Platelet Proteomics and Tissue Metabolomics Investigation for the Mechanism of Aspirin Eugenol Ester on Preventive Thrombosis Mechanism in a Rat Thrombosis Model
Abstract
1. Introduction
2. Results
2.1. Identification and Quantitative Evaluation of Proteins
2.2. DEPs in Rat Platelet
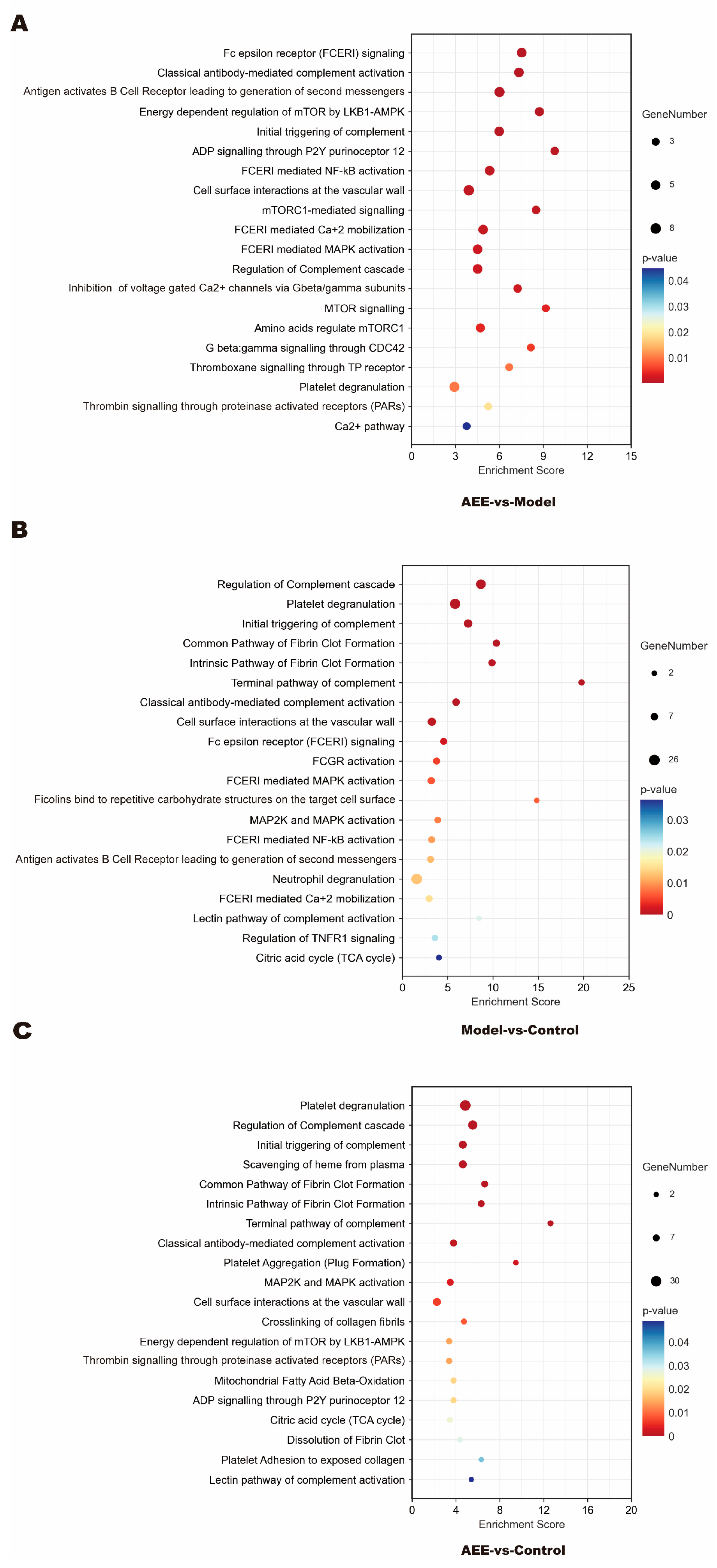
2.3. Reactome Pathway Enrichment of DEPs
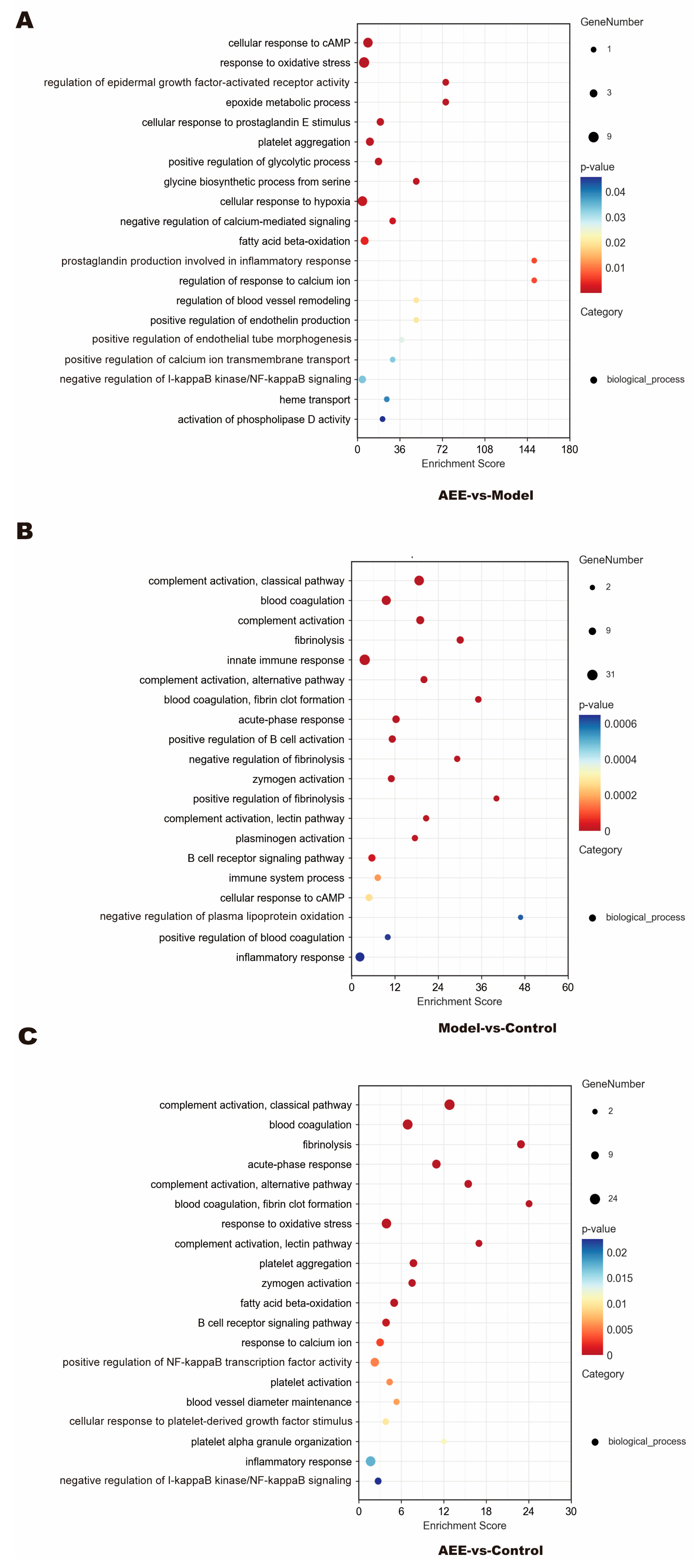
2.4. GO Enrichment of DEPs
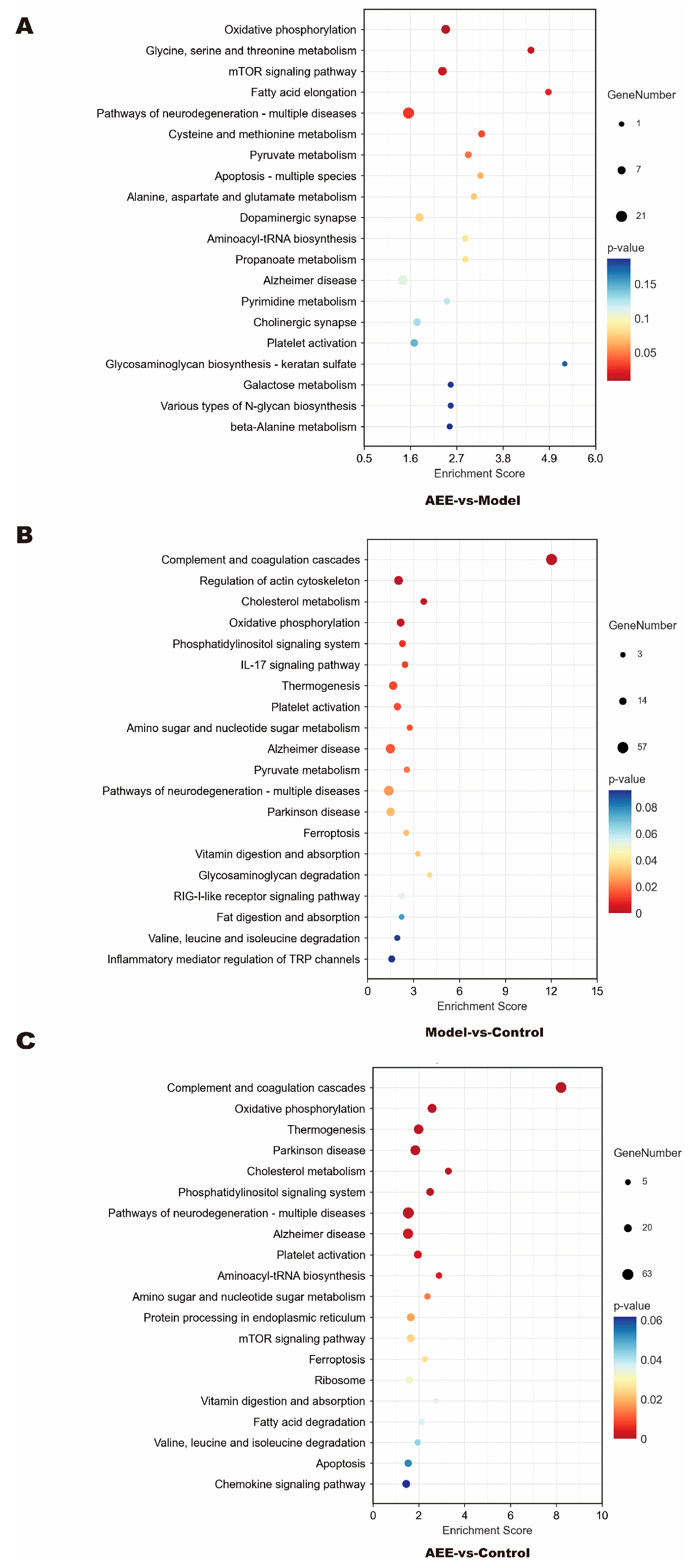
2.5. KEGG Pathway Enrichment of DEPs
2.6. PPI Analysis
2.7. Metabonomics Analysis Results
2.7.1. Analysis of Heart and Abdominal Aorta Metabolites
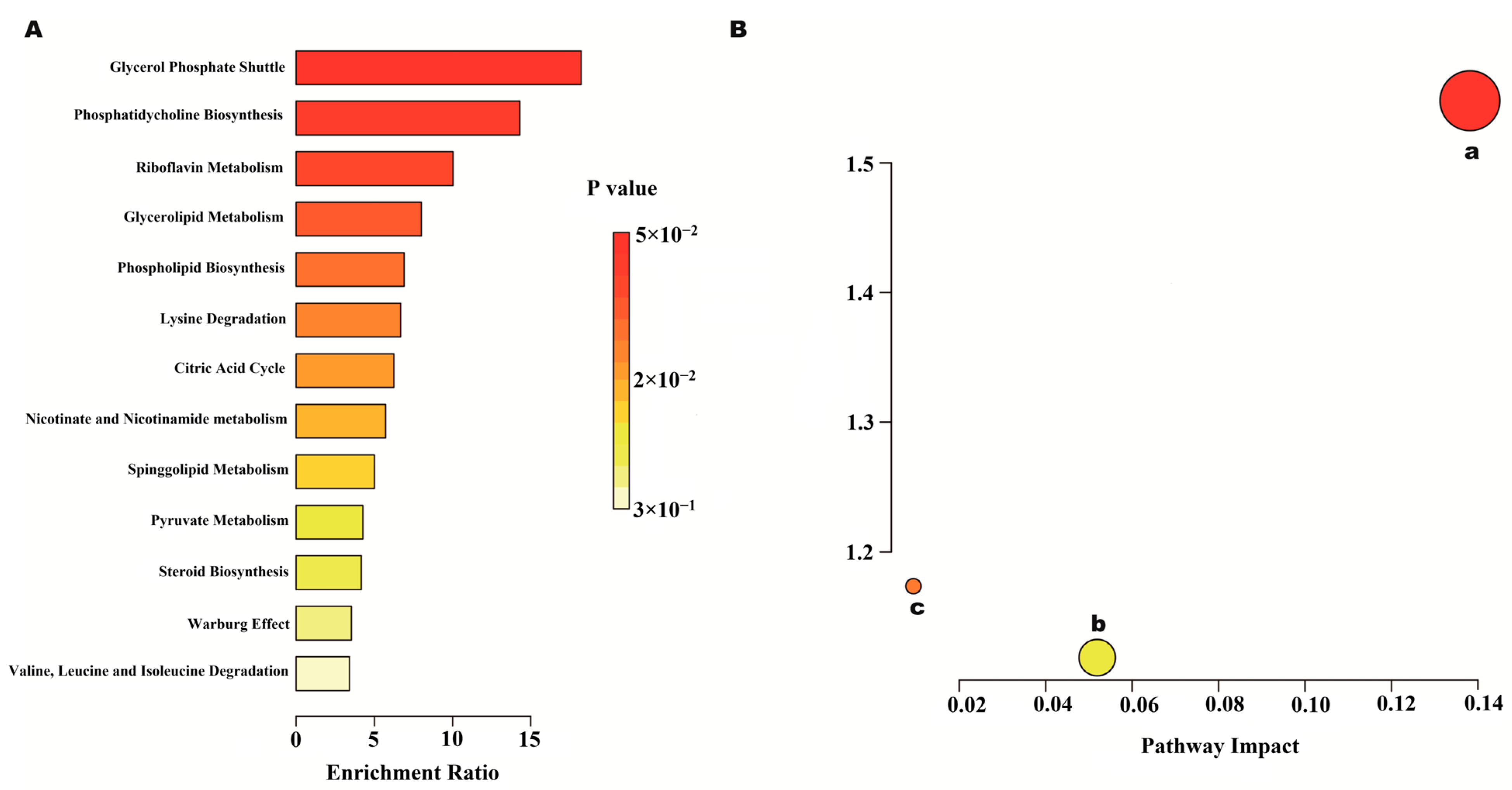
2.7.2. Metabolic Pathway and Function Analysis
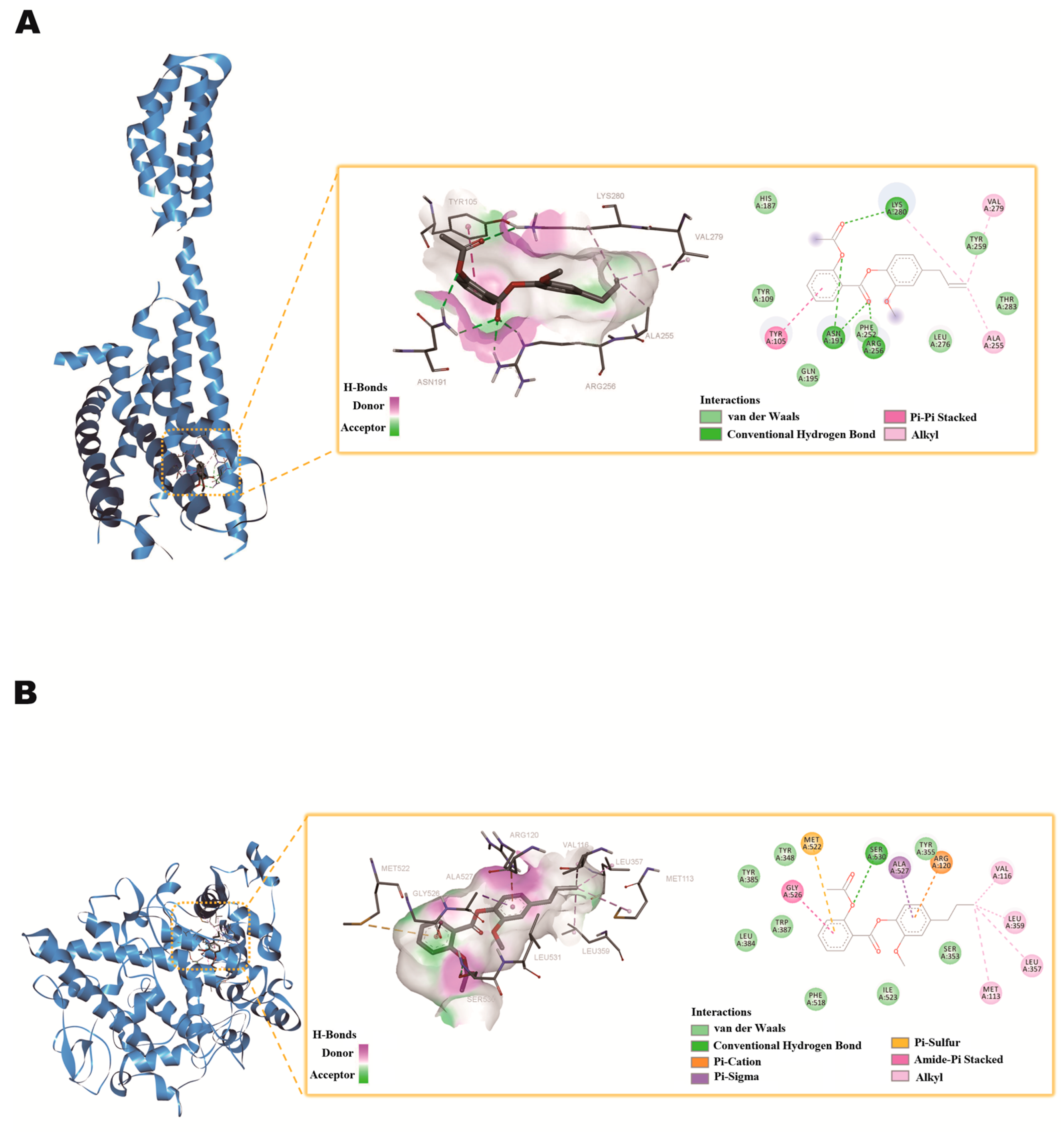
2.8. Molecular Docking Studies
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animal Experiment and Study Design
4.3. Proteomics Analysis
4.3.1. Platelet Collection and Preparation
4.3.2. Chromatography Conditions
4.3.3. Mass Spectrometry Conditions
4.3.4. Protein Identification and Bioinformatics Analysis
4.4. Metabolomics Analysis
4.4.1. Rat Heart and Abdominal Aorta Sample Preparation
4.4.2. UPLC-QTOF-MS/MS Conditions
4.4.3. Metabolomics Data Analysis
4.5. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Li, X.; Guo, T.; Feng, Q.; Bai, T.; Wu, L.; Liu, Y.; Zheng, X.; Jia, J.; Pei, J.; Wu, S.; et al. Progress of thrombus formation and research on the structure-activity relationship for antithrombotic drugs. Eur. J. Med. Chem. 2022, 228, 114035. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll of Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P. Arterial thrombosis--insidious, unpredictable and deadly. Nat. Med. 2011, 17, 1423–1436. [Google Scholar] [CrossRef]
