Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
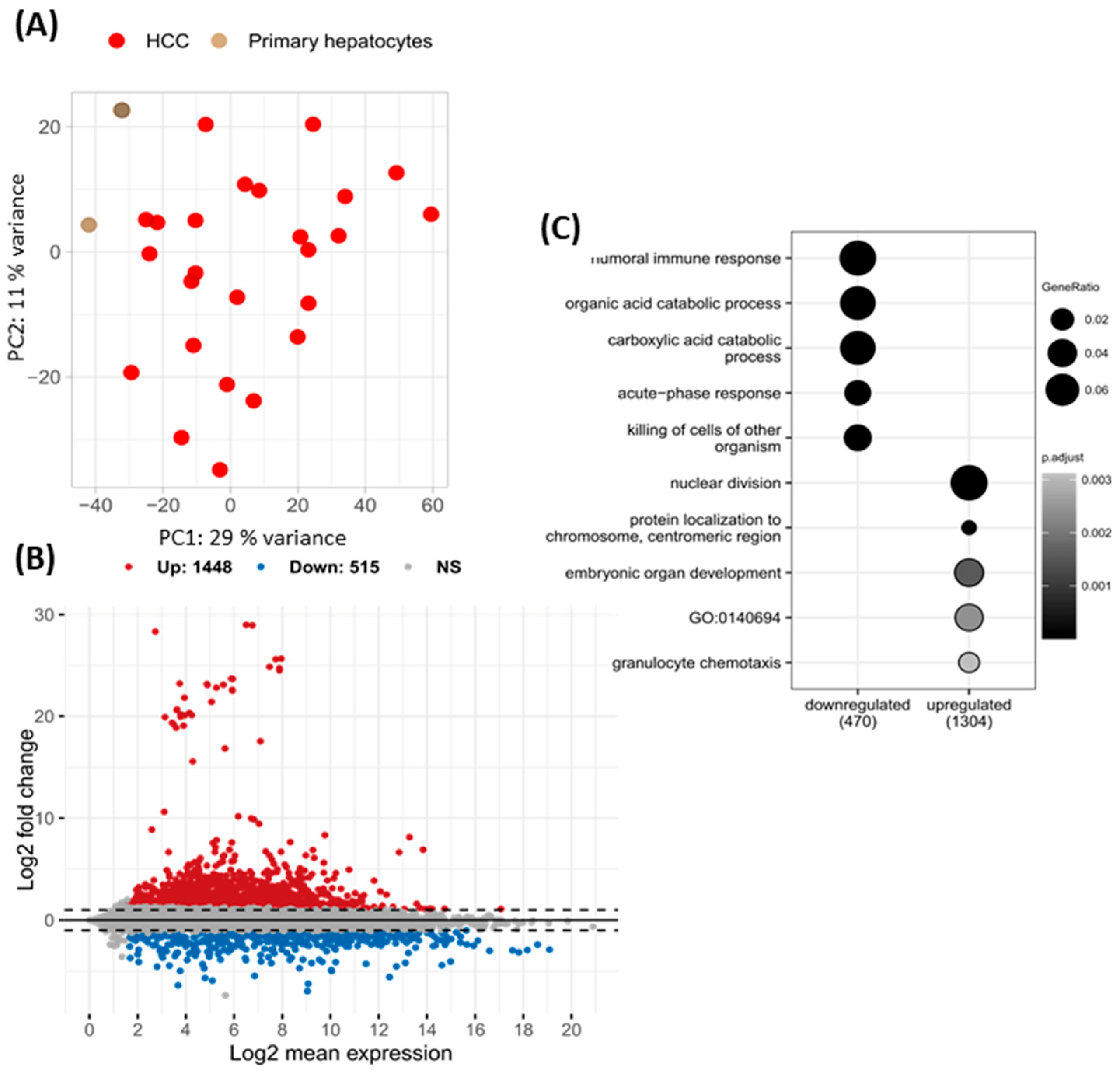
2.1. Differentially Expressed Genes in HCC of Viral Etiology and Primary Hepatocytes
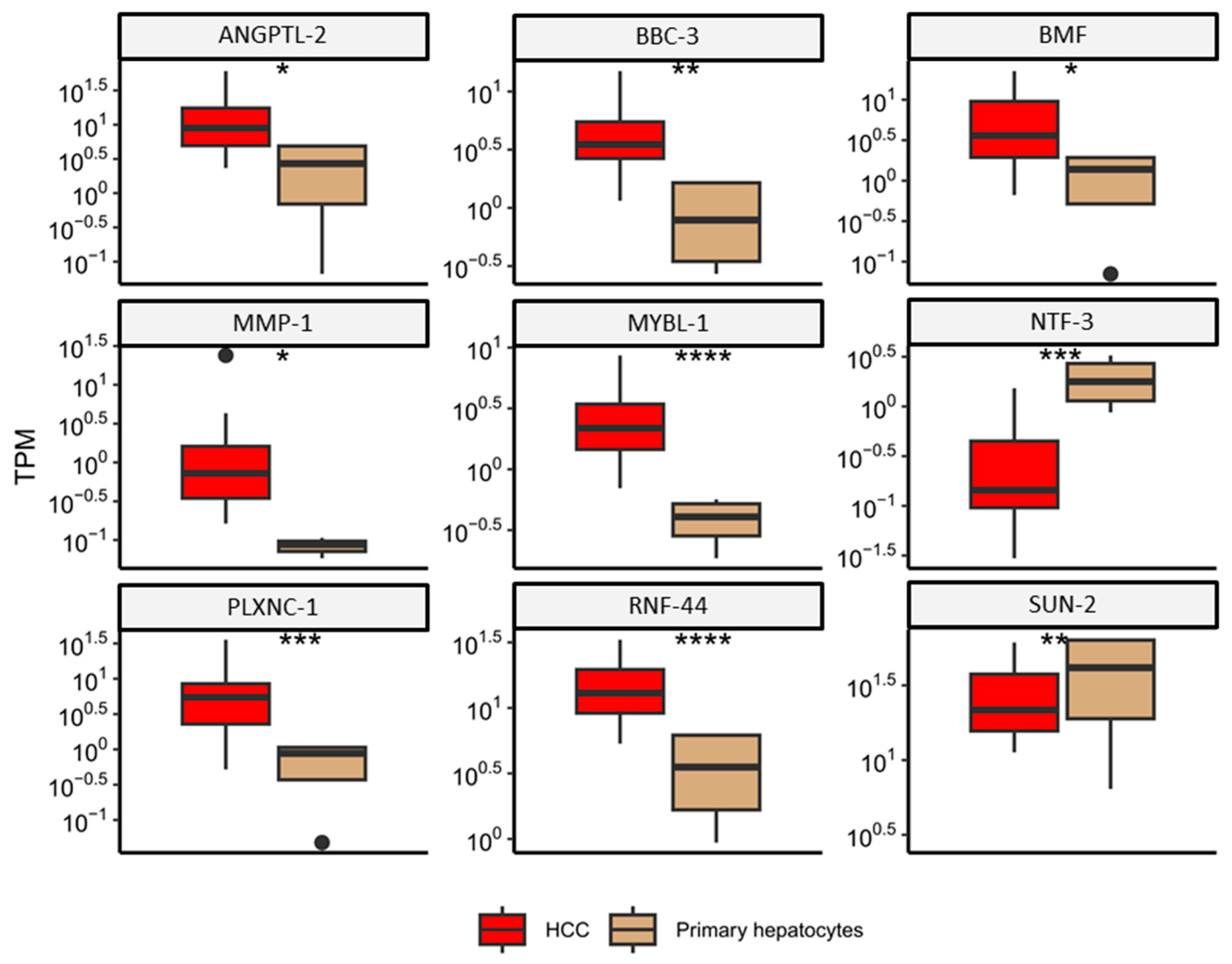
2.2. miRNA-Regulated Genes Characteristic for HCC
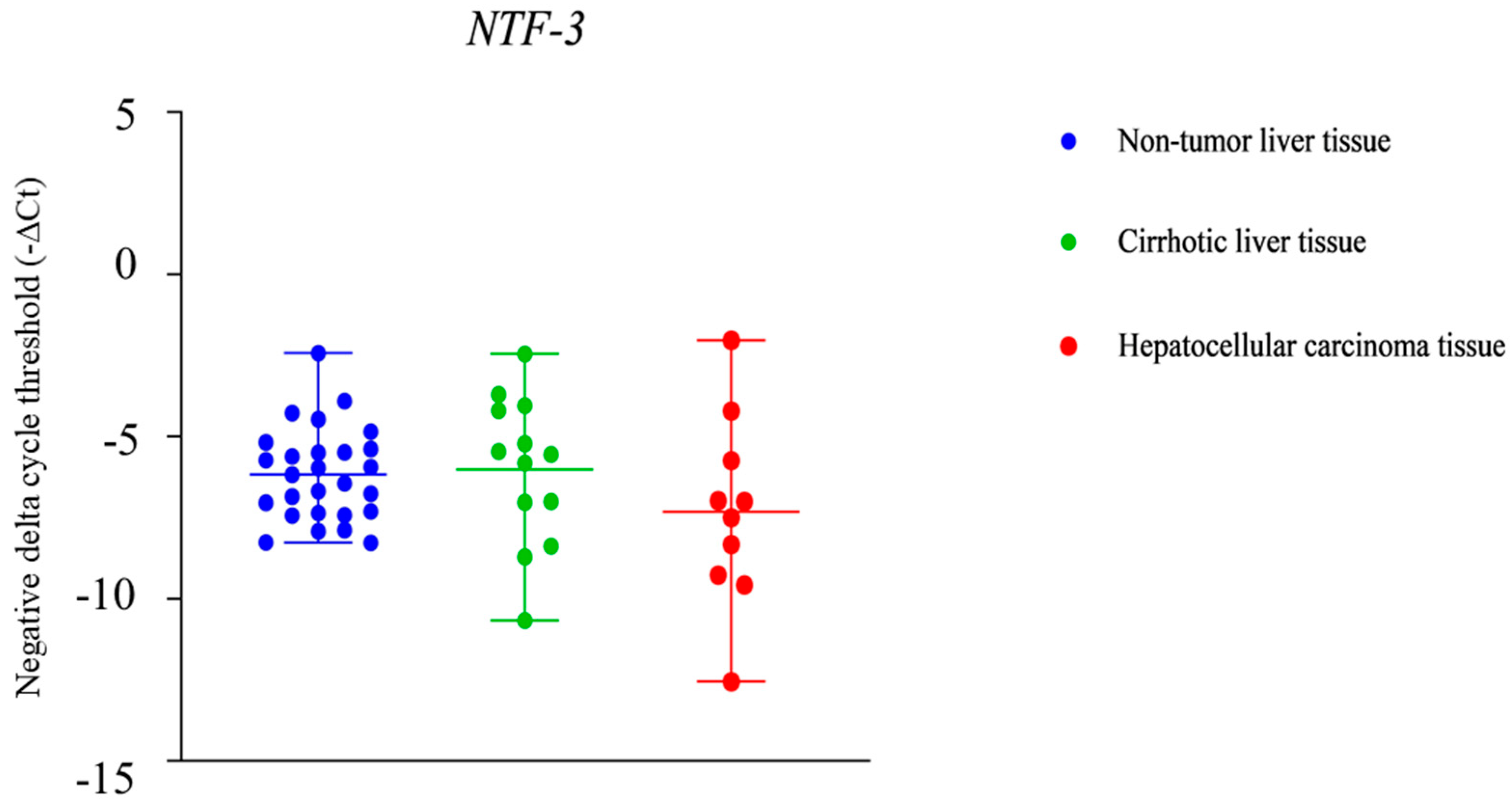
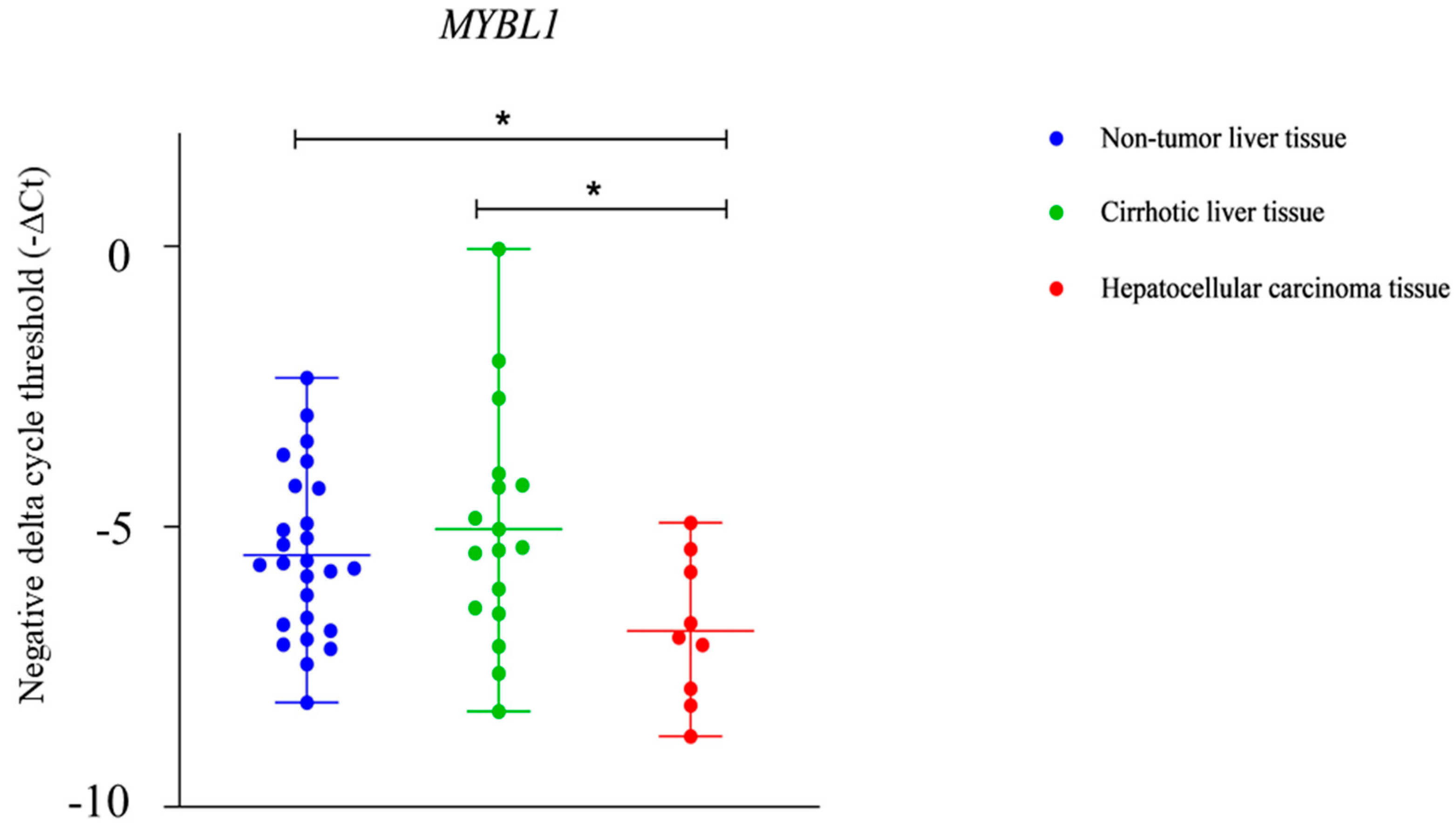
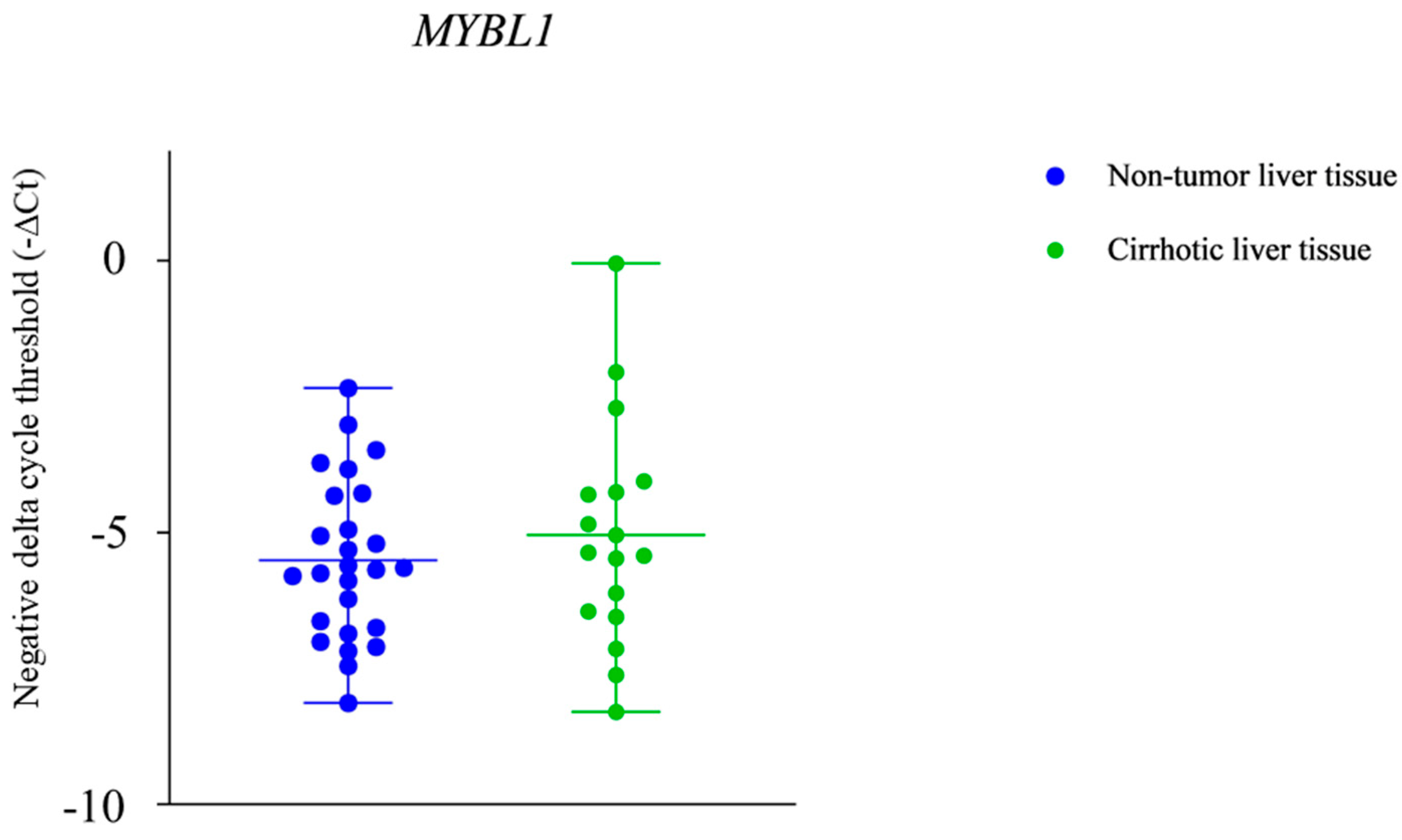
2.3. NTF-3 and MYBL1 Expression in Samples from Patients with Alcohol-Associated HCC, Cirrhotic Tissue, and Non-Tumor Liver Tissue
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis
4.1.1. Publicly Available Datasets
4.1.2. Quality Control and Mapping
4.1.3. Principal Component Analysis (PCA)
4.1.4. Differential Expression Analysis between HCC of Viral Etiology and Primary Hepatocytes
4.2. HCC Patients’ Samples
4.2.1. Materials
Tissue Specimens
4.2.2. Methods
Ribonucleic Acid (RNA) Isolation, Reverse Transcription, qPCR
4.2.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Who Classification of Tumours: Digestive System Tumours; World Health Organization (WHO): Lyon, France, 2019. [Google Scholar]
- Srivastava, A.; Allende, D.S.; Goldblum, J.R. Gastrointestinal and Liver Pathology: A Volume in the Series: Foundations in Diagnostic Pathology; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Christopher, D.M.F. Diagnostic Histopathology of Tumors; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Hosseini, N.; Shor, J.; Szabo, G. Alcoholic Hepatitis: A Review. Alcohol Alcohol. 2019, 54, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Das, U.N. Beneficial role of bioactive lipids in the pathobiology, prevention, and management of HBV, HCV and alcoholic hepatitis, NAFLD, and liver cirrhosis: A review. J. Adv. Res. 2018, 17, 17–29. [Google Scholar] [CrossRef]
- Mohr, R.; Özdirik, B.; Lambrecht, J.; Demir, M.; Eschrich, J.; Geisler, L.; Hellberg, T.; Loosen, S.H.; Luedde, T.; Tacke, F.; et al. From Liver Cirrhosis to Cancer: The Role of Micro-RNAs in Hepatocarcinogenesis. Int. J. Mol. Sci. 2021, 22, 1492. [Google Scholar] [CrossRef]
- Oura, K.; Morishita, A.; Masaki, T. Molecular and Functional Roles of MicroRNAs in the Progression of Hepatocellular carcinoma—A Review. Int. J. Mol. Sci. 2020, 21, 8362. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Li, L.; Hu, H.Q.; Meng, X.M.; Huang, C.; Zhang, L.; Qin, J.; Li, J. MicroRNAs in alcoholic liver disease: Recent advances and future applications. J. Cell Physiol. 2018, 234, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Cucarull, B.; Tutusaus, A.; Rider, P.; Hernáez-Alsina, T.; Cuño, C.; García de Frutos, P.; Colell, A.; Marí, M.; Morales, A. Hepatocellular carcinoma: Molecular Pathogenesis and Therapeutic Advances. Cancers 2022, 14, 621. [Google Scholar] [CrossRef]
- Llovet, J.M.; Pinyol, R.; Kelley, R.K.; El-Khoueiry, A.; Reeves, H.L.; Wang, X.W.; Gores, G.J.; Villanueva, A. Molecular pathogenesis and systemic therapies for Hepatocellular carcinoma. Nat. Cancer 2022, 3, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Harish, D.R.; Sampat, G.H.; Roy, S.; Jalalpure, S.S.; Khanal, P.; Gujarathi, S.S.; Hegde, H.V. System Biology Investigation Revealed Lipopolysaccharide and Alcohol-Induced Hepatocellular carcinoma Resembled Hepatitis B Virus Immunobiology and Pathogenesis. Int. J. Mol. Sci. 2023, 24, 11146. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Barrena, M.G.; Arechederra, M.; Colyn, L.; Berasain, C.; Avila, M.A. Epigenetics in Hepatocellular carcinoma development and therapy: The tip of the iceberg. JHEP Rep. 2020, 2, 100167. [Google Scholar] [CrossRef] [PubMed]
- Toh, T.B.; Lim, J.J.; Chow, E.K.H. Epigenetics of Hepatocellular carcinoma. Clin. Transl. Med. 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Morishita, A.; Oura, K.; Tadokoro, T.; Fujita, K.; Tani, J.; Masaki, T. MicroRNAs in the Pathogenesis of Hepatocellular carcinoma: A Review. Cancers 2021, 13, 514. [Google Scholar] [CrossRef]
- He, X.X.; Guo, A.Y.; Xu, C.R.; Chang, Y.; Xiang, G.Y.; Gong, J.; Dan, Z.L.; Tian, D.A.; Liao, J.Z.; Lin, J.S. Bioinformatics analysis identifies miR-221 as a core regulator in Hepatocellular carcinoma and its silencing suppresses tumor properties. Oncol. Rep. 2014, 32, 1200–1210. [Google Scholar] [CrossRef]
- Wang, X.; Liao, X.; Huang, K.; Zeng, X.; Liu, Z.; Zhou, X.; Yu, T.; Yang, C.; Yu, L.; Wang, Q.; et al. Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for Hepatocellular carcinoma. J. Cancer 2019, 10, 2520–2533. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.; Alhelf, M.; Morcos, G.; Elsharkawy, A. miRNA-101-1 and miRNA-221 expressions and their polymorphisms as biomarkers for early diagnosis of Hepatocellular carcinoma. Infect. Genet. Evol. 2017, 51, 173–181. [Google Scholar] [CrossRef]
- Loureiro, D.; Tout, I.; Narguet, S.; Benazzouz, S.M.; Mansouri, A.; Asselah, T. miRNAs as Potential Biomarkers for Viral Hepatitis B and C. Viruses 2020, 12, 1440. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Szabo, G. MicroRNA Signature in Alcoholic Liver Disease. Int. J. Hepatol. 2012, 2012, 498232. [Google Scholar] [CrossRef]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; An, Q.; Niu, B.; Lu, X.; Zhang, N.; Cao, X. Role of miR-221/222 in Tumor Development and the Underlying Mechanism. J. Oncol. 2019, 2019, 7252013. [Google Scholar] [CrossRef]
- Khalaf, A.M.; Fuentes, D.; Morshid, A.I.; Burke, M.R.; Kaseb, A.O.; Hassan, M.; Hazle, J.D.; Elsayes, K.M. Role of Wnt/β-catenin signaling in Hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 2018, 5, 61–73. [Google Scholar] [CrossRef]
- Ha, H.L.; Shin, H.J.; Feitelson, M.A.; Yu, D.Y. Oxidative stress and antioxidants in hepatic pathogenesis. World J. Gastroenterol. 2010, 16, 6035–6043. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, R.; Yu, H.; Liu, J.; Zheng, S.; Li, Y.; Ye, L. NTF3 Correlates With Prognosis and Immune Infiltration in Hepatocellular carcinoma. Front. Med. 2021, 8, 795849. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, J.; Liu, B.; Zhao, M.; Xing, E.; Dang, C. miRNA-222 promotes liver cancer cell proliferation, migration and invasion and inhibits apoptosis by targeting BBC3. Int. J. Mol. Med. 2018, 42, 141–148. [Google Scholar] [CrossRef]
- Scheau, C.; Badarau, I.A.; Costache, R.; Caruntu, C.; Mihai, G.L.; Didilescu, A.C.; Constantin, C.; Neagu, M. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular carcinoma. Anal. Cell. Pathol. 2019, 2019, 9423907. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wu, Y.; Yu, Y.; Li, Y.; Shen, J.; Zhang, R. MYBL1 induces transcriptional activation of ANGPT2 to promote tumor angiogenesis and confer sorafenib resistance in human Hepatocellular carcinoma. Cell Death Dis. 2022, 13, 727. [Google Scholar] [CrossRef]
- Odabas, G.; Cetin, M.; Turhal, S.; Baloglu, H.; Sayan, A.E.; Yagci, T. Plexin C1 Marks Liver Cancer Cells with Epithelial Phenotype and Is Overexpressed in Hepatocellular carcinoma. Cell Death Dis. 2022, 13, 4040787. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y.; Huang, H.M.; Li, H.D.; Bu, F.T.; Pan, X.Y.; Yang, Y.; Li, W.X.; Li, X.F.; Huang, C.; et al. SUN2: A potential therapeutic target in cancer. Oncol. Lett. 2018, 17, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Chalazonitis, A. Neurotrophin-3 in the development of the enteric nervous system. Prog. Brain Res. 2004, 146, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.X.; Liu, T.; Yang, J.L.; Liu, F.; Chang, L.; Che, G.L.; Lai, S.Y.; Jiang, Y.M. Low expression of NTF3 is associated with unfavorable prognosis in Hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2020, 13, 2280–2288. [Google Scholar]
- Liot, G.; Gabriel, C.; Cacquevel, M.; Ali, C.; MacKenzie, E.T.; Buisson, A.; Vivien, D. Neurotrophin-3-induced PI-3 kinase/Akt signaling rescues cortical neurons from apoptosis. Exp. Neurol. 2004, 187, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Thiele, C.J.; Li, Z. Neurotrophin Signaling in Cancer. In Handbook of Neurotoxicity; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1825–1847. [Google Scholar] [CrossRef]
- Ramsay, R.G.; Gonda, T.J. MYB function in normal and cancer cells. Nat. Rev. Cancer 2008, 8, 523–534. [Google Scholar] [CrossRef]
- Oh, I.H.; Reddy, E.P. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 1999, 18, 3017–3033. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.A.; Ramsay, R.G. MYB: An old oncoprotein with new roles. BioEssays 1995, 17, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Bolcun-Filas, E.; Bannister, L.A.; Barash, A.; Schimenti, K.J.; Hartford, S.A.; Eppig, J.J.; Handel, M.A.; Shen, L.; Schimenti, J.C. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 2011, 138, 3319–3330. [Google Scholar] [CrossRef] [PubMed]
- Facchinetti, V.; Loffarelli, L.; Schreek, S.; Oelgeschläger, M.; Lüscher, B.; Introna, M.; Golay, J. Regulatory domains of the A-Myb transcription factor and its interaction with the CBP/p300 adaptor molecules. Biochem. J. 1997, 324, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Golay, J.; Facchinetti, V.; Ying, G.; Introna, M. The A-myb transcription factor in neoplastic and normal B cells. Leuk. Lymphoma 1997, 26, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Blondy, S.; Christou, N.; David, V.; Verdier, M.; Jauberteau, M.O.; Mathonnet, M.; Perraud, A. Neurotrophins and their involvement in digestive cancers. Cell Death Dis. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, H.; Yin, M.; Cheng, Z.; Jiang, P.; Feng, M.; Liao, B.; Liu, Z. Neurotrophin3 promotes Hepatocellular carcinoma apoptosis through the JNK and P38 MAPK pathways. Int. J. Biol. Sci. 2022, 18, 5963–5977. [Google Scholar] [CrossRef]
- Guo, Y.; Hu, J.; Zhao, Z.; Zhong, G.; Gong, J.; Cai, D. Identification of a Prognostic Model Based on 2-Gene Signature and Analysis of Corresponding Tumor Microenvironment in Alcohol-Related Hepatocellular carcinoma. Front. Oncol. 2021, 11, 719355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kang, C.; Li, N.; Liu, X.; Zhang, J.; Gao, F.; Dai, L. Identification of special key genes for alcohol-related Hepatocellular carcinoma through bioinformatic analysis. PeerJ 2019, 7, e6375. [Google Scholar] [CrossRef] [PubMed]
- Louie, E.; Chen, X.F.; Coomes, A.; Ji, K.; Tsirka, S.; Chen, E.I. Neurotrophin-3 modulates breast cancer cells and the microenvironment to promote the growth of breast cancer brain metastasis. Oncogene 2013, 32, 4064–4077. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Numata, M.; Tsukioka, Y.; Futagami, F.; Kayahara, M.; Kitagawa, H.; Nagakawa, T.; Yamamoto, M.; Wakayama, T.; Kitamura, Y.; et al. Neurotrophin-3 expression in human pancreatic cancers. J. Pathol. 1997, 181, 405–412. [Google Scholar] [CrossRef]
- Ricci, A.; Greco, S.; Mariotta, S.; Felici, L.; Bronzetti, E.; Cavazzana, A.; Cardillo, G.; Amenta, F.; Bisetti, A.; Barbolini, G. Neurotrophins and neurotrophin receptors in human lung cancer. Am. J. Respir. Cell Mol. Biol. 2001, 25, 439–446. [Google Scholar] [CrossRef]
- Prakash, Y.; Thompson, M.A.; Meuchel, L.; Pabelick, C.M.; Mantilla, C.B.; Zaidi, S.; Martin, R.J. Neurotrophins in lung health and disease. Expert Rev. Respir. Med. 2010, 4, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Bouzas-Rodriguez, J.; Cabrera, J.R.; Delloye-Bourgeois, C.; Ichim, G.; Delcros, J.G.; Raquin, M.A.; Rousseau, R.; Combaret, V.; Bénard, J.; Tauszig-Delamasure, S.; et al. Neurotrophin-3 production promotes human neuroblastoma cell survival by inhibiting TrkC-induced apoptosis. J. Clin. Investig. 2010, 120, 850–858. [Google Scholar] [CrossRef]
- Ziebold, U.; Klempnauer, K.H. Linking Myb to the cell cycle: Cyclin-dependent phosphorylation and regulation of A-Myb activity. Oncogene 1997, 15, 1011–1019. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, B.; Liu, Y.; Zhao, Z.; Liu, Q.; Wang, X.; Xie, Y.; Liu, Y.; Liu, Y.; Yang, Y.; Long, J.; et al. MYB Proto-oncogene-like 1-TWIST1 Axis Promotes Growth and Metastasis of Hepatocellular carcinoma Cells. Mol. Ther. Oncolytics 2020, 18, 58–69. [Google Scholar] [CrossRef]
- Player, A.; Cunningham, S.; Philio, D.; Roy, R.; Haynes, C.; Dixon, C.; Thirston, L.; Ibikunle, F.; Boswell, T.A.; Alnakhalah, A.; et al. Characterization of MYBL1 Gene in Triple-Negative Breast Cancers and the Genes’ Relationship to Alterations Identified at the Chromosome 8q Loci. Int. J. Mol. Sci. 2024, 25, 2539. [Google Scholar] [CrossRef] [PubMed]
- Brayer, K.J.; Frerich, C.A.; Kang, H.; Ness, S.A. Recurrent Fusions in MYB and MYBL1 Define a Common, Transcription Factor-Driven Oncogenic Pathway in Salivary Gland Adenoid cystic carcinoma. Cancer Discov. 2016, 6, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Ramkissoon, L.A.; Horowitz, P.M.; Craig, J.M.; Ramkissoon, S.H.; Rich, B.E.; Schumacher, S.E.; McKenna, A.; Lawrence, M.S.; Bergthold, G.; Brastianos, P.K.; et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc. Natl. Acad. Sci. USA 2013, 110, 8188–8193. [Google Scholar] [CrossRef]
- Persson, M.; Andersson, M.K.; Mitani, Y.; Brandwein-Weber, M.S.; Frierson, H.F., Jr.; Moskaluk, C.; Fonseca, I.; Ferrarotto, R.; Boecker, W.; Loening, T.; et al. Rearrangements, Expression, and Clinical Significance of MYB and MYBL1 in Adenoid cystic carcinoma: A Multi-Institutional Study. Cancers 2022, 14, 3691. [Google Scholar] [CrossRef]
- Gorbatenko, A.; Søkilde, R.; Sorensen, E.E.; Newie, I.; Persson, H.; Morancho, B.; Arribas, J.; Litman, T.; Rovira, C.; Pedersen, S.F. HER2 and p95HER2 differentially regulate miRNA expression in MCF-7 breast cancer cells and downregulate MYB proteins through miR-221/222 and miR-503. Sci. Rep. 2019, 9, 3352. [Google Scholar] [CrossRef] [PubMed]
- Sala, A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur. J. Cancer 2005, 41, 2479–2484. [Google Scholar] [CrossRef]
- Taylor, D.; Badiani, P.; Weston, K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996, 10, 2732–2744. [Google Scholar] [CrossRef] [PubMed]
- Ryall, S.; Tabori, U.; Hawkins, C. A comprehensive review of paediatric low-grade diffuse glioma: Pathology, molecular genetics and treatment. Brain Tumor Pathol. 2017, 34, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, S.; Hirato, J.; Yokoo, H. Molecular genetics of ependymomas and pediatric diffuse gliomas: A short review. Brain Tumor Pathol. 2014, 31, 229–233. [Google Scholar] [CrossRef]
- Sala, A.; Casella, I.; Grasso, L.; Bellon, T.; Reed, J.C.; Miyashita, T.; Peschle, C. Apoptotic response to oncogenic stimuli: Cooperative and antagonistic interactions between c-myb and the growth suppressor p53. Cancer Res. 1996, 56, 1991–1996. [Google Scholar] [PubMed]
- Shu, G.S.; Lv, F.; Yang, Z.L.; Miao, X.Y. Immunohistochemical study of PUMA, c-Myb and p53 expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Int. J. Clin. Oncol. 2012, 18, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular carcinoma. Cell 2017, 169, 1327–1341.e23. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC: A Quality Control Tool for High Throughput Sequence Data 2023. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 July 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Carbonetto, P.; Gerard, D.; Lu, M.; Sun, L.; Willwerscheid, J.; Xiao, N. ashr: Methods for Adaptive Shrinkage, Using Empirical Bayes. R Package Version 2.2-54. 2022. Available online: https://CRAN.R-project.org/package=ashr (accessed on 10 July 2024).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 10 July 2024).
- Dahn, M.L.; Dean, C.A.; Jo, D.B.; Coyle, K.M.; Marcato, P. Human-specific GAPDH qRT-PCR is an accurate and sensitive method of xenograft metastasis quantification. Mol. Ther.-Methods Clin. Dev. 2020, 20, 398–408. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer | Primer Sequence |
|---|---|---|
| MYBL-1 | MYBL-1 F | 5′-CGTGGAGGCAAACGCTGTGTTA-3′ |
| MYBL1 R | 5′-GGTGGATTTGATAGGAGAAGCAG-3′ | |
| NTF-3 | NTF-3 F | 5′-CAAGCAGATGGTGGACGTTAAGG-3′ |
| NTF-3 R | 5′-TCGCAGCAGTTCGGTGTCCATT-3′ | |
| GAPDH | GAPDH F | 5′-TCAAGGCTGAGAACGGGAAG-3′ |
| GAPDH R | 5′-CGCCCCACTTGATTTTGGAG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrović, A.; Štancl, P.; Gršković, P.; Hančić, S.; Karlić, R.; Gašparov, S.; Korać, P. Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma. Int. J. Mol. Sci. 2024, 25, 10558. https://doi.org/10.3390/ijms251910558
Petrović A, Štancl P, Gršković P, Hančić S, Karlić R, Gašparov S, Korać P. Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2024; 25(19):10558. https://doi.org/10.3390/ijms251910558
Chicago/Turabian StylePetrović, Andreja, Paula Štancl, Paula Gršković, Suzana Hančić, Rosa Karlić, Slavko Gašparov, and Petra Korać. 2024. "Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma" International Journal of Molecular Sciences 25, no. 19: 10558. https://doi.org/10.3390/ijms251910558
APA StylePetrović, A., Štancl, P., Gršković, P., Hančić, S., Karlić, R., Gašparov, S., & Korać, P. (2024). Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma. International Journal of Molecular Sciences, 25(19), 10558. https://doi.org/10.3390/ijms251910558







