Integrated Metabolomics and Transcriptomics Analyses Reveal the Regulatory Mechanisms of Anthocyanin and Carotenoid Accumulation in the Peel of Coffea arabica
Abstract
1. Introduction
2. Results
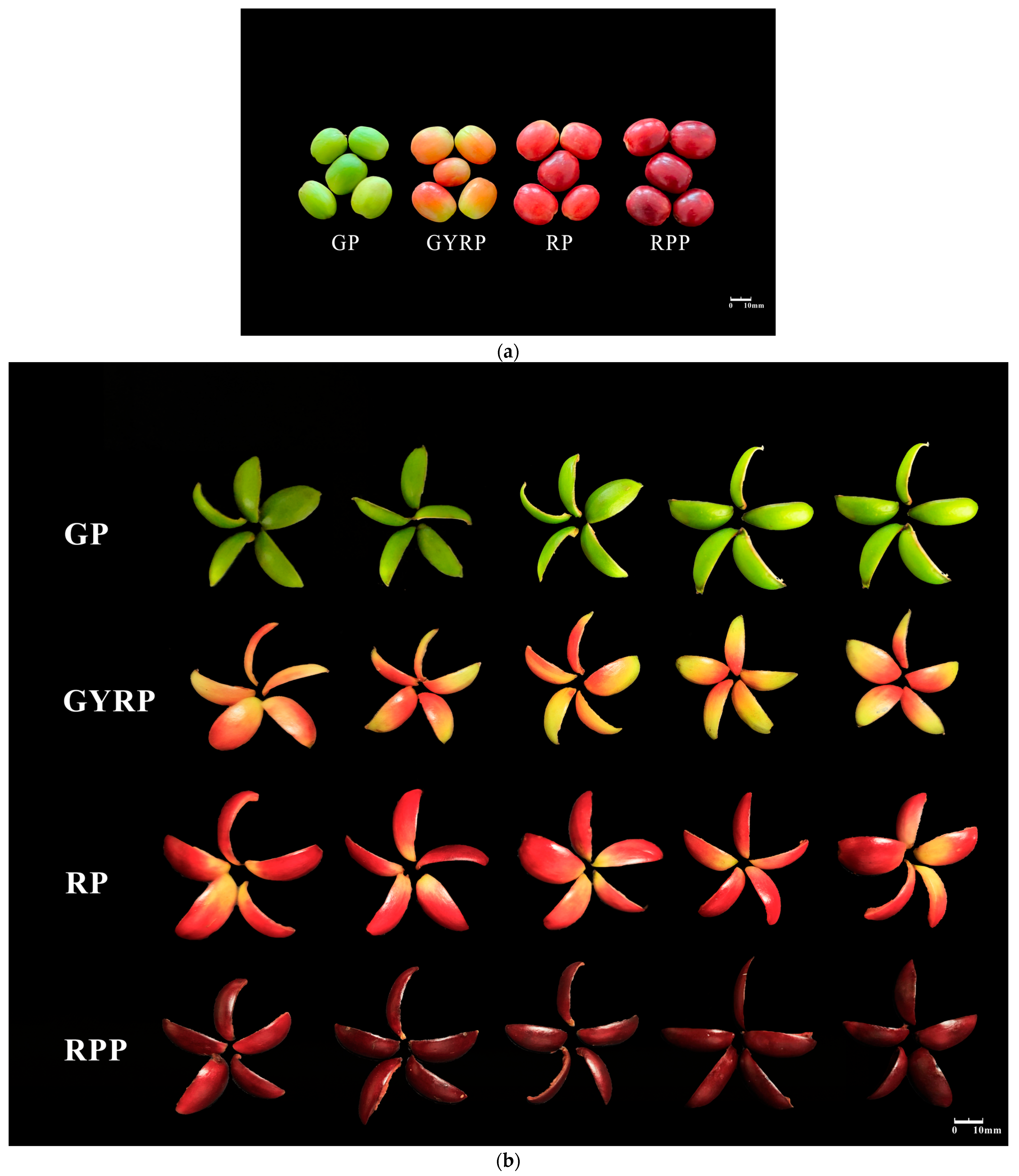
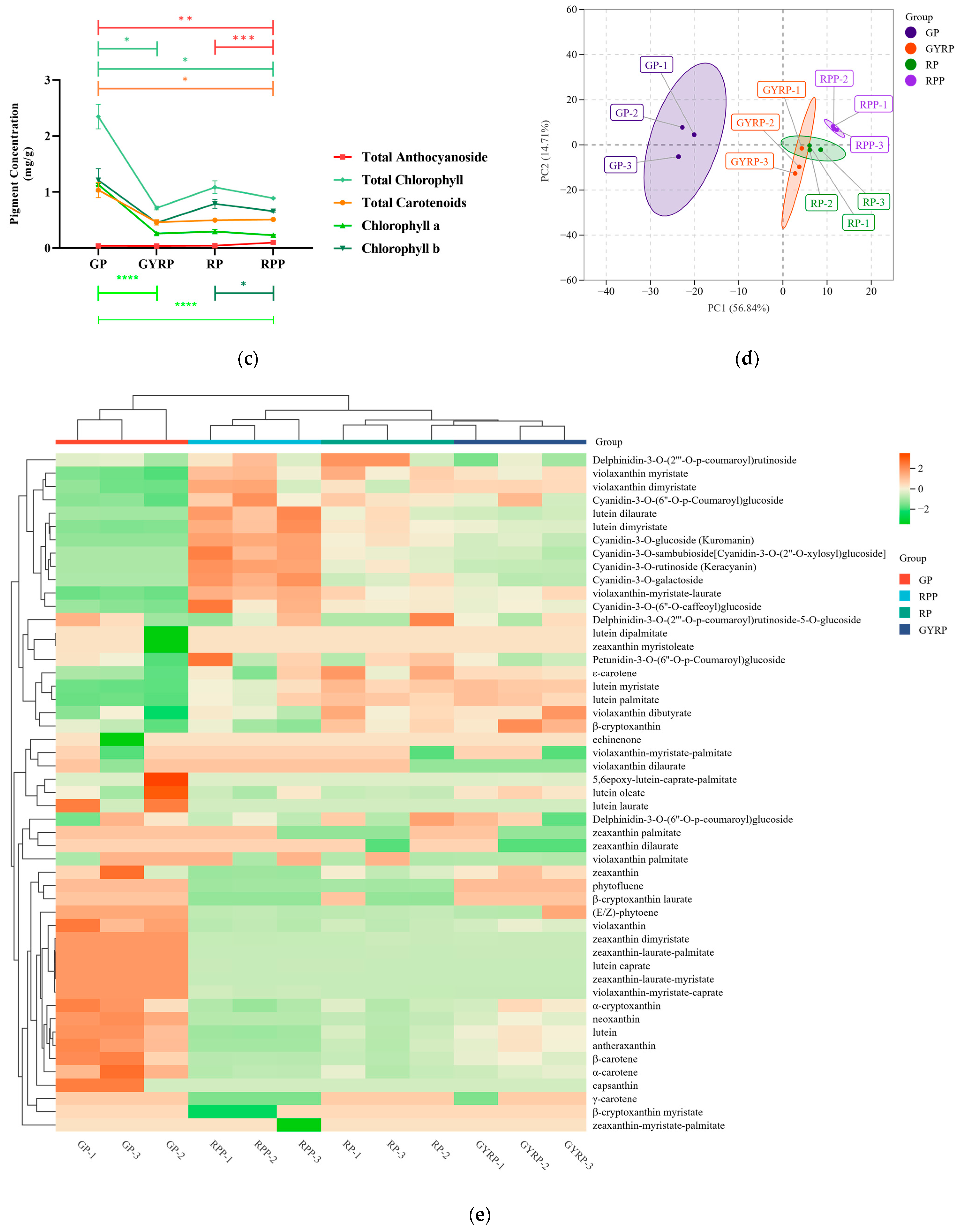
2.1. Changes in Total Pigment Content Related to the Maturation Process of Arabica Coffee Fruit Peels
2.2. Accumulation Differences in Anthocyanin and Carotenoid Metabolism
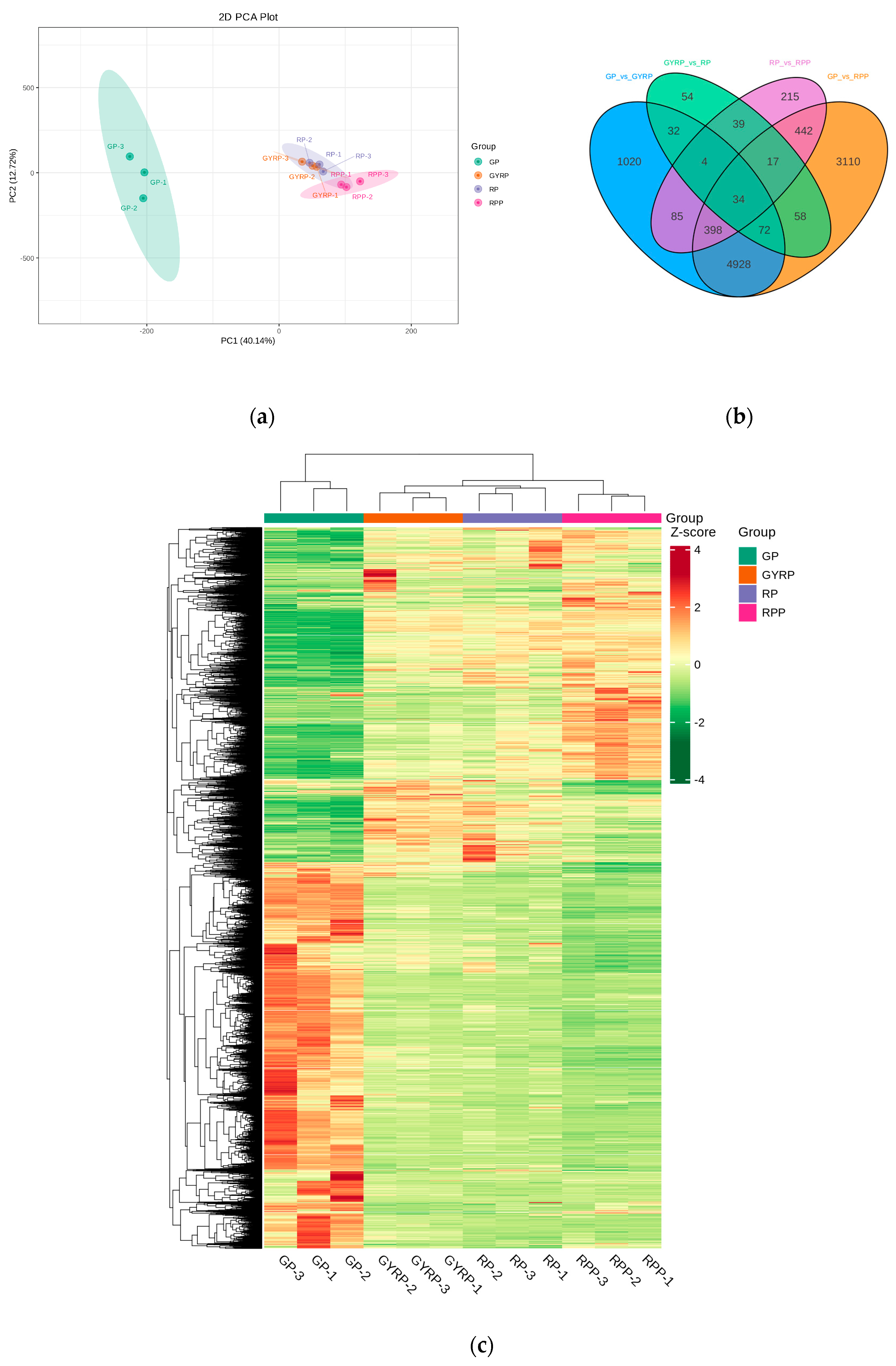
2.3. Transcriptional Atlas of Coffee Berry Peel during Ripening Process
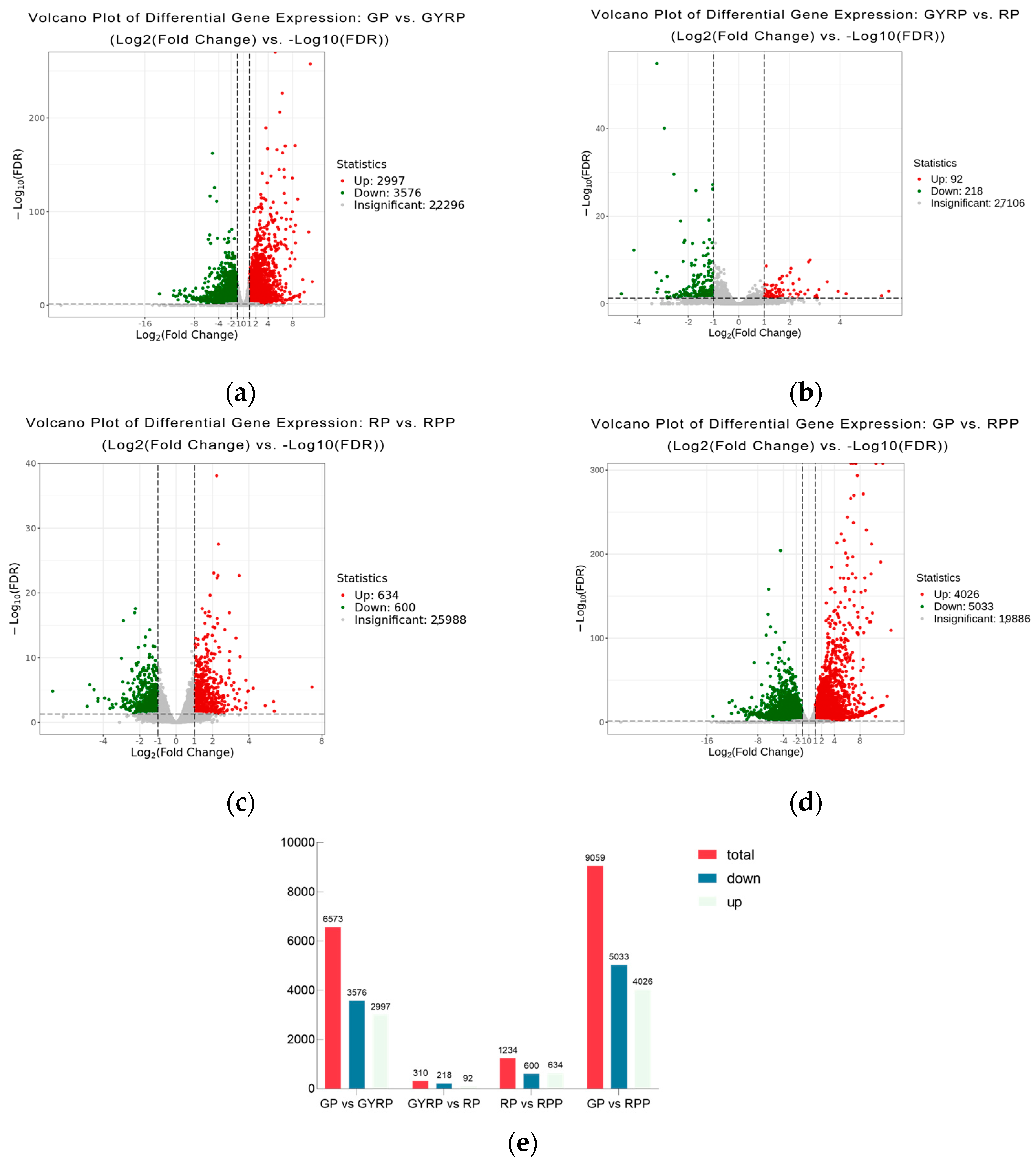
