Distinct Non-Human Leukocyte Antigen Antibody Signatures Correlate with Endothelial Crossmatch Status in Lung and Renal Transplant Recipients
Abstract
:1. Introduction
2. Results
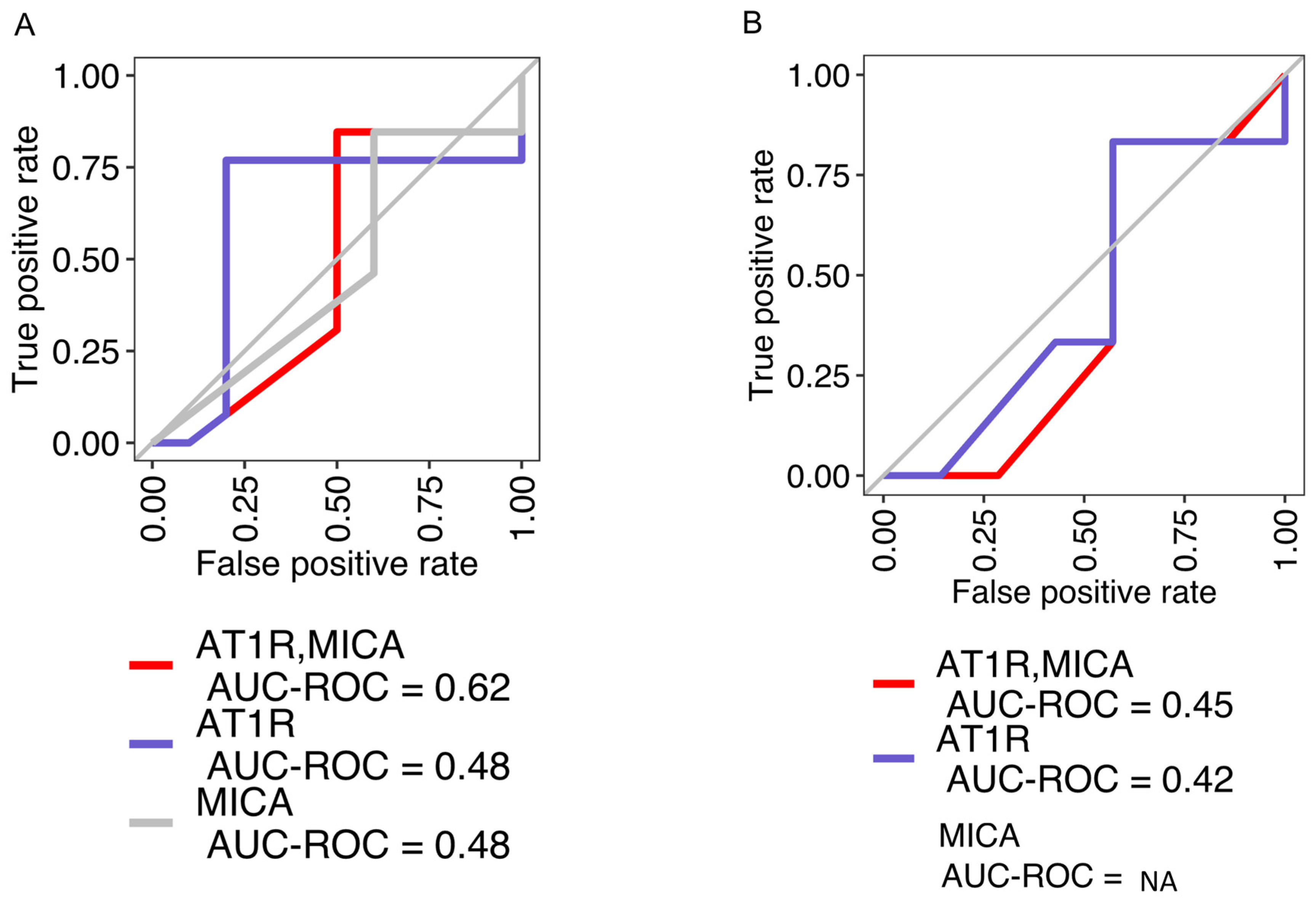
2.1. Concordance among AT1R, MICA, and Positive Endothelial Crossmatch Is Limited
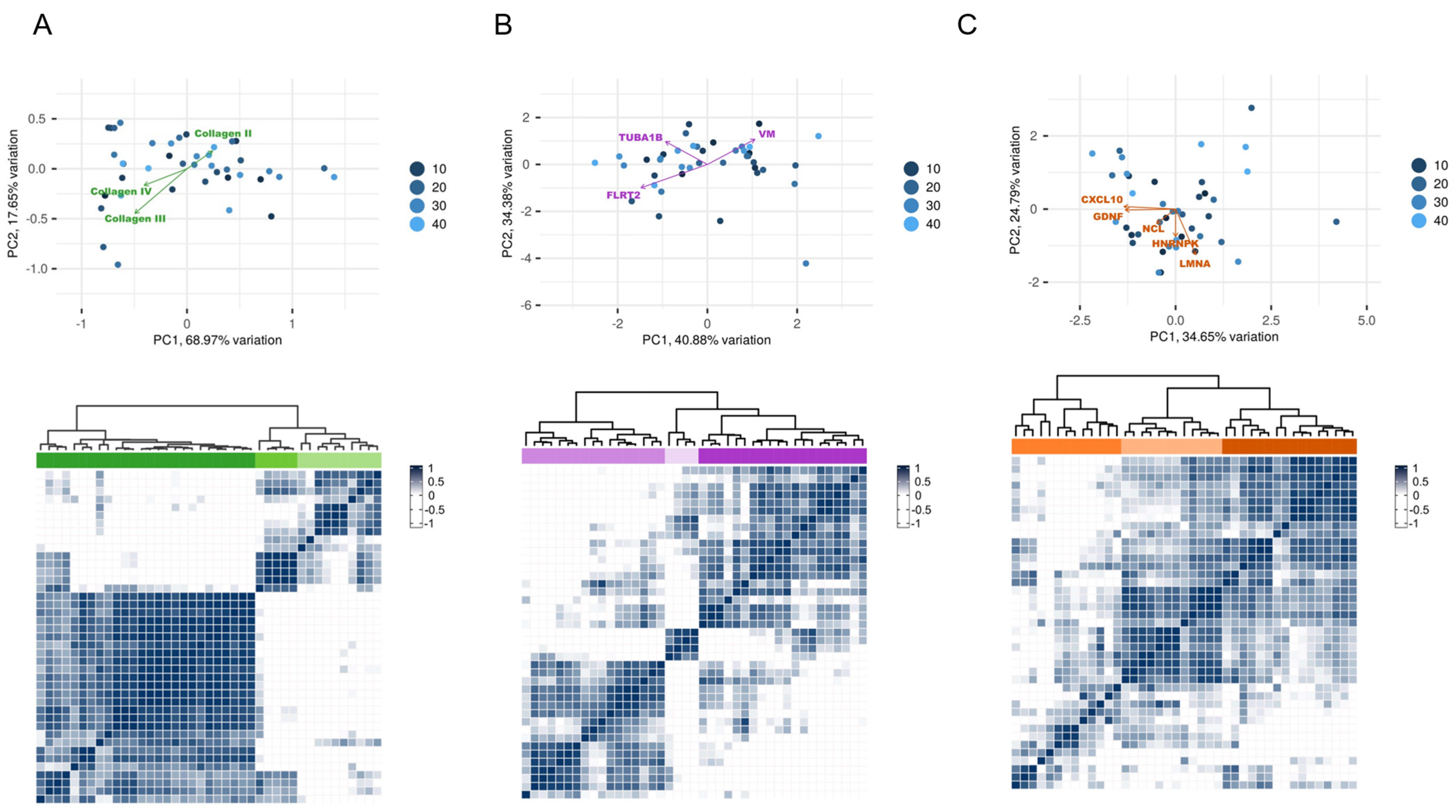
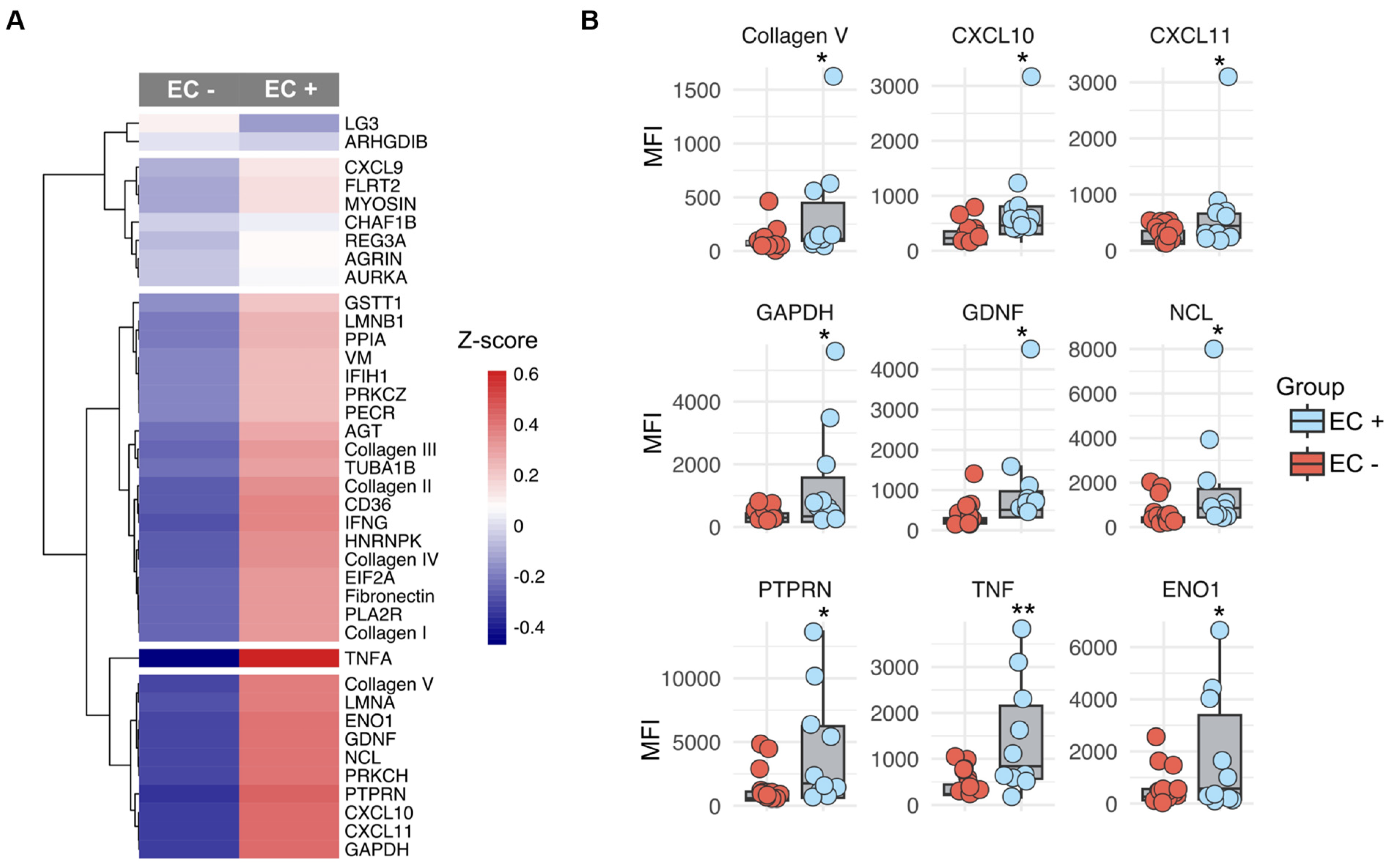
2.2. Identification of Non-HLA Antibody Clusters in Post-Transplant Patients
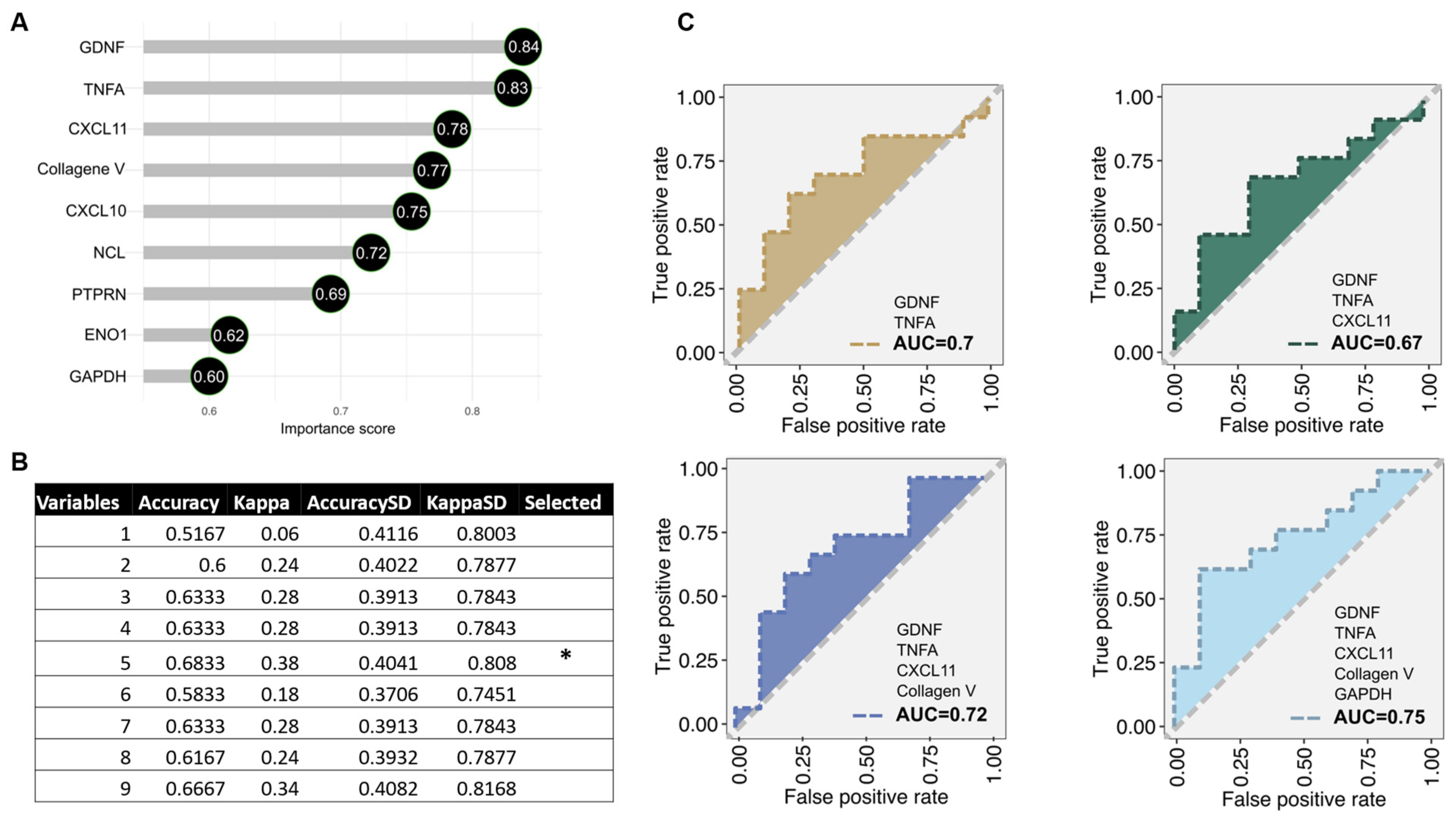
2.3. Antibody Subtypes as Predictors for Endothelial Crossmatch Subtypes
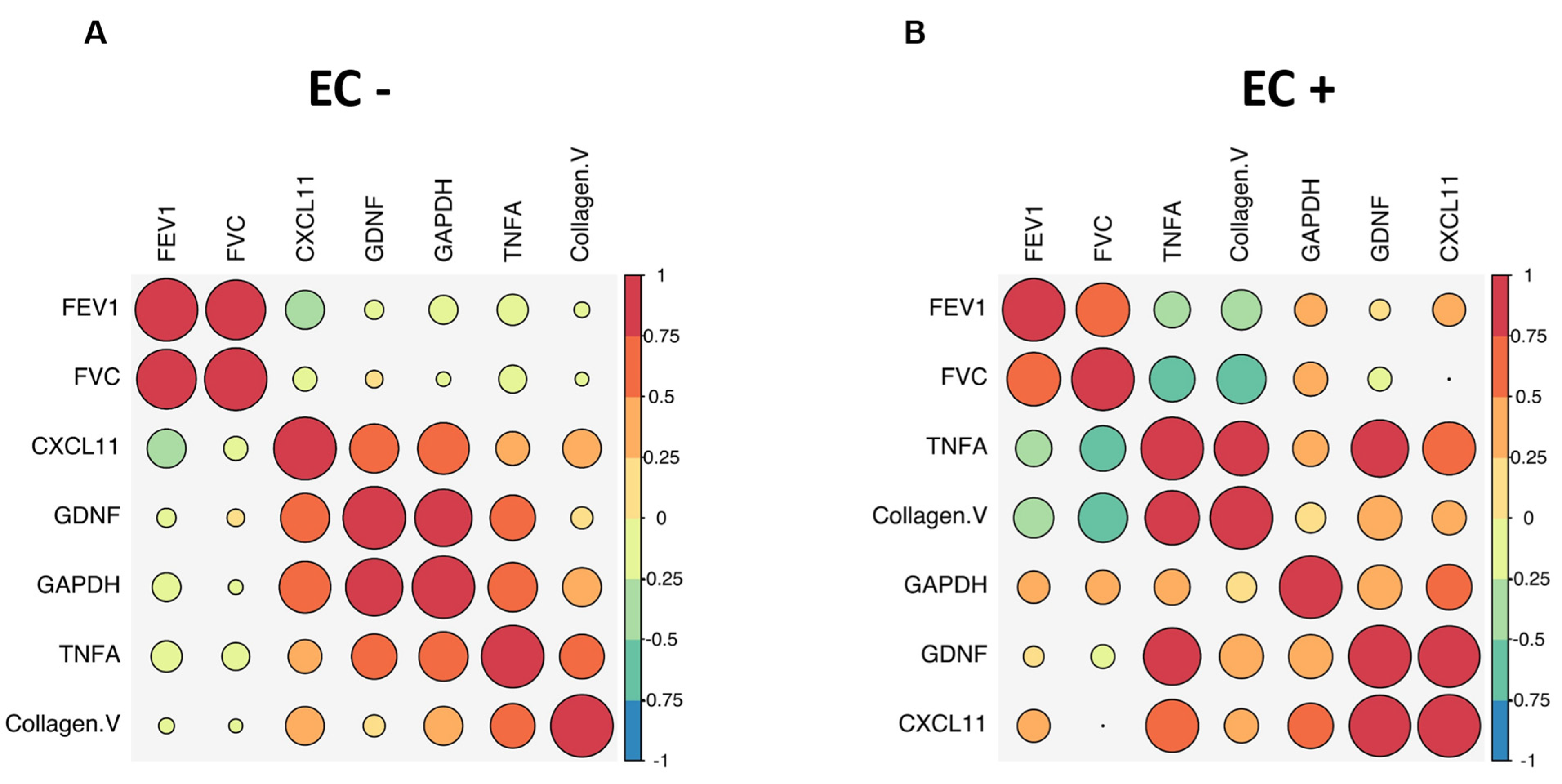
2.4. Distinct Antibody Signature Correlates Inversely with Lung Function in LTRs
3. Discussion
4. Materials and Methods
4.1. Human Sample Collection
4.2. Sample Processing and Autoantibody Testing
4.3. Quality Control and Analyses
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leard, L.E.; Holm, A.M.; Valapour, M.; Glanville, A.R.; Attawar, S.; Aversa, M.; Campos, S.V.; Christon, L.M.; Cypel, M.; Dellgren, G.; et al. Consensus document for the selection of lung transplant candidates: An update from the International Society for Heart and Lung Transplantation. J. Heart Lung Transpl. 2021, 40, 1349–1379. [Google Scholar] [CrossRef]
- Abate, D.A.; Watanabe, S.; Mocarski, E.S. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 2004, 78, 10995–11006. [Google Scholar] [CrossRef] [PubMed]
- Glanville, A.R.; Verleden, G.M.; Todd, J.L.; Benden, C.; Calabrese, F.; Gottlieb, J.; Hachem, R.R.; Levine, D.; Meloni, F.; Palmer, S.M.; et al. Chronic lung allograft dysfunction: Definition and update of restrictive allograft syndrome-A consensus report from the Pulmonary Council of the ISHLT. J. Heart Lung Transpl. 