase in Human Diseases. Antioxidants 2017, 6, 17. [Google Scholar] [CrossRef]
- Fernandes, S.G.; Dsouza, R.; Pandya, G.; Kirtonia, A.; Tergaonkar, V.; Lee, S.Y.; Garg, M.; Khattar, E. Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential. Cancers 2020, 12, 1901. [Google Scholar] [CrossRef]
- Akincilar, S.C.; Chan, C.H.T.; Ng, Q.F.; Fidan, K.; Tergaonkar, V. Non-Canonical Roles of Canonical Telomere Binding Proteins in Cancers. Cell Mol. Life Sci. 2021, 78, 4235–4257. [Google Scholar] [CrossRef] [PubMed]
- Fice, H.; Robaire, B. Telomere Dynamics Throughout Spermatogenesis. Genes 2019, 10, 525. [Google Scholar] [CrossRef] [PubMed]
- Thilagavathi, J.; Mishra, S.S.; Kumar, M.; Vemprala, K.; Deka, D.; Dhadwal, V.; Dada, R. Analysis of Telomere Length in Couples Experiencing Idiopathic Recurrent Pregnancy Loss. J. Assist. Reprod. Genet. 2013, 30, 793–798. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, F.; Dai, S.; Zhang, N.; Zhao, W.; Bai, R.; Sun, Y. Sperm Telomere Length Is Positively Associated with the Quality of Early Embryonic Development. Hum. Reprod. 2015, 30, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Tan, Y.; Qiu, X.; Luo, H.; Li, Y.; Li, R.; Yang, X. Sperm Telomere Length as a Novel Biomarker of Male Infertility and Embryonic Development: A Systematic Review and Meta-Analysis. Front. Endocrinol. 2023, 13, 1079966. [Google Scholar] [CrossRef]
- Su, L.; Mruk, D.D.; Cheng, C.Y. Drug Transporters, the Blood-Testis Barrier, and Spermatogenesis. J. Endocrinol. 2011, 208, 207–223. [Google Scholar] [CrossRef]
- Boussouar, F.; Benahmed, M. Lactate and Energy Metabolism in Male Germ Cells. Trends Endocrinol. Metab. 2004, 15, 345–350. [Google Scholar] [CrossRef]
- Rato, L.; Alves, M.G.; Socorro, S.; Duarte, A.I.; Cavaco, J.E.; Oliveira, P.F. Metabolic Regulation Is Important for Spermatogenesis. Nat. Rev. Urol. 2012, 9, 330–338. [Google Scholar] [CrossRef]
- Bajpai, M.; Gupta, G.; Setty, B.S. Changes in Carbohydrate Metabolism of Testicular Germ Cells during Meiosis in the Rat. Eur. J. Endocrinol. 1998, 138, 322–327. [Google Scholar] [CrossRef]
- Voigt, A.L.; Thiageswaran, S.; De Lima E Martins Lara, N.; Dobrinski, I. Metabolic Requirements for Spermatogonial Stem Cell Establishment and Maintenance In Vivo and In Vitro. Int. J. Mol. Sci. 2021, 22, 1998. [Google Scholar] [CrossRef]
- Nakamura, M.; Fujiwara, A.; Yasumasu, I.; Okinaga, S.; Arai, K. Regulation of Glucose Metabolism by Adenine Nucleotides in Round Spermatids from Rat Testes. J. Biol. Chem. 1982, 257, 13945–13950. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, A.J.; Bustamante, X.; Bertinat, R.; Werner, E.; Rauch, M.C.; Concha, I.I.; Reyes, J.G.; Slebe, J.C. Expression of Key Substrate Cycle Enzymes in Rat Spermatogenic Cells: Fructose 1,6 Bisphosphatase and 6 Phosphofructose 1-Kinase. J. Cell Physiol. 2007, 212, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Anifandis, G.; Samara, M.; Simopoulou, M.; Messini, C.I.; Chatzimeletiou, K.; Thodou, E.; Daponte, A.; Georgiou, I. Insights into the Role of Telomeres in Human Embryological Parameters. Opinions Regarding IVF. JDB 2021, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Berneau, S.C.; Shackleton, J.; Nevin, C.; Altakroni, B.; Papadopoulos, G.; Horne, G.; Brison, D.R.; Murgatroyd, C.; Povey, A.C.; Carroll, M. Associations of Sperm Telomere Length with Semen Parameters, Clinical Outcomes and Lifestyle Factors in Human Normozoospermic Samples. Andrology 2020, 8, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Beygi, Z.; Forouhari, S.; Mahmoudi, E.; Hayat, S.M.G.; Nourimand, F. Role of Oxidative Stress and Antioxidant Supplementation in Male Fertility. CMM 2021, 21, 265–282. [Google Scholar] [CrossRef]
- Ahmed, W.; Lingner, J. PRDX1 and MTH1 Cooperate to Prevent ROS-Mediated Inhibition of Telomerase. Genes Dev. 2018, 32, 658–669. [Google Scholar] [CrossRef]
- Liu, L.; Bailey, S.M.; Okuka, M.; Muñoz, P.; Li, C.; Zhou, L.; Wu, C.; Czerwiec, E.; Sandler, L.; Seyfang, A.; et al. Telomere Lengthening Early in Development. Nat. Cell Biol. 2007, 9, 1436–1441. [Google Scholar] [CrossRef]
- Bender, H.S.; Murchison, E.P.; Pickett, H.A.; Deakin, J.E.; Strong, M.A.; Conlan, C.; McMillan, D.A.; Neumann, A.A.; Greider, C.W.; Hannon, G.J.; et al. Extreme Telomere Length Dimorphism in the Tasmanian Devil and Related Marsupials Suggests Parental Control of Telomere Length. PLoS ONE 2012, 7, e46195. [Google Scholar] [CrossRef][Green Version]
- Eisenhauer, K.M.; Gerstein, R.M.; Chiu, C.-P.; Conti, M.; Hsueh, A.J.W. Telomerase Activity in Female and Male Rat Germ Cells Undergoing Meiosis and in Early Embryos1. Biol. Reprod. 1997, 56, 1120–1125. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Telomeres and Telomerase: Implications for Cancer and Aging. Radiat. Res. 2001, 155, 188–193. [Google Scholar] [CrossRef]
- Liu, L.; Blasco, M.A.; Trimarchi, J.R.; Keefe, D.L. An Essential Role for Functional Telomeres in Mouse Germ Cells during Fertilization and Early Development. Dev. Biol. 2002, 249, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Siderakis, M.; Tarsounas, M. Telomere Regulation and Function during Meiosis. Chromosome Res. 2007, 15, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Keefe, D.L.; Franco, S.; Liu, L.; Trimarchi, J.; Cao, B.; Weitzen, S.; Agarwal, S.; Blasco, M.A. Telomere Length Predicts Embryo Fragmentation after in Vitro Fertilization in Women—Toward a Telomere Theory of Reproductive Aging in Women. Am. J. Obstet. Gynecol. 2005, 192, 1256–1260. [Google Scholar] [CrossRef]
- Liu, L.; Franco, S.; Spyropoulos, B.; Moens, P.B.; Blasco, M.A.; Keefe, D.L. Irregular Telomeres Impair Meiotic Synapsis and Recombination in Mice. Proc. Natl. Acad. Sci. USA 2004, 101, 6496–6501. [Google Scholar] [CrossRef] [PubMed]
- Treff, N.R.; Su, J.; Taylor, D.; Scott, R.T. Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy. PLoS Genet. 2011, 7, e1002161. [Google Scholar] [CrossRef]
- Keefe, D.L. Telomeres and Genomic Instability during Early Development. Eur. J. Med. Genet. 2020, 63, 103638. [Google Scholar] [CrossRef]
- Yamada-Fukunaga, T.; Yamada, M.; Hamatani, T.; Chikazawa, N.; Ogawa, S.; Akutsu, H.; Miura, T.; Miyado, K.; Tarín, J.J.; Kuji, N.; et al. Age-Associated Telomere Shortening in Mouse Oocytes. Reprod. Biol. Endocrinol. 2013, 11, 108. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Ye, X.; Liu, K.; Huang, J.; Wang, L.; Ji, G.; Liu, N.; Tang, X.; Baltz, J.M.; et al. Delay in Oocyte Aging in Mice by the Antioxidant N-Acetyl-l-Cysteine (NAC). Hum. Reprod. 2012, 27, 1411–1420. [Google Scholar] [CrossRef]
- Kinugawa, C.; Murakami, T.; Okamura, K.; Yajima, A. Telomerase Activity in Normal Ovaries and Premature Ovarian Failure. Tohoku J. Exp. Med. 2000, 190, 231–238. [Google Scholar] [CrossRef]
- Lavranos, T.C.; Mathis, J.M.; Latham, S.E.; Kalionis, B.; Shay, J.W.; Rodgers, R.J. Evidence for Ovarian Granulosa Stem Cells: Telomerase Activity and Localization of the Telomerase Ribonucleic Acid Component in Bovine Ovarian Follicles1. Biol. Reprod. 1999, 61, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Russo, V.; Berardinelli, P.; Martelli, A.; Giacinto, O.D.; Nardinocchi, D.; Fantasia, D.; Barboni, B. Expression of Telomerase Reverse Transcriptase Subunit (TERT) and Telomere Sizing in Pig Ovarian Follicles. J. Histochem. Cytochem. 2006, 54, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Y.; Nakamura, Y.; Umayahara, K.; Harada, A.; Takayama, H.; Sugino, N.; Kato, H. Changes in Telomerase Activity in Experimentally Induced Atretic Follicles of Immature Rats. Endocr. J. 2002, 49, 589–595. [Google Scholar] [CrossRef]
- Bayne, S.; Li, H.; Jones, M.E.E.; Pinto, A.R.; Van Sinderen, M.; Drummond, A.; Simpson, E.R.; Liu, J.-P. Estrogen Deficiency Reversibly Induces Telomere Shortening in Mouse Granulosa Cells and Ovarian Aging In Vivo. Protein Cell 2011, 2, 333–346. [Google Scholar] [CrossRef]
- Goto, H.; Iwata, H.; Takeo, S.; Nisinosono, K.; Murakami, S.; Monji, Y.; Kuwayama, T. Effect of Bovine Age on the Proliferative Activity, Global DNA Methylation, Relative Telomere Length and Telomerase Activity of Granulosa Cells. Zygote 2013, 21, 256–264. [Google Scholar] [CrossRef]
- Endo, M.; Kimura, K.; Kuwayama, T.; Monji, Y.; Iwata, H. Effect of Estradiol during Culture of Bovine Oocyte–Granulosa Cell Complexes on the Mitochondrial DNA Copies of Oocytes and Telomere Length of Granulosa Cells. Zygote 2014, 22, 431–439. [Google Scholar] [CrossRef]
- Liu, J.-P.; Li, H. Telomerase in the Ovary. Reproduction 2010, 140, 215–222. [Google Scholar] [CrossRef]
- Butts, S.; Riethman, H.; Ratcliffe, S.; Shaunik, A.; Coutifaris, C.; Barnhart, K. Correlation of Telomere Length and Telomerase Activity with Occult Ovarian Insufficiency. J. Clin. Endocrinol. Metab. 2009, 94, 4835–4843. [Google Scholar] [CrossRef]
- Alemany, M. Estrogens and the Regulation of Glucose Metabolism. WJD 2021, 12, 1622–1654. [Google Scholar] [CrossRef]
- Klinge, C.M. Estrogenic Control of Mitochondrial Function. Redox Biol. 2020, 31, 101435. [Google Scholar] [CrossRef]
- Sreerangaraja Urs, D.B.; Wu, W.-H.; Komrskova, K.; Postlerova, P.; Lin, Y.-F.; Tzeng, C.-R.; Kao, S.-H. Mitochondrial Function in Modulating Human Granulosa Cell Steroidogenesis and Female Fertility. Int. J. Mol. Sci. 2020, 21, 3592. [Google Scholar] [CrossRef]