- Blanch-Ruiz, M.A.; Ortega-Luna, R.; Martínez-Cuesta, M.; Álvarez, Á. The neutrophil secretome as a crucial link between inflammation and thrombosis. Int. J. Mol. Sci. 2021, 22, 4170. [Google Scholar] [CrossRef] [PubMed]
- Thon, J.N.; Italiano, J.E. Platelets: Production, morphology and ultrastructure. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 3–22. [Google Scholar]
- Sellers, S.L.; Gulsin, G.S.; Zaminski, D.; Bing, R.; Latib, A.; Sathananthan, J.; Pibarot, P.; Bouchareb, R. Platelets: Implications in aortic valve stenosis and bioprosthetic valve dysfunction from pathophysiology to clinical care. J. Am. Coll. Cardiol. Basic Trans. Sci. 2021, 6, 1007–1020. [Google Scholar]
- George, J.N. Platelets. Lancet 2000, 355, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
- Machlus, K.R.; Italiano, J.E., Jr. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 2013, 201, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lv, K.; Sharda, A.; Deng, J.; Zeng, W.; Zhang, C.; Hu, Q.; Jin, P.; Yao, G.; Xu, X.; et al. Anti-thrombotic effects mediated by dihydromyricetin involve both platelet inhibition and endothelial protection. Pharmacol. Res. 2021, 167, 105540. [Google Scholar] [CrossRef] [PubMed]
- Furie, B.; Furie, B.C. Mechanisms of thrombus formation. N. Engl. J. Med. 2008, 359, 938–949. [Google Scholar] [CrossRef]
- Plow, E.F.; Wang, Y.; Simon, D.I. The search for new antithrombotic mechanisms and therapies that may spare hemostasis. Blood 2018, 131, 1899–1902. [Google Scholar] [CrossRef]
- Bjarnason, I.; Scarpignato, C.; Holmgren, E.; Olszewski, M.; Rainsford, K.D.; Lanas, A. Mechanisms of damage to the gastrointestinal tract from nonsteroidal anti-inflammatory drugs. Gastroenterology 2018, 154, 500–514. [Google Scholar] [CrossRef]
- Balamuthusamy, S.; Arora, R. Hematologic adverse effects of clopidogrel. Am. J. Ther. 2007, 14, 106–112. [Google Scholar] [CrossRef]
- Yang, M.; Huo, X.; Miao, Z.; Wang, Y. Platelet glycoprotein IIb/IIIa receptor inhibitor tirofiban in acute ischemic stroke. Drugs 2019, 79, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Habibi, Z.; Luijkx, J.; Gho, B.C.G.; Ilhan, M.; Veenstra, L.F.; Ruiters, L.A.W.; Stein, M.; van’t Hof, A.W.J.; Rasoul, S. Platelet glycoprotein IIb/IIIa inhibitor tirofiban in clopidogrel-naïve patients undergoing elective percutaneous coronary intervention. Netherlands Heart J. 2023, 31, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Harloff, A.; Schlachetzki, F. Rivaroxaban for stroke prevention after embolic stroke of undetermined source. N. Engl. J. Med. 2018, 379, 986–987. [Google Scholar]
- Byon, W.; Garonzik, S.; Boyd, R.A.; Frost, C.E. Apixaban: A clinical pharmacokinetic and pharmacodynamic review. Clin. Pharmacokinet. 2019, 58, 1265–1279. [Google Scholar] [CrossRef]
- Chaieb, K.; Hajlaoui, H.; Zmantar, T.; Kahla-Nakbi, A.B.; Rouabhia, M.; Mahdouani, K.; Bakhrouf, A. The chemical composition and biological activity of clove essential oil, Eugenia caryophyllata (Syzigium aromaticum L. Myrtaceae): A short review. Phytother. Res. 2007, 21, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Taher, Y.A.; Samud, A.M.; El-Taher, F.E.; Ben-Hussin, G.; Elmezogi, J.S.; Al-Mehdawi, B.F.; Salem, H.A. Experimental evaluation of anti-inflammatory, antinociceptive and antipyretic activities of clove oil in mice. Libyan J. Med. 2015, 10, 28685. [Google Scholar] [CrossRef]