2019, 38, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Loupy, A.; Lefaucheur, C. Antibody-Mediated Rejection of Solid-Organ Allografts. N. Engl. J. Med. 2018, 379, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Levine, D.J.; Glanville, A.R.; Aboyoun, C.; Belperio, J.; Benden, C.; Berry, G.J.; Hachem, R.; Hayes, D., Jr.; Neil, D.; Reinsmoen, N.L.; et al. Antibody-mediated rejection of the lung: A consensus report of the International Society for Heart and Lung Transplantation. J. Heart Lung Transpl. 2016, 35, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Verleden, S.E.; Vanaudenaerde, B.M.; Emonds, M.P.; Van Raemdonck, D.E.; Neyrinck, A.P.; Verleden, G.M.; Vos, R. Donor-specific and -nonspecific HLA antibodies and outcome post lung transplantation. Eur. Respir. J. 2017, 50, 1701248. [Google Scholar] [CrossRef]
- Butler, C.L.; Hickey, M.J.; Jiang, N.; Zheng, Y.; Gjertson, D.; Zhang, Q.; Rao, P.; Fishbein, G.A.; Cadeiras, M.; Deng, M.C.; et al. Discovery of non-HLA antibodies associated with cardiac allograft rejection and development and validation of a non-HLA antigen multiplex panel: From bench to bedside. Am. J. Transpl. 2020, 20, 2768–2780. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Hu, J.; Luo, Q.; Ding, X.; Luo, W.; Zhuang, Q.; Zou, Y. Acute Antibody-Mediated Rejection in Presence of MICA-DSA and Successful Renal Re-Transplant with Negative-MICA Virtual Crossmatch. PLoS ONE 2015, 10, e0127861. [Google Scholar] [CrossRef] [PubMed]
- Hachem, R.R. The impact of non-HLA antibodies on outcomes after lung transplantation and implications for therapeutic approaches. Hum. Immunol. 2019, 80, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, P.I.; Ozawa, M.; Castro, R. Four-year follow-up of a prospective trial of HLA and MICA antibodies on kidney graft survival. Am. J. Transpl. 2007, 7, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, E.; Calabrese, F.; Schiavon, M.; Feltracco, P.; Seveso, M.; Carollo, C.; Loy, M.; Cardillo, M.; Rea, F. Immediate and Catastrophic Antibody-Mediated Rejection in a Lung Transplant Recipient with Anti-Angiotensin II Receptor Type 1 and Anti-Endothelin-1 Receptor Type A Antibodies. Am. J. Transpl. 2017, 17, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Reinsmoen, N.L.; Mirocha, J.; Ensor, C.R.; Marrari, M.; Chaux, G.; Levine, D.J.; Zhang, X.; Zeevi, A. A 3-Center Study Reveals New Insights Into the Impact of Non-HLA Antibodies on Lung Transplantation Outcome. Transplantation 2017, 101, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Saini, D.; Weber, J.; Ramachandran, S.; Phelan, D.; Tiriveedhi, V.; Liu, M.; Steward, N.; Aloush, A.; Hachem, R.; Trulock, E.; et al. Alloimmunity-induced autoimmunity as a potential mechanism in the pathogenesis of chronic rejection of human lung allografts. J. Heart Lung Transpl. 2011, 30, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.M.; Sigdel, T.K.; Delville, M.; Hsieh, S.C.; Dai, H.; Bagnasco, S.; Montgomery, R.A.; Sarwal, M.M. Endothelial cell antibodies associated with novel targets and increased rejection. J. Am. Soc. Nephrol. 2015, 26, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Lammerts, R.G.M.; Altulea, D.; Hepkema, B.G.; Sanders, J.S.; van den Born, J.; Berger, S.P. Antigen and Cell-Based Assays for the Detection of Non-HLA Antibodies. Front. Immunol. 2022, 13, 864671. [Google Scholar] [CrossRef] [PubMed]
- Alheim, M.; AlMahri, A.; Nilsson, J.; Tyden, G.; Holgersson, J. The outcome of the endothelial precursor cell crossmatch test in lymphocyte crossmatch positive and negative patients evaluated for living donor kidney transplantation. Hum. Immunol. 2013, 74, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Zitzner, J.R.; Shah, S.; Jie, C.; Wegner, W.; Tambur, A.R.; Friedewald, J.J. A prospective study evaluating the role of donor-specific anti-endothelial crossmatch (XM-ONE assay) in predicting living donor kidney transplant outcome. Hum. Immunol. 2013, 74, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Breimer, M.E.; Rydberg, L.; Jackson, A.M.; Lucas, D.P.; Zachary, A.A.; Melancon, J.K.; Von Visger, J.; Pelletier, R.; Saidman, S.L.; Williams, W.W., Jr.; et al. Multicenter evaluation of a novel endothelial cell crossmatch test in kidney transplantation. Transplantation 2009, 87, 549–556. [Google Scholar] [CrossRef]
- Zhang, Q.; Cecka, J.M.; Gjertson, D.W.; Ge, P.; Rose, M.L.; Patel, J.K.; Ardehali, A.; Kobashigawa, J.A.; Fishbein, M.C.; Reed, E.F. HLA and MICA: Targets of antibody-mediated rejection in heart transplantation. Transplantation 2011, 91, 1153–1158. [Google Scholar] [CrossRef]
- Regele, H. Non-HLA antibodies in kidney allograft rejection: Convincing concept in need of further evidence. Kidney Int. 2011, 79, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Kauke, T.; Kaczmarek, I.; Dick, A.; Schmoeckel, M.; Deutsch, M.A.; Beiras-Fernandez, A.; Reichart, B.; Spannagl, M. Anti-MICA antibodies are related to adverse outcome in heart transplant recipients. J. Heart Lung Transpl. 2009, 28, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Vinson, A.D.; Rampolla-Selles, R.; Cooper, E.S.; Alquist, C.R. Non-Human Leukocyte Antigen Antibody-Mediated Lung Transplant Rejection: The Other Anti-A. Ochsner J. 2018, 18, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Stastny, P.; Susal, C.; Dohler, B.; Opelz, G. Antibodies against MICA antigens and kidney-transplant rejection. N. Engl. J. Med. 2007, 357, 1293–1300. [Google Scholar] [CrossRef]
- Nowanska, K.; Wisnicki, K.; Kuriata-Kordek, M.; Krajewska, M.; Banasik, M. The role of endothelin II type A receptor (ETAR) in transplant injury. Transpl. Immunol. 2022, 70, 101505. [Google Scholar] [CrossRef] [PubMed]
- Son, B.S.; Lee, H.J.; Cho, W.H.; So, M.W.; Park, J.M.; Yeo, H.J. Association of positive pre-transplant angiotensin II type 1 receptor antibodies with clinical outcomes in lung transplant recipients. Transpl. Immunol. 2023, 80, 101901. [Google Scholar] [CrossRef] [PubMed]
- Ciszek, M.; Foroncewicz, B.; Mucha, K.; Zochowska, D.; Ziarkiewicz-Wroblewska, B.; Krawczyk, M.; Paczek, L. Anti-HLA and anti-MICA antibodies in liver transplant recipients: Effect on long-term graft survival. Clin. Dev. Immunol. 2013, 2013, 828201. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, L.J.; Hickey, M.J.; Chan, A.P.; Venick, R.S.; Farmer, D.G.; Busuttil, R.W.; Reed, E.F.; McDiarmid, S.V. Angiotensin II Type-1 Receptor Antibodies Are Associated with Active Allograft Dysfunction Following Pediatric Liver Transplantation. Transplantation 2020, 104, 2547–2556. [Google Scholar] [CrossRef]
- Zhang, Q.; Reed, E.F. The importance of non-HLA antibodies in transplantation. Nat. Rev. Nephrol. 2016, 12, 484–495. [Google Scholar] [CrossRef]
- Christian, S.; Pilch, J.; Akerman, M.E.; Porkka, K.; Laakkonen, P.; Ruoslahti, E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell. Biol. 2003, 163, 871–878. [Google Scholar] [CrossRef]
- Jia, T.; Vaganay, E.; Carpentier, G.; Coudert, P.; Guzman-Gonzales, V.; Manuel, R.; Eymin, B.; Coll, J.L.; Ruggiero, F. A collagen Valpha1-derived fragment inhibits FGF-2 induced-angiogenesis by modulating endothelial cells plasticity through its heparin-binding site. Matrix Biol. 2020, 94, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, J.; Guo, J.; Li, Y.; Lian, S.; Guo, W.; Yang, H.; Kong, F.; Zhen, L.; Guo, L.; et al. Progress in the biological function of alpha-enolase. Anim. Nutr. 2016, 2, 12–17. [Google Scholar] [CrossRef]
- Li, Y.Q.; Ngo, A.; Hoffmann, P.; Ferrante, A.; Hii, C.S. Regulation of endothelial cell survival and death by the MAP kinase/ERK kinase kinase 3-glyceraldehyde-3-phosphate dehydrogenase signaling axis. Cell. Signal. 2019, 58, 20–33. [Google Scholar] [CrossRef]
- Leeming, D.J.; Karsdal, M.A. Chapter 5-Type V Collagen. In Biochemistry of Collagens, Laminins and Elastin; Karsdal, M.A., Ed.; Academic Press: London, UK, 2016; pp. 43–48. [Google Scholar] [CrossRef]
- Dragun, D. Humoral responses directed against non-human leukocyte antigens in solid-organ transplantation. Transplantation 2008, 86, 1019–1025. [Google Scholar] [CrossRef]
- Michielsen, L.A.; van Zuilen, A.D.; Krebber, M.M.; Verhaar, M.C.; Otten, H.G. Clinical value of non-HLA antibodies in kidney transplantation: Still an enigma? Transpl. Rev. 2016, 30, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Fukami, N.; Ramachandran, S.; Saini, D.; Walter, M.; Chapman, W.; Patterson, G.A.; Mohanakumar, T. Antibodies to MHC class I induce autoimmunity: Role in the pathogenesis of chronic rejection. J. Immunol. 2009, 182, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Burlingham, W.J.; Love, R.B.; Jankowska-Gan, E.; Haynes, L.D.; Xu, Q.; Bobadilla, J.L.; Meyer, K.C.; Hayney, M.S.; Braun, R.K.; Greenspan, D.S.; et al. IL-17-dependent cellular immunity to collagen type V predisposes to obliterative bronchiolitis in human lung transplants. J. Clin. Investig. 2007, 117, 3498–3506. [Google Scholar] [CrossRef] [PubMed]
- Hachem, R.R.; Tiriveedhi, V.; Patterson, G.A.; Aloush, A.; Trulock, E.P.; Mohanakumar, T. Antibodies to K-alpha 1 tubulin and collagen V are associated with chronic rejection after lung transplantation. Am. J. Transpl. 2012, 12, 2164–2171. [Google Scholar] [CrossRef] [PubMed]
- Alunno, A.; Bistoni, O.; Pratesi, F.; La Paglia, G.M.C.; Puxeddu, I.; Migliorini, P.; Gerli, R. Anti-citrullinated alpha enolase antibodies, interstitial lung disease and bone erosion in rheumatoid arthritis. Rheumatology 2018, 57, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Murtas, C.; Bruschi, M.; Candiano, G.; Moroni, G.; Magistroni, R.; Magnano, A.; Bruno, F.; Radice, A.; Furci, L.; Argentiero, L.; et al. Coexistence of different circulating anti-podocyte antibodies in membranous nephropathy. Clin. J. Am. Soc. Nephrol. 2012, 7, 1394–1400. [Google Scholar] [CrossRef] [PubMed]