- Yung, Y.; Maydan, S.A.; Bart, Y.; Orvieto, R.; Aizer, A. Human Granulosa Cells of Poor Ovarian Responder Patients Display Telomeres Shortening. J. Assist. Reprod. Genet. 2023, 40, 1943–1947. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Kong, F.; Luan, Y.; Sun, C.; Wang, J.; Zhang, L.; Jiang, B.; Qi, T.; Zhao, J.; Zheng, C.; et al. Differential Shortening Rate of Telomere Length in the Development of Human Fetus. Biochem. Biophys. Res. Commun. 2013, 442, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Liu, H.; Gu, X.; Boots, C.; Moley, K.H.; Wang, Q. Metabolic Control of Oocyte Development: Linking Maternal Nutrition and Reproductive Outcomes. Cell. Mol. Life Sci. 2015, 72, 251–271. [Google Scholar] [CrossRef] [PubMed]
- Lounas, A.; Breton, Y.; Lebrun, A.; Laflamme, I.; Vernoux, N.; Savage, J.; Tremblay, M.-È.; Pelletier, M.; Germain, M.; Richard, F.J. The Follicle-Stimulating Hormone Triggers Rapid Changes in Mitochondrial Structure and Function in Porcine Cumulus Cells. Sci. Rep. 2024, 14, 436. [Google Scholar] [CrossRef]
- Ozturk, S.; Sozen, B.; Demir, N. Telomere Length and Telomerase Activity during Oocyte Maturation and Early Embryo Development in Mammalian Species. Mol. Hum. Reprod. 2014, 20, 15–30. [Google Scholar] [CrossRef]
- Gorshinova, V.K.; Tsvirkun, D.V.; Sukhanova, I.A.; Tarasova, N.V.; Volodina, M.A.; Marey, M.V.; Smolnikova, V.U.; Vysokikh, M.Y.; Sukhikh, G.T. Cumulus Cell Mitochondrial Activity in Relation to Body Mass Index in Women Undergoing Assisted Reproductive Therapy. BBA Clin. 2017, 7, 141–146. [Google Scholar] [CrossRef]
- Turner, S.; Wong, H.P.; Rai, J.; Hartshorne, G.M. Telomere Lengths in Human Oocytes, Cleavage Stage Embryos and Blastocysts. Mol. Hum. Reprod. 2010, 16, 685–694. [Google Scholar] [CrossRef]
- Le, R.; Huang, Y.; Zhang, Y.; Wang, H.; Lin, J.; Dong, Y.; Li, Z.; Guo, M.; Kou, X.; Zhao, Y.; et al. Dcaf11 Activates Zscan4-Mediated Alternative Telomere Lengthening in Early Embryos and Embryonic Stem Cells. Cell Stem Cell 2021, 28, 732–747.e9. [Google Scholar] [CrossRef]
- Dan, J.; Zhou, Z.; Wang, F.; Wang, H.; Guo, R.; Keefe, D.L.; Liu, L. Zscan4 Contributes to Telomere Maintenance in Telomerase-Deficient Late Generation Mouse ESCs and Human ALT Cancer Cells. Cells 2022, 11, 456. [Google Scholar] [CrossRef]
- Kordowitzki, P.; López De Silanes, I.; Guío-Carrión, A.; Blasco, M.A. Dynamics of Telomeric Repeat-Containing RNA Expression in Early Embryonic Cleavage Stages with Regards to Maternal Age. Aging 2020, 12, 15906–15917. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, R.; Sharpley, M.S.; Chi, F.; Braas, D.; Zhou, Y.; Kim, R.; Clark, A.T.; Banerjee, U. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell 2017, 168, 210–223.e11. [Google Scholar] [CrossRef] [PubMed]
- Schaetzlein, S.; Lucas-Hahn, A.; Lemme, E.; Kues, W.A.; Dorsch, M.; Manns, M.P.; Niemann, H.; Rudolph, K.L. Telomere Length Is Reset during Early Mammalian Embryogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 8034–8038. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Morimoto, N.; Yamanaka, M.; Matsumoto, H.; Yamochi, T.; Goto, H.; Inoue, M.; Nakaoka, Y.; Shibahara, H.; Morimoto, Y. Quantitative and Qualitative Changes of Mitochondria in Human Preimplantation Embryos. J. Assist. Reprod. Genet. 2017, 34, 573–580. [Google Scholar] [CrossRef]
- Wang, C.; Gu, Y.; Zhou, J.; Zang, J.; Ling, X.; Li, H.; Hu, L.; Xu, B.; Zhang, B.; Qin, N.; et al. Leukocyte Telomere Length in Children Born Following Blastocyst-Stage Embryo Transfer. Nat. Med. 2022, 28, 2646–2653. [Google Scholar] [CrossRef]
- Ge, J.; Li, C.; Sun, H.; Xin, Y.; Zhu, S.; Liu, Y.; Tang, S.; Han, L.; Huang, Z.; Wang, Q. Telomere Dysfunction in Oocytes and Embryos From Obese Mice. Front. Cell Dev. Biol. 2021, 9, 617225. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, K.; Liu, Y.; Fu, Y.; Gao, S.; Gong, P.; Wang, H.; Zhou, Z.; Zeng, M.; Wu, Z.; et al. Telomere Heterogeneity Linked to Metabolism and Pluripotency State Revealed by Simultaneous Analysis of Telomere Length and RNA-Seq in the Same Human Embryonic Stem Cell. BMC Biol. 2017, 15, 114. [Google Scholar] [CrossRef]
- Tsogtbaatar, E.; Landin, C.; Minter-Dykhouse, K.; Folmes, C.D.L. Energy Metabolism Regulates Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Schellnegger, M.; Hofmann, E.; Carnieletto, M.; Kamolz, L.-P. Unlocking Longevity: The Role of Telomeres and Its Targeting Interventions. Front. Aging 2024, 5, 1339317. [Google Scholar] [CrossRef]
- Weng, N.; Palmer, L.D.; Levine, B.L.; Lane, H.C.; June, C.H.; Hodes, R.J. Tales of Tails: Regulation of Telomere Length and Telomerase Activity during Lymphocyte Development, Differentiation, Activation, and Aging. Immunol. Rev. 1997, 160, 43–54. [Google Scholar] [CrossRef]
- Corrado, M.; Pearce, E.L. Targeting Memory T Cell Metabolism to Improve Immunity. J. Clin. Investig. 2022, 132, e148546. [Google Scholar] [CrossRef] [PubMed]
- Quinn, K.M.; Palchaudhuri, R.; Palmer, C.S.; La Gruta, N.L. The Clock Is Ticking: The Impact of Ageing on T Cell Metabolism. Clin. Transl. Immunol. 2019, 8, e01091. [Google Scholar] [CrossRef] [PubMed]
- Mosoyan, G.; Kraus, T.; Ye, F.; Eng, K.; Crispino, J.D.; Hoffman, R.; Iancu-Rubin, C. Imetelstat, a Telomerase Inhibitor, Differentially Affects Normal and Malignant Megakaryopoiesis. Leukemia 2017, 31, 2458–2467. [Google Scholar] [CrossRef]
- Platzbecker, U.; Santini, V.; Fenaux, P.; Sekeres, M.A.; Savona, M.R.; Madanat, Y.F.; Díez-Campelo, M.; Valcárcel, D.; Illmer, T.; Jonášová, A.; et al. Imetelstat in Patients with Lower-Risk Myelodysplastic Syndromes Who Have Relapsed or Are Refractory to Erythropoiesis-Stimulating Agents (IMerge): A Multinational, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2024, 403, 249–260. [Google Scholar] [CrossRef]
- Borrell, B. Lawsuit Challenges Anti-Ageing Claims. Nature 2012, 488, 18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sengupta, S.; Sobo, M.; Lee, K.; Senthil Kumar, S.; White, A.R.; Mender, I.; Fuller, C.; Chow, L.M.L.; Fouladi, M.; Shay, J.W.; et al. Induced Telomere Damage to Treat Telomerase Expressing Therapy-Resistant Pediatric Brain Tumors. Mol. Cancer Ther. 2018, 17, 1504–1514. [Google Scholar] [CrossRef]
- Zeng, X.; Hernandez-Sanchez, W.; Xu, M.; Whited, T.L.; Baus, D.; Zhang, J.; Berdis, A.J.; Taylor, D.J. Administration of a Nucleoside Analog Promotes Cancer Cell Death in a Telomerase-Dependent Manner. Cell Rep. 2018, 23, 3031–3041. [Google Scholar] [CrossRef] [PubMed]
- Moye, A.L.; Porter, K.C.; Cohen, S.B.; Phan, T.; Zyner, K.G.; Sasaki, N.; Lovrecz, G.O.; Beck, J.L.; Bryan, T.M. Telomeric G-Quadruplexes Are a Substrate and Site of Localization for Human Telomerase. Nat. Commun. 2015, 6, 7643. [Google Scholar] [CrossRef]
- Miglietta, G.; Russo, M.; Capranico, G. G-Quadruplex–R-Loop Interactions and the Mechanism of Anticancer G-Quadruplex Binders. Nucleic Acids Res. 2020, 48, 11942–11957. [Google Scholar] [CrossRef]
- Salvati, E.; Leonetti, C.; Rizzo, A.; Scarsella, M.; Mottolese, M.; Galati, R.; Sperduti, I.; Stevens, M.F.G.; D’Incalci, M.; Blasco, M.; et al. Telomere Damage Induced by the G-Quadruplex Ligand RHPS4 Has an Antitumor Effect. J. Clin. Investig. 2007, 117, 3236–3247. [Google Scholar] [CrossRef]
- Chaithanya, V.; Kumar, J.; Leela, K.V.; Murugesan, R.; Angelin, M.; Satheesan, A. Impact of Telomere Attrition on Diabetes Mellitus and Its Complications. Diabetes Epidemiol. Manag. 2023, 12, 100174. [Google Scholar] [CrossRef]
- Cheng, F.-F.; Liu, Y.-L.; Du, J.; Lin, J.-T. Metformin’s Mechanisms in Attenuating Hallmarks of Aging and Age-Related Disease. Aging Dis. 2022, 13, 970. [Google Scholar] [CrossRef] [PubMed]
- Herman, R.; Kravos, N.A.; Jensterle, M.; Janež, A.; Dolžan, V. Metformin and Insulin Resistance: A Review of the Underlying Mechanisms behind Changes in GLUT4-Mediated Glucose Transport. Int. J. Mol. Sci. 2022, 23, 1264. [Google Scholar] [CrossRef] [PubMed]
- Vidaček, N.Š.; Nanić, L.; Ravlić, S.; Sopta, M.; Gerić, M.; Gajski, G.; Garaj-Vrhovac, V.; Rubelj, I. Telomeres, Nutrition, and Longevity: Can We Really Navigate Our Aging? J. Gerontol. Ser. A 2018, 73, 39–47. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubtsova, M.P.; Nikishin, D.A.; Vyssokikh, M.Y.; Koriagina, M.S.; Vasiliev, A.V.; Dontsova, O.A. Telomere Reprogramming and Cellular Metabolism: Is There a Link? Int. J. Mol. Sci. 2024, 25, 10500. https://doi.org/10.3390/ijms251910500
Rubtsova MP, Nikishin DA, Vyssokikh MY, Koriagina MS, Vasiliev AV, Dontsova OA. Telomere Reprogramming and Cellular Metabolism: Is There a Link? International Journal of Molecular Sciences. 2024; 25(19):10500. https://doi.org/10.3390/ijms251910500
Chicago/Turabian StyleRubtsova, Maria P., Denis A. Nikishin, Mikhail Y. Vyssokikh, Maria S. Koriagina, Andrey V. Vasiliev, and Olga A. Dontsova. 2024. "Telomere Reprogramming and Cellular Metabolism: Is There a Link?" International Journal of Molecular Sciences 25, no. 19: 10500. https://doi.org/10.3390/ijms251910500
APA StyleRubtsova, M. P., Nikishin, D. A., Vyssokikh, M. Y., Koriagina, M. S., Vasiliev, A. V., & Dontsova, O. A. (2024). Telomere Reprogramming and Cellular Metabolism: Is There a Link? International Journal of Molecular Sciences, 25(19), 10500. https://doi.org/10.3390/ijms251910500