- Mnafgui, K.; Hajji, R.; Derbali, F.; Gammoudi, A.; Khabbabi, G.; Ellefi, H.; Allouche, N.; Kadri, A.; Gharsallah, N. Anti-inflammatory, antithrombotic and cardiac remodeling preventive effects of eugenol in isoproterenol-induced myocardial infarction in wistar rat. Cardiovasc. Toxicol. 2016, 16, 336–344. [Google Scholar] [CrossRef]
- Huang, M.Z.; Yang, Y.J.; Liu, X.W.; Qin, Z.; Li, J.Y. Aspirin eugenol ester reduces H2O2-induced oxidative stress of HUVECs via mitochondria-lysosome axis. Oxid. Med. Cell. Longev. 2019, 2019, 8098135. [Google Scholar] [CrossRef]
- Broekman, M.J.; Eiroa, A.M.; Marcus, A.J. Inhibition of human platelet reactivity by endothelium-derived relaxing factor from human umbilical vein endothelial cells in suspension: Blockade of aggregation and secretion by an aspirin-insensitive mechanism. Blood 1991, 78, 1033–1040. [Google Scholar] [CrossRef]
- Barale, C.; Buracco, S.; Cavalot, F.; Frascaroli, C.; Guerrasio, A.; Russo, I. Glucagon-like peptide 1-related peptides increase nitric oxide effects to reduce platelet activation. Thromb. Haemost. 2017, 117, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Yang, Y.; Liu, X.; Qin, Z.; Li, J. Aspirin eugenol ester attenuates oxidative injury of vascular endothelial cells by regulating NOS and Nrf2 signalling pathways. Br. J. Pharmacol. 2019, 176, 906–918. [Google Scholar] [CrossRef]
- Ma, N.; Yang, G.-Z.; Liu, X.-W.; Yang, Y.-J.; Mohamed, I.; Liu, G.-R.; Li, J.-Y. Impact of Aspirin Eugenol Ester on Cyclooxygenase-1, Cyclooxygenase-2, C-Reactive Protein, Prothrombin and Arachidonate 5-Lipoxygenase in Healthy Rats. Iran J. Pharm.Res. 2017, 16, 1443–1451. [Google Scholar]
- Shen, D.S.; Yang, Y.J.; Kong, X.J.; Ma, N.; Liu, X.W.; Li, S.H.; Jiao, Z.H.; Qin, Z.; Huang, M.Z.; Li, J.Y. Aspirin eugenol ester inhibits agonist-induced platelet aggregation in vitro by regulating PI3K/Akt, MAPK and Sirt 1/CD40L pathways. Eur. J. Pharmacol. 2019, 852, 1–13. [Google Scholar] [CrossRef]
- Ma, N.; Liu, X.W.; Yang, Y.J.; Shen, D.S.; Zhao, X.L.; Mohamed, I.; Kong, X.J.; Li, J.Y. Evaluation on antithrombotic effect of aspirin eugenol ester from the view of platelet aggregation, hemorheology, TXB2/6-keto-PGF1α and blood biochemistry in rat model. BMC Vet. Res. 2016, 12, 108. [Google Scholar] [CrossRef] [PubMed]
- Cavalli-Sforza, L.L. The human genome diversity project: Past, present and future. Nat. Rev. Genetics. 2005, 6, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Ma, N.; Fan, L.; Ge, W.; Zhang, Z.; Liu, X.; Li, J.; Yang, Y. Multi-Omics Approaches for Liver Reveal the Thromboprophylaxis Mechanism of Aspirin Eugenol Ester in Rat Thrombosis Model. Int. J. Mol. Sci. 2024, 25, 2141. [Google Scholar] [CrossRef]
- Ma, N.; Yang, Y.; Liu, X.; Li, S.; Qin, Z.; Li, J. Plasma metabonomics and proteomics studies on the anti-thrombosis mechanism of aspirin eugenol ester in rat tail thrombosis model. J. Proteom. 2020, 215, 103631. [Google Scholar] [CrossRef] [PubMed]
- Larsen, G.L.; Henson, P.M. Mediators of inflammation. Annu. Rev. Immunology. 1983, 1, 335–359. [Google Scholar] [CrossRef]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.A.; Vieira-De-Abreu, A.; Rowley, J.W.; Franks, Z.G.; Rondina, M.T.; Kraiss, L.W.; Majersik, J.J.; Zimmerman, G.A.; Weyrich, A. Abstract 354: Clots Are Potent Triggers of Inflammatory Cell Gene Expression: Indications for Timely Fibrinolysis. Arter. Thromb. Vasc. Biol. 2017, 37. [Google Scholar] [CrossRef]
- Esmon, C.T.; Xu, J.; Lupu, F. Innate immunity and coagulation. J. Thromb. Haemostasis. 2011, 9, 182–188. [Google Scholar] [CrossRef]
- Rondina, M.T.; Weyrich, A.S.; Zimmerman, G.A. Platelets as cellular effectors of inflammation in vascular diseases. Circ. Res. 2013, 112, 1506–1519. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.M.; Ito, N. A Critical review of the toxicological effects of carrageenan and processed eucheuma seaweed on the gastrointestinal tract. Crit. Rev. Toxicol. 2002, 32, 413–444. [Google Scholar] [CrossRef]
- Bian, C.; Ji, L.; Qu, H.; Wang, Z. A Novel Polysaccharide from AuriculariaAuricula Alleviates Thrombosis Induced by Carrageenan in Mice. Molecules 2022, 27, 4831. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Y.; Zhao, D.; Yang, S.; Zhang, S.; Wei, Z.; Wang, Y.; Qian, K.; Zhao, B.; Zhu, Y.; et al. LongShengZhi Capsule reduces carrageenan-induced thrombosis by reducing activation of platelets and endothelial cells. Pharmacol. Res. 2019, 144, 167–180. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, J.; Wan, H.; Shao, C.; Chen, T.; Yang, J.; He, Y.; Wan, H. The combination of danhong injection plus tissue plasminogen activator ameliorates mouse tail thrombosis-induced by κ-carrageenan. Phytomedicine 2022, 104, 154320. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.H.; Lee, S.Y.; Chae, J.Y.; Kim, J.W.; Kim, J.-H.; Jeong, H.G. Antarctic Krill Oil from Euphausia superba Ameliorates Carrageenan-Induced Thrombosis in a Mouse Model. Int. J. Mol. Sci. 2023, 24, 17440. [Google Scholar] [CrossRef] [PubMed]
- Gawaz, M.; Langer, H.; May, A.E. Platelets in inflammation and atherogenesis. J. Clin. Investig. 2005, 115, 3378–3384. [Google Scholar] [CrossRef]
- Barrett, T.J.; Schlegel, M.; Zhou, F.; Gorenchtein, M.; Bolstorff, J.; Moore, K.J.; Fisher, E.A.; Berger, J.S. Platelet regulation of myeloid suppressor of cytokine signaling 3 accelerates atherosclerosis. Sci. Transl. Med. 2019, 11, eaax0481. [Google Scholar] [CrossRef]
- Weyrich, A.S.; Denis, M.M.; Schwertz, H.; Tolley, N.D.; Foulks, J.; Spencer, E.; Kraiss, L.W.; Albertine, K.H.; McIntyre, T.M.; Zimmerman, G.A. mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets. Blood 2007, 109, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.F.; Hunter, R.W.; Hers, I. Protein kinase C and P2Y12 take center stage in thrombin-mediated activation of mammalian target of rapamycin complex 1 in human platelets. J. Thromb. Haemost. 2014, 12, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Aslan, J.E.; Tormoen, G.W.; Loren, C.P.; Pang, J.; McCarty, O.J.T. S6K1 and mTOR regulate Rac1-driven platelet activation and aggregation. Blood 2011, 118, 3129–3136. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, X.; Fan, X.; Xiao, M.; Yang, D.; Liang, B.; Dai, M.; Shan, L.; Lu, J.; Lin, Z.; et al. mTORC1 promotes aging-related venous thrombosis in mice via elevation of platelet volume and activation. Blood 2016, 128, 615–624. [Google Scholar] [CrossRef]
- Raslova, H.; Baccini, V.; Loussaief, L.; Comba, B.; Larghero, J.; Debili, N.; Vainchenker, W. Mammalian target of rapamycin (mTOR) regulates both proliferation of megakaryocyte progenitors and late stages of megakaryocyte differentiation. Blood 2006, 107, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, G.; Cao, X. The Function and Regulation of Platelet P2Y12 Receptor. Cardiovasc. Drugs Ther. 2023, 37, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Tunströmer, K.; Faxälv, L.; Larsson, P.; Lindahl, T.L.; Boknäs, N. Thrombus remodelling by reversible and irreversible P2Y (12) inhibitors. Platelets 2023, 34, 2157805. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Ma, J.; Liu, Y.; Chen, Z.; Xiao, N.; Lu, Y.; Fu, Y.; Yang, C.; Li, M.; Wu, S.; et al. iProX in 2021: Connecting proteomics data sharing with big data. Nucleic Acids Res. 2021, 50, D1522–D1527. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Q.; Fan, L.-P.; Feng, J.; Zhang, Z.-J.; Liu, X.-W.; Qin, Z.; Li, J.-Y.; Yang, Y.-J. Platelet Proteomics and Tissue Metabolomics Investigation for the Mechanism of Aspirin Eugenol Ester on Preventive Thrombosis Mechanism in a Rat Thrombosis Model. Int. J. Mol. Sci. 2024, 25, 10747. https://doi.org/10.3390/ijms251910747
Tao Q, Fan L-P, Feng J, Zhang Z-J, Liu X-W, Qin Z, Li J-Y, Yang Y-J. Platelet Proteomics and Tissue Metabolomics Investigation for the Mechanism of Aspirin Eugenol Ester on Preventive Thrombosis Mechanism in a Rat Thrombosis Model. International Journal of Molecular Sciences. 2024; 25(19):10747. https://doi.org/10.3390/ijms251910747
Chicago/Turabian StyleTao, Qi, Li-Ping Fan, Ji Feng, Zhi-Jie Zhang, Xi-Wang Liu, Zhe Qin, Jian-Yong Li, and Ya-Jun Yang. 2024. "Platelet Proteomics and Tissue Metabolomics Investigation for the Mechanism of Aspirin Eugenol Ester on Preventive Thrombosis Mechanism in a Rat Thrombosis Model" International Journal of Molecular Sciences 25, no. 19: 10747. https://doi.org/10.3390/ijms251910747
APA StyleTao, Q., Fan, L.-P., Feng, J., Zhang, Z.-J., Liu, X.-W., Qin, Z., Li, J.-Y., & Yang, Y.-J. (2024). Platelet Proteomics and Tissue Metabolomics Investigation for the Mechanism of Aspirin Eugenol Ester on Preventive Thrombosis Mechanism in a Rat Thrombosis Model. International Journal of Molecular Sciences, 25(19), 10747. https://doi.org/10.3390/ijms251910747



