Novel CD44-Targeted Albumin Nanoparticles: An Innovative Approach to Improve Breast Cancer Treatment
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of Cationic cysHSA
2.2. Synthesis and Characterization of FNPs
2.3. Drug Delivery Studies
2.4. Safety and Toxicity of DOX@FNPs in Breast Cancer Cell Lines
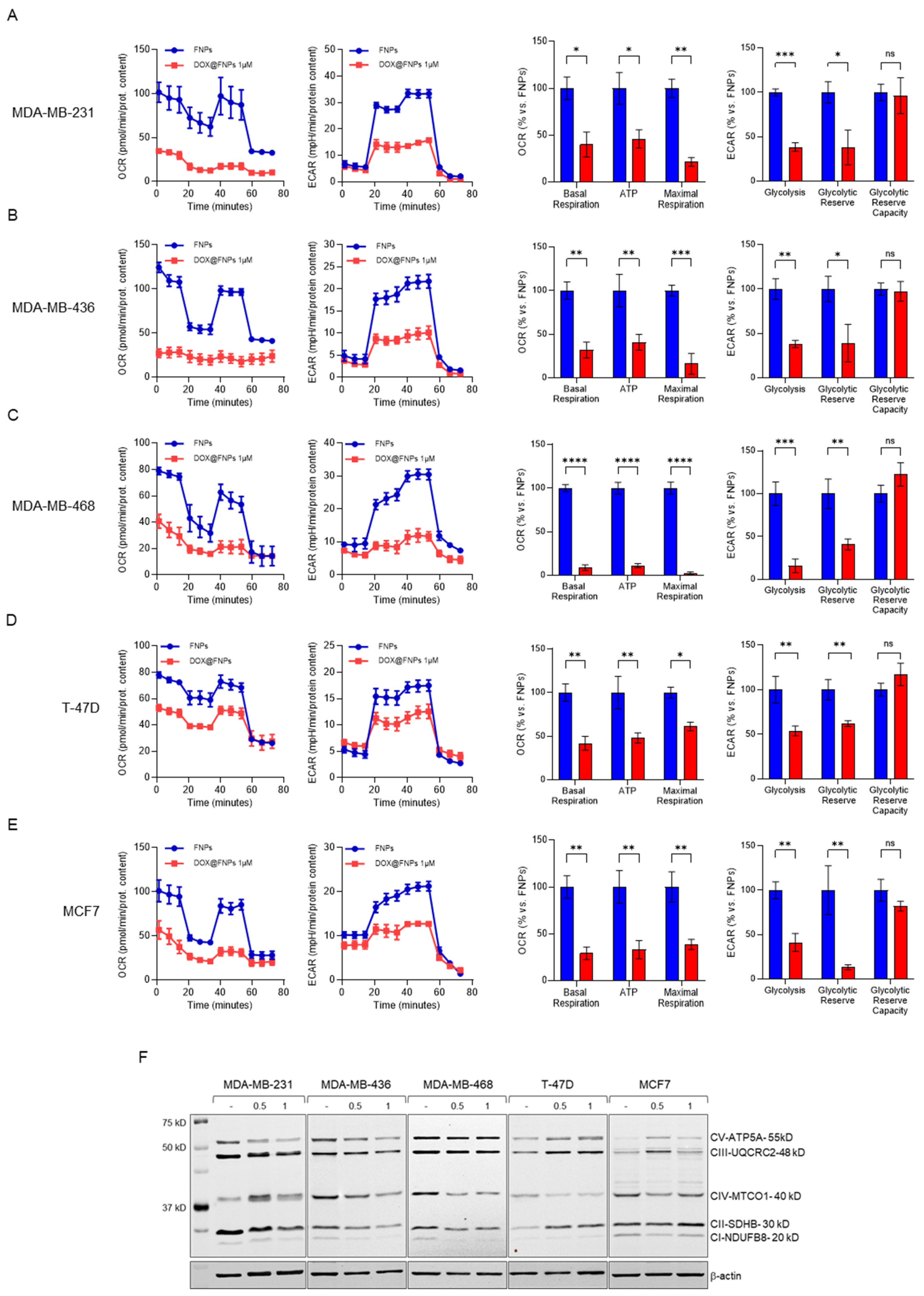
2.5. DOX@FNPs Affected Cellular Metabolism by Targeting CD44
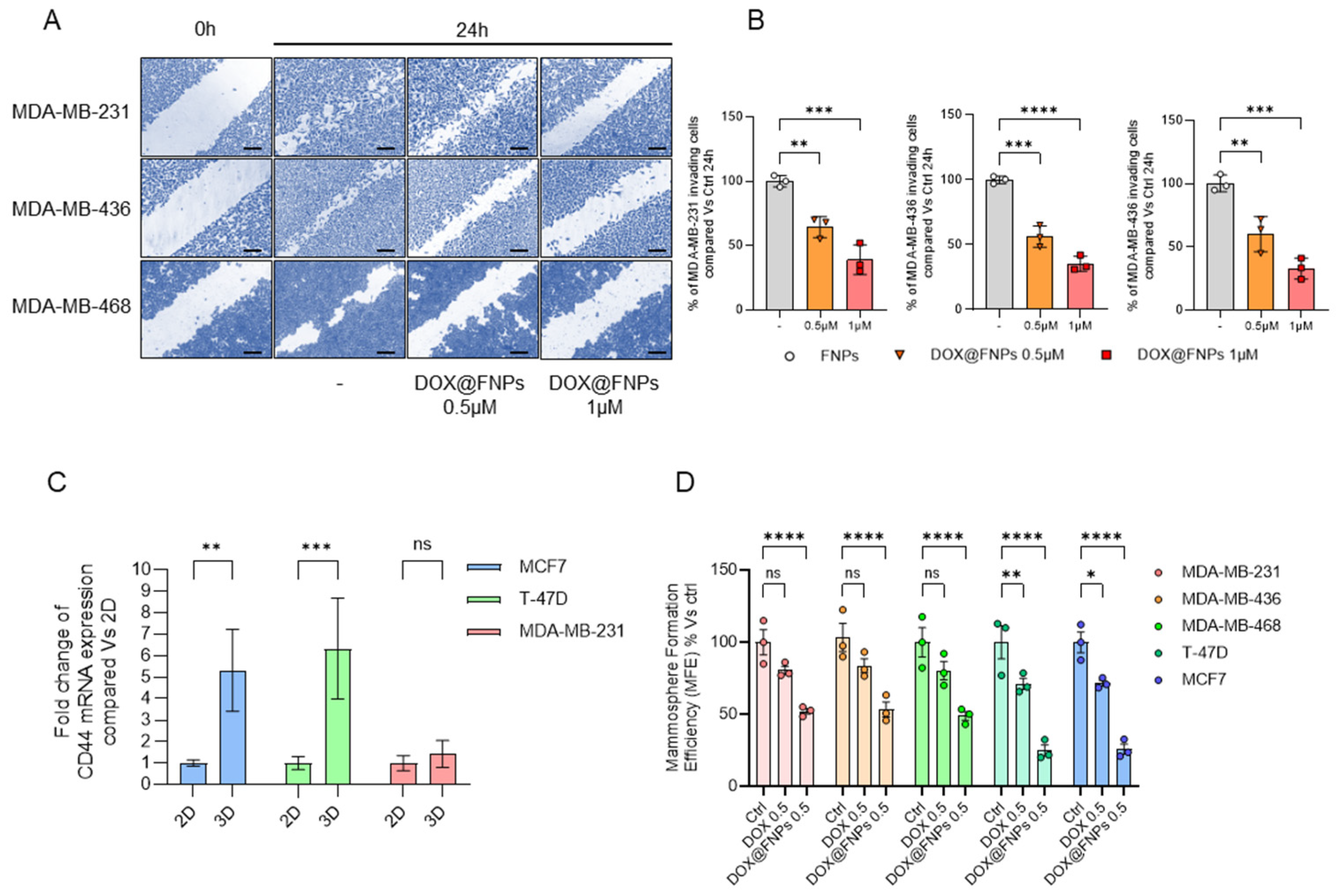
2.6. DOX@FNPs Inhibited 2D Migration and 3D Tumor Formation
3. Materials and Methods
3.1. Synthesis and Characterization of cysHSA Conjugate
3.2. Preparation of FNPs and DOX@FNPs
3.3. DOX Loading and Release Experiments
3.4. Cell Cultures
3.5. Viability Assay
3.6. Metabolic Flux Analysis with the Seahorse XFe96
3.7. Western Blotting
3.8. Wound-Healing Scratch Assay
3.9. Mammosphere Formation Efficiency
3.10. qPCR Analysis of CD44 Expression
3.11. Three-Dimensional Spheroid Formation Assay
3.12. cBioPortal Analysis
3.13. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The Ever-Increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.M.; Cao, Z.; Prettner, K.; Kuhn, M.; Yang, J.T.; Jiao, L.R.; Wang, Z.R.; Li, W.M.; Geldsetzer, P.; Baernighausen, T.; et al. Estimates and Projections of the Global Economic Cost of 29 Cancers in 204 Countries and Territories from 2020 to 2050. JAMA Oncol. 2023, 9, 465–472. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.Y.A.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA—Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Y.; Gong, M.Y.; Wang, Y.; Yang, Y.; Liu, S.; Zeng, Q.B. Global Trends and Forecasts of Breast Cancer Incidence and Deaths. Sci. Data 2023, 10, 334. [Google Scholar] [CrossRef] [PubMed]
- Abed, H.F.; Abuwatfa, W.H.; Husseini, G.A. Redox-Responsive Drug Delivery Systems: A Chemical Perspective. Nanomaterials 2022, 12, 3183. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Veerapandian, M.; Ramasundaram, S.; Jerome, P.; Chellasamy, G.; Govindaraju, S.; Yun, K.; Oh, T.H. Drug Delivery Application of Functional Nanomaterials Synthesized Using Natural Sources. J. Funct. Biomater. 2023, 14, 426. [Google Scholar] [CrossRef]
- Mahani, M.; Bahmanpouri, M.; Khakbaz, F.; Divsar, F. Doxorubicin-Loaded Polymeric Micelles Decorated with Nitrogen-Doped Carbon Dots for Targeted Breast Cancer Therapy. J. Drug Deliv. Sci. Technol. 2023, 79, 104055. [Google Scholar] [CrossRef]
- Dristant, U.; Mukherjee, K.; Saha, S.; Maity, D. An Overview of Polymeric Nanoparticles-Based Drug Delivery System in Cancer Treatment. Technol. Cancer Res. Treat. 2023, 22, 15330338231152083. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.T.; Wang, X.; Liu, Y.; Yang, G.Z.; Falconer, R.J.; Zhao, C.X. Lipid Nanoparticles for Drug Delivery. Adv. NanoBiomed Res. 2022, 2, 2100109. [Google Scholar] [CrossRef]
- Sharifi, M.; Cho, W.C.; Ansariesfahani, A.; Tarharoudi, R.; Malekisarvar, H.; Sari, S.; Bloukh, S.H.; Edis, Z.; Amin, M.; Gleghorn, J.P.; et al. An Updated Review on Epr-Based Solid Tumor Targeting Nanocarriers for Cancer Treatment. Cancers 2022, 14, 2868. [Google Scholar] [CrossRef] [PubMed]
- Caro, C.; Avasthi, A.; Paez-Muñoz, J.M.; Leal, M.P.; García-Martín, M.L. Passive Targeting of High-Grade Gliomas the Epr Effect: A Closed Path for Metallic Nanoparticles? Biomater. Sci. 2021, 9, 7984–7995. [Google Scholar] [CrossRef]
- Soleymani, M.; Poorkhani, A.; Khalighfard, S.; Velashjerdi, M.; Khori, V.; Khodayari, S.; Khodayari, H.; Dehghan, M.; Alborzi, N.; Agah, S.; et al. Folic Acid-Conjugated Dextran-Coated Zn Mn FeO Nanoparticles as Systemically Delivered Nano Heaters with Self-Regulating Temperature for Magnetic Hyperthermia Therapy of Liver Tumors. Sci. Rep. 2023, 13, 13560. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, P.; Sarker, S.; Ghosh, A.; Gupta, P.; Das, S.; Ahir, M.; Bhattacharya, S.; Chattopadhyay, S.; Ghosh, S.; Adhikary, A. Transferrin-Decorated Thymoquinone-Loaded Peg-Plga Nanoparticles Exhibit Anticarcinogenic Effect in Non-Small Cell Lung Carcinoma the Modulation of Mir-34a and Mir-16. Biomater. Sci. 2019, 7, 4325–4344. [Google Scholar] [CrossRef] [PubMed]
- Palanikumar, L.; Al-Hosani, S.; Kalmouni, M.; Nguyen, V.P.; Ali, L.; Pasricha, R.; Barrera, F.N.; Magzoub, M. Ph-Responsive High Stability Polymeric Nanoparticles for Targeted Delivery of Anticancer Therapeutics. Commun. Biol. 2020, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.F.; Liu, R.; Wang, Y.S.; Hu, C.A.; Yu, W.Q.; Zhou, Y.; Wang, L.; Zhang, M.J.; Gao, H.L.; Gao, X. Dual-Responsive Nanoparticles with Transformable Shape and Reversible Charge for Amplified Chemo-Photodynamic Therapy of Breast Cancer. Acta Pharm. Sin. B 2022, 12, 3354–3366. [Google Scholar] [CrossRef]
- Yang, J.H.; Jia, C.Y.; Yang, J.S. Designing Nanoparticle-Based Drug Delivery Systems for Precision Medicine. Internatl. J. Med. Sci. 2021, 18, 2943–2949. [Google Scholar] [CrossRef]
- Curcio, M.; Diaz-Gomez, L.; Cirillo, G.; Nicoletta, F.P.; Leggio, A.; Iemma, F. Dual-Targeted Hyaluronic Acid/Albumin Micelle-Like Nanoparticles for the Vectorization of Doxorubicin. Pharmaceutics 2021, 13, 304. [Google Scholar] [CrossRef]
- Kesharwani, P.; Chadar, R.; Sheikh, A.; Rizg, W.Y.; Safhi, A.Y. Cd44-Targeted Nanocarrier for Cancer Therapy. Front. Pharmacol. 2022, 12, 800481. [Google Scholar] [CrossRef] [PubMed]
- Suksiriworapong, J.; Pongprasert, N.; Bunsupa, S.; Taresco, V.; Crucitti, V.C.; Janurai, T.; Phruttiwanichakun, P.; Sakchaisri, K.; Wongrakpanich, A. Cd44-Targeted Lipid Polymer Hybrid Nanoparticles Enhance Anti-Breast Cancer Effect of Extracts. Pharmaceutics 2023, 15, 1771. [Google Scholar] [CrossRef]
- Gomari, M.M.; Farsimadan, M.; Rostami, N.; Mahmoudi, Z.; Fadaie, M.; Farhani, I.; Tarighi, P. Cd44 Polymorphisms and Its Variants, as an Inconsistent Marker in Cancer Investigations. Mutat. Res.-Rev. Mutat. Res. 2021, 787, 108374. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kang, M.S.; Jeong, W.Y.; Han, D.W.; Kim, K.S. Hyaluronic Acid-Based Theranostic Nanomedicines for Targeted Cancer Therapy. Cancers 2020, 12, 940. [Google Scholar] [CrossRef] [PubMed]
- Puluhulawa, L.E.; Joni, I.M.; Elamin, K.M.; Mohammed, A.F.A.; Muchtaridi, M.; Wathoni, N. Chitosan-Hyaluronic Acid Nanoparticles for Active Targeting in Cancer Therapy. Polymers 2022, 14, 3410. [Google Scholar] [CrossRef]
- Farzan, M.; Varshosaz, J.; Mirian, M.; Minaiyan, M.; Pezeshki, A. Hyaluronic Acid-Conjugated Gliadin Nanoparticles for Targeted Delivery of Usnic Acid in Breast Cancer: An In Vitro/In Vivo Translational Study. J. Drug Deliv. Sci. Technol. 2023, 84, 104459. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Hou, D.; Marigo, C.C.; Bonelli, J.; Rocas, P.; Cheng, F.Z.; Yang, X.Q.; Rocas, J.; Hamberg, N.M.; Han, J.Y. Redox-Responsive Polyurethane-Polyurea Nanoparticles Targeting to Aortic Endothelium and Atherosclerosis. iScience 2022, 25, 105390. [Google Scholar] [CrossRef]
- Mollazadeh, S.; Mackiewicz, M.; Yazdimamaghani, M. Recent Advances in the Redox-Responsive Drug Delivery Nanoplatforms: A Chemical Structure and Physical Property Perspective. Mater. Sci. Eng. C-Mater. Biol. Appl. 2021, 118, 111536. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Luo, S.Y.; Mukerabigwi, J.F.; Mei, J.; Zhang, Y.N.; Wang, S.F.; Xiao, W.; Huang, X.Y.; Cao, Y. Targeted Nanoparticles from Xyloglucan-Doxorubicin Conjugate Loaded with Doxorubicin against Drug Resistance. RSC Adv. 2016, 6, 26137–26146. [Google Scholar] [CrossRef]
- Gou, Y.; Miao, D.D.; Zhou, M.; Wang, L.J.; Zhou, H.Y.; Su, G.X. Bio-Inspired Protein-Based Nanoformulations for Cancer Theranostics. Front. Pharmacol. 2018, 9, 421. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Bahrami, S.; Ravari, S.B.; Zangabad, P.S.; Mirshekari, H.; Bozorgomid, M.; Shahreza, S.; Sori, M.; Hamblin, M.R. Albumin Nanostructures as Advanced Drug Delivery Systems. Expert Opin. Drug Deliv. 2016, 13, 1609–1623. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.T.; Lee, R.J.; Meng, F.C.; Wang, G.Y.; Zheng, X.L.; Dong, S.Y.; Teng, L.S. Microfluidic Self-Assembly of High Cabazitaxel Loading Albumin Nanoparticles. Nanoscale 2020, 12, 16928–16933. [Google Scholar] [CrossRef]
- Saleh, T.; Soudi, T.; Shojaosadati, S.A. Redox Responsive Curcumin-Loaded Human Serum Albumin Nanoparticles: Preparation, Characterization and Evaluation. Int. J. Biol. Macromol. 2018, 114, 759–766. [Google Scholar] [CrossRef]
- Yasuda, K.; Maeda, H.; Kinoshita, R.; Minayoshi, Y.; Mizuta, Y.; Nakamura, Y.; Imoto, S.; Nishi, K.; Yamasaki, K.; Sakuragi, M.; et al. Encapsulation of an Antioxidant in Redox-Sensitive Self-Assembled Albumin Nanoparticles for the Treatment of Hepatitis. ACS Nano 2023, 17, 16668–16681. [Google Scholar] [CrossRef] [PubMed]
- Varga, N.; Seres, L.; Kovács, N.A.; Turcsányi, A.; Juhász, A.; Csapó, E. Serum Albumin/Hyaluronic Acid Nanoconjugate: Evaluation of Concentration-Dependent Structural Changes to Form an Efficient Drug Carrier Particle. Int. J. Biol. Macromol. 2022, 220, 1523–1531. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, X.; Xu, Y.Y.; Han, X.R.; Wang, M.; Gong, T.; Zhang, Z.R.; Kao, W.J.; Fu, Y. Hierarchical Assembly of Hyaluronan Coated Albumin Nanoparticles for Pancreatic Cancer Chemoimmunotherapy. Nanoscale 2019, 11, 16476–16487. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yang, C.X.; Gao, F. The State of Cd44 Activation in Cancer Progression and Therapeutic Targeting. FEBS J. 2022, 289, 7970–7986. [Google Scholar] [CrossRef]
- Tamada, M.; Nagano, O.; Tateyama, S.; Ohmura, M.; Yae, T.; Ishimoto, T.; Sugihara, E.; Onishi, N.; Yamamoto, T.; Yanagawa, H.; et al. Modulation of Glucose Metabolism by Cd44 Contributes to Antioxidant Status and Drug Resistance in Cancer Cells. Cancer Res. 2012, 72, 1438–1448. [Google Scholar] [CrossRef]
- Fischer, D.; Bieber, T.; Brüsselbach, S.; Elsässer, H.P.; Kissel, T. Cationized Human Serum Albumin as a Non-Viral Vector System for Gene Delivery? Characterization of Complex Formation with Plasmid DNA and Transfection Efficiency. Int. J. Pharm. 2001, 225, 97–111. [Google Scholar] [CrossRef]
- Oganesyan, V.; Damschroder, M.M.; Cook, K.E.; Li, Q.; Gao, C.S.; Wu, H.R.; Dall, W.F. Structural Insights into Neonatal Fc Receptor-Based Recycling Mechanisms. J. Biol. Chem. 2014, 289, 7812–7824. [Google Scholar] [CrossRef]
- Kang, H.; Rho, S.; Stiles, W.R.; Hu, S.; Baek, Y.; Hwang, D.W.; Kashiwagi, S.; Kim, M.S.; Choi, H.S. Size-Dependent Epr Effect of Polymeric Nanoparticles on Tumor Targeting. Adv. Healthc. Mater. 2020, 9, e1901223. [Google Scholar] [CrossRef]
- Lazzari, S.; Moscatelli, D.; Codari, F.; Salmona, M.; Morbidelli, M.; Diomede, L. Colloidal Stability of Polymeric Nanoparticles in Biological Fluids. J. Nanopart. Res. 2012, 14, 920. [Google Scholar] [CrossRef] [PubMed]
- Hervault, A.; Dunn, A.E.; Lim, M.; Boyer, C.; Mott, D.; Maenosono, S.; Thanh, N.T.K. Doxorubicin Loaded Dual Ph- and Thermo-Responsive Magnetic Nanocarrier for Combined Magnetic Hyperthermia and Targeted Controlled Drug Delivery Applications. Nanoscale 2016, 8, 12152–12161. [Google Scholar] [CrossRef]
- Curcio, M.; Paoli, A.; Cirillo, G.; Di Pietro, S.; Forestiero, M.; Giordano, F.; Mauro, L.; Amantea, D.; Di Bussolo, V.; Nicoletta, F.P.; et al. Combining Dextran Conjugates with Stimuli-Responsive and Folate-Targeting Activity: A New Class of Multifunctional Nanoparticles for Cancer Therapy. Nanomaterials 2021, 11, 1108. [Google Scholar] [CrossRef]
- Nicoletto, R.E.; Ofner, C.M. Cytotoxic Mechanisms of Doxorubicin at Clinically Relevant Concentrations in Breast Cancer Cells. Cancer Chemother. Pharmacol. 2022, 89, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Onafuye, H.; Pieper, S.; Mulac, D.; Cinatl, J.; Wass, M.N.; Langer, K.; Michaelis, M. Doxorubicin-Loaded Human Serum Albumin Nanoparticles Overcome Transporter-Mediated Drug Resistance in Drug-Adapted Cancer Cells. Beilstein J. Nanotechnol. 2019, 10, 1707–1715. [Google Scholar] [CrossRef]
- Kumbham, S.; Paul, M.; Itoo, A.; Ghosh, B.; Biswas, S. Oleanolic Acid-Conjugated Human Serum Albumin Nanoparticles Encapsulating Doxorubicin as Synergistic Combination Chemotherapy in Oropharyngeal Carcinoma and Melanoma. Int. J. Pharm. 2022, 614, 121479. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, S.P.H.; Yazdimamaghani, M.; Ghandehari, H. Glutathione-Sensitive Hollow Mesoporous Silica Nanoparticles for Controlled Drug Delivery. J. Control. Release 2018, 282, 62–75. [Google Scholar] [CrossRef]
- Yang, G.-G.; Su, X.-X.; Liang, B.B.; Pan, Z.Y.; Cao, Q.; Mao, Z.W. A Platinum-Ruthenium Hybrid Prodrug with Multi-Enzymatic Activities for Chemo-Catalytic Therapy of Hypoxic Tumors. Chem. Sci. 2022, 13, 11360–11367. [Google Scholar] [CrossRef]
- Magazzù, A.; Marcuello, C. Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review. Nanomaterials 2023, 13, 963. [Google Scholar] [CrossRef]
- Fraczkowska, K.; Bacia, M.; Przybylo, M.; Drabik, D.; Kaczorowska, A.; Rybka, J.; Stefanko, E.; Drobczynski, S.; Masajada, J.; Podbielska, H.; et al. Alterations of Biomechanics in Cancer and Normal Cells Induced by Doxorubicin. Biomed. Pharmacother. 2018, 97, 1195–1203. [Google Scholar] [CrossRef]
- Haripriyaa, M.; Suthindhiran, K. Pharmacokinetics of Nanoparticles: Current Knowledge, Future Directions and Its Implications in Drug Delivery. Future J. Pharm. Sci. 2023, 9, 113. [Google Scholar] [CrossRef]
- Valcourt, D.M.; Kapadia, C.H.; Scully, M.A.; Dang, M.N.; Day, E.S. Best Practices for Preclinical in Vivo Testing of Cancer Nanomedicines. Adv. Healthc. Mater. 2020, 9, e2000110. [Google Scholar] [CrossRef]
- Provencher, S.W. A Constrained Regularization Method for Inverting Data Represented by Linear Algebraic or Integral Equations. Comput. Phys. Commun. 1982, 27, 213–227. [Google Scholar] [CrossRef]
- Armentano, B.; Curcio, R.; Brindisi, M.; Mancuso, R.; Rago, V.; Ziccarelli, I.; Frattaruolo, L.; Fiorillo, M.; Dolce, V.; Gabriele, B.; et al. 5-(Carbamoylmethylene)-Oxazolidin-2-Ones as a Promising Class of Heterocycles Inducing Apoptosis Triggered by Increased Ros Levels and Mitochondrial Dysfunction in Breast and Cervical Cancer. Biomedicines 2020, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, M.; Sanchez-Alvarez, R.; Sotgia, F.; Lisanti, M.P. The Er-Alpha Mutation Y537s Confers Tamoxifen-Resistance Via Enhanced Mitochondrial Metabolism, Glycolysis and Rho-Gdi/Pten Signaling: Implicating Tigar in Somatic Resistance to Endocrine Therapy. Aging 2018, 10, 4000–4023. [Google Scholar] [CrossRef] [PubMed]
- Frattaruolo, L.; Lauria, G.; Aiello, F.; Carullo, G.; Curcio, R.; Fiorillo, M.; Campiani, G.; Dolce, V.; Cappello, A.R. Exploiting Glycyrrhiza glabra L. (Licorice) Flavanones: Licoflavanone’s Impact on Breast Cancer Cell Bioenergetics. Int. J. Mol. Sci. 2024, 25, 7907. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Rizza, P.; Donà, A.; Nigro, A.; Ricci, E.; Fiorillo, M.; Perrotta, I.; Lanzino, M.; Giordano, C.; Bonofiglio, D.; et al. Foxo3a as a Positive Prognostic Marker and a Therapeutic Target in Tamoxifen-Resistant Breast Cancer. Cancers 2019, 11, 1858. [Google Scholar] [CrossRef] [PubMed]
- Abasalizadeh, F.; Moghaddam, S.V.; Alizadeh, E.; Akbari, E.; Kashani, E.; Fazljou, S.M.B.; Torbati, M.; Akbarzadeh, A. Alginate-Based Hydrogels as Drug Delivery Vehicles in Cancer Treatment and Their Applications in Wound Dressing and 3d Bioprinting. J. Biol. Eng. 2020, 14, 8. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cirillo, G.; Cappello, A.R.; Curcio, M.; Fiorillo, M.; Frattaruolo, L.; Avena, P.; Scorzafave, L.; Dolce, V.; Nicoletta, F.P.; Iemma, F. Novel CD44-Targeted Albumin Nanoparticles: An Innovative Approach to Improve Breast Cancer Treatment. Int. J. Mol. Sci. 2024, 25, 10560. https://doi.org/10.3390/ijms251910560
Cirillo G, Cappello AR, Curcio M, Fiorillo M, Frattaruolo L, Avena P, Scorzafave L, Dolce V, Nicoletta FP, Iemma F. Novel CD44-Targeted Albumin Nanoparticles: An Innovative Approach to Improve Breast Cancer Treatment. International Journal of Molecular Sciences. 2024; 25(19):10560. https://doi.org/10.3390/ijms251910560
Chicago/Turabian StyleCirillo, Giuseppe, Anna Rita Cappello, Manuela Curcio, Marco Fiorillo, Luca Frattaruolo, Paola Avena, Ludovica Scorzafave, Vincenza Dolce, Fiore Pasquale Nicoletta, and Francesca Iemma. 2024. "Novel CD44-Targeted Albumin Nanoparticles: An Innovative Approach to Improve Breast Cancer Treatment" International Journal of Molecular Sciences 25, no. 19: 10560. https://doi.org/10.3390/ijms251910560