2.4. Comparative Analysis of Differentially Expressed Genes
2.5. Dynamic Changes in Enzymes Related to Anthocyanin and Carotenoid Biosynthesis
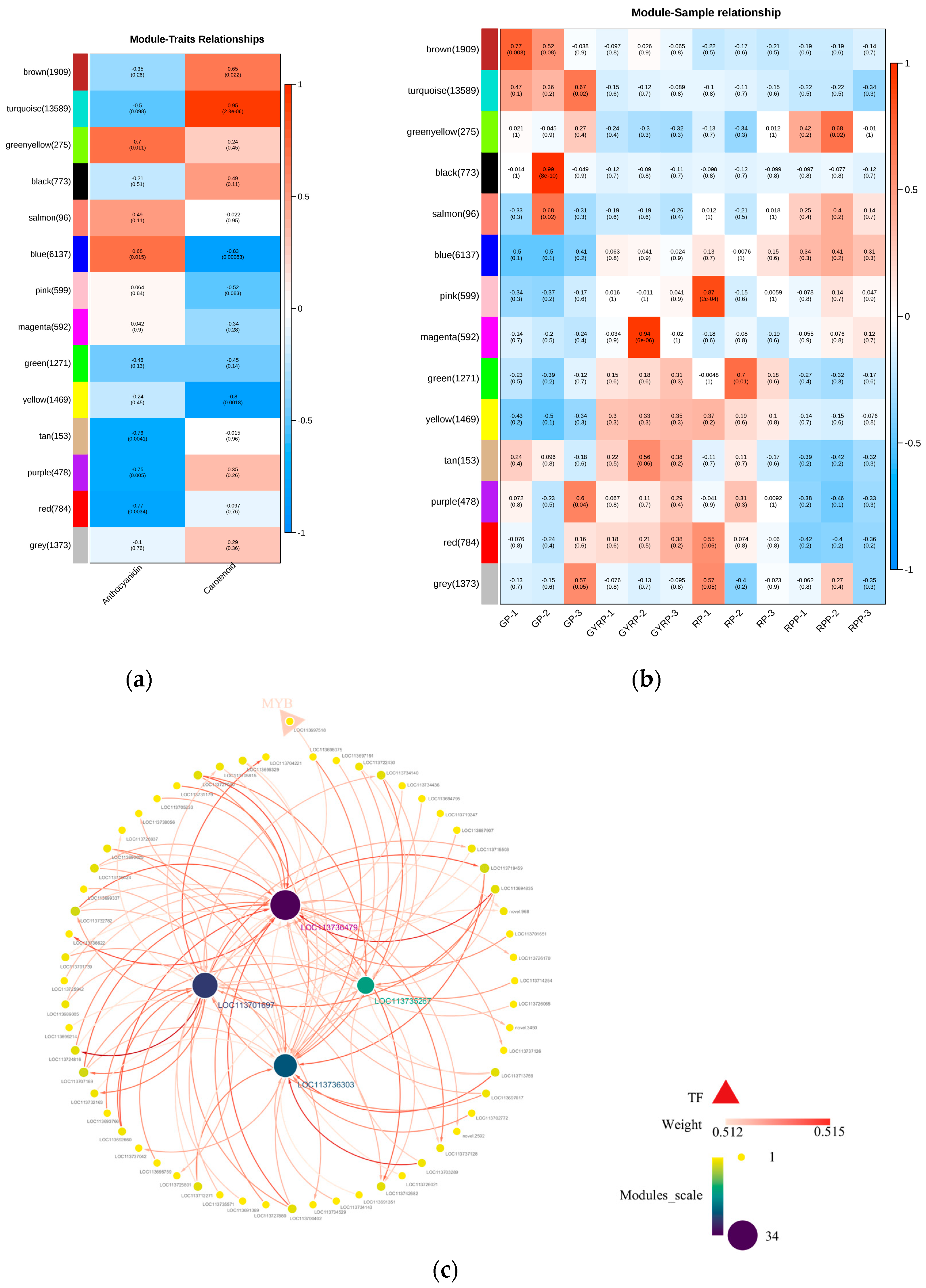
2.6. WGCNA-Based Network Analysis
2.7. Validation of RNA-Seq Data by qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Total Anthocyanins, Carotenoids, and Chlorophyll Content
4.3. Sample Preparation and Metabolite Extraction
4.4. Metabolomic Data Analysis
4.5. RNA Extraction, Quantification, and Sequencing
4.6. Transcriptome Data Analysis
4.7. Validation of RNA-Seq by Quantitative PCR