- Steers, N.J.; Li, Y.; Drace, Z.; D’Addario, J.A.; Fischman, C.; Liu, L.; Xu, K.; Na, Y.J.; Neugut, Y.D.; Zhang, J.Y.; et al. Genomic Mismatch at LIMS1 Locus and Kidney Allograft Rejection. N. Engl. J. Med. 2019, 380, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Nath, D.S.; Ilias Basha, H.; Tiriveedhi, V.; Alur, C.; Phelan, D.; Ewald, G.A.; Moazami, N.; Mohanakumar, T. Characterization of immune responses to cardiac self-antigens myosin and vimentin in human cardiac allograft recipients with antibody-mediated rejection and cardiac allograft vasculopathy. J. Heart Lung Transpl. 2010, 29, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, D.; Wang, X.; Wang, X.; Sha, J.C.; Gao, Q. Mechanisms underlying the production of chemokine CXCL11 in the reaction of renal tubular epithelial cells with CD4+ and CD8+ T cells. Transpl. Immunol. 2021, 65, 101337. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, R.; Chiu, S.; Raparia, K.; Garcha, P.; Farver, C.; Budev, M.; Tambur, A.R.; DeCamp, M.M.; Budinger, S.; Perlman, H.; et al. Humoral Human Lung Allograft Rejection by Tissue-Restricted Non-HLA Antibodies. Ann. Thorac. Surg. 2016, 102, e339–e341. [Google Scholar] [CrossRef]


| ECXM+ (n = 11) | ECXM− (n = 14) | |
|---|---|---|
| Age-years (Mean (SD)) | 53.8 (13.73) | 57.5 (10.05) |
| Gender (% male) | 36% | 64% |
| FEV1 (L) Mean (SD) | 1.58 (0.59) | 1.64 (0.82) |
| FVC (L) Mean (SD) | 2.38 (0.58) | 2.38 (0.60) |
| Duration from transplant (Mean (SD)) | 1725 (2389) | 1355(2086) |
| Immunosuppression | ||
| T, M, P | 54.5% | 35.7% |
| T, A, P | 36.3% | 21.4% |
| T, S, P | 0% | 7% |
| T, P | 9% | 3.7% |
| LSAUT Kit | Lot | Lot |
|---|---|---|
| LSAUT1 | 002 | 007 |
| LSAUT2 | 002 | 003 |
| Specificity | 95% Cutoff MFI | 95% Cutoff MFI |
| ENO1 | 4218 | 4218 |
| FLRT2 | 688 | 688 |
| VM | 820 | 820 |
| TUBA1B | 1987 | 1987 |
| CD36 | 1591 | 1591 |
| IFIH1 | 3870 | 3870 |
| MYOSIN | 9341 | 9341 |
| AGT | 1641 | 1641 |
| PTPRN | 3042 | 3042 |
| AURKA | 4892 | 4892 |
| CHAF1B | 11,722 | 11,722 |
| PPIA | 3292 | 3292 |
| EIF2A | 6901 | 6901 |
| GSTT1 | 6136 | 6136 |
| LMNA | 6633 | 6633 |
| PRKCZ | 9104 | 9104 |
| PECR | 4120 | 4120 |
| PRKCH | 1048 | 1048 |
| LMNB | 2065 | 2065 |
| CXCL11 | 309 | 309 |
| CXCL10 | 285 | 285 |
| CXCL9 | 575 | 575 |
| AGRN | 504 | 504 |
| ARHGDIB | 3918 | 3918 |
| GDNF | 1004 | 1004 |
| HNRNPK | 845 | 845 |
| IFNG | 498 | 498 |
| NCL | 3669 | 3669 |
| REG3A | 86 | 86 |
| GAPDH | 508 | 508 |
| TNFA | 5331 | 5331 |
| PLA2R | 195 | 195 |
| LG3 | 3801 | 4154 |
| LSAUT Kit | Lot | Lot | Lot |
|---|---|---|---|
| LSAUT3 | 002 | 004 | 005 |
| Specificity | 95% Cutoff MFI | 95% Cutoff MFI | 95% Cutoff MFI |
| Collagen I | 375 | 375 | 375 |
| Collagen II | 155 | 155 | 155 |
| Collagen III | 128 | 128 | 128 |
| Collagen IV | 71 | 71 | 71 |
| Collagen V | 187 | 187 | 187 |
| Fibronectin | 141 | 141 | 141 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhamdan, F.; Coppolino, A.; Sheikh, A.; Miele, A.; Lee, S.; Gasiewski, A.; Brescia, P.; Wood, I.; Venkat, A.; Thaniyavarn, T.; et al. Distinct Non-Human Leukocyte Antigen Antibody Signatures Correlate with Endothelial Crossmatch Status in Lung and Renal Transplant Recipients. Int. J. Mol. Sci. 2024, 25, 10562. https://doi.org/10.3390/ijms251910562
Alhamdan F, Coppolino A, Sheikh A, Miele A, Lee S, Gasiewski A, Brescia P, Wood I, Venkat A, Thaniyavarn T, et al. Distinct Non-Human Leukocyte Antigen Antibody Signatures Correlate with Endothelial Crossmatch Status in Lung and Renal Transplant Recipients. International Journal of Molecular Sciences. 2024; 25(19):10562. https://doi.org/10.3390/ijms251910562
Chicago/Turabian StyleAlhamdan, Fahd, Antonio Coppolino, Adil Sheikh, Anna Miele, Stefi Lee, Allison Gasiewski, Peter Brescia, Isabelle Wood, Arvin Venkat, Tany Thaniyavarn, and et al. 2024. "Distinct Non-Human Leukocyte Antigen Antibody Signatures Correlate with Endothelial Crossmatch Status in Lung and Renal Transplant Recipients" International Journal of Molecular Sciences 25, no. 19: 10562. https://doi.org/10.3390/ijms251910562