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Avelino, J.; Allinne, C.; Cerda, R.; Willocquet, L.; Savary, S. Multiple-Disease System in Coffee: From Crop Loss Assessment to Sustainable Management. Annu. Rev. Phytopathol. 2018, 56, 611–635. [Google Scholar] [CrossRef] [PubMed]
- ICO. Coffee Market Report December 2023; International Coffee Organization: London, UK, 2023; Available online: https://www.icocoffee.org/documents/cy2023-24/cmr-1223-e.pdf (accessed on 23 March 2024).
- van Dam, R.A.-O.; Hu, F.B.; Willett, W.C. Coffee, Caffeine, and Health. N. Engl. J. Med. 2020, 383, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Asamenew, G.; Kim, H.W.; Lee, M.K.; Lee, S.H.; Lee, S.; Cha, Y.S.; Lee, S.H.; Yoo, S.M.; Kim, J.B. Comprehensive characterization of hydroxycinnamoyl derivatives in green and roasted coffee beans: A new group of methyl hydroxycinnamoyl quinate. Food Chem. X 2019, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Bonilla-Hermosa, V.A.; Duarte, W.F.; Schwan, R.F. Utilization of coffee by-products obtained from semi-washed process for production of value-added compounds. Bioresour. Technol. 2014, 166, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Murthy, P.S.; Manjunatha, M.; Sulochannama, G.; Naidu, M.M. Extraction, characterization and bioactivity of coffee anthocyanins. Eur. J. Biol. Sci. 2012, 4, 13–19. [Google Scholar]
- Heeger, A.; Kosińska-Cagnazzo, A.; Cantergiani, E.; Andlauer, W. Bioactives of coffee cherry pulp and its utilisation for production of Cascara beverage. Food Chem. 2017, 221, 969–975. [Google Scholar] [CrossRef]
- Bertrand, B.; Villegas Hincapie, A.M.; Marie, L.; Breitler, J.-C. Breeding for the main agricultural farming of arabica coffee. Front. Sustain. Food Syst. 2021, 5, 709901. [Google Scholar] [CrossRef]
- Macheiner, L.; Schmidt, A.; Mayer, H.K. A novel basis for monitoring the coffee roasting process: Isomerization reactions of 3-caffeoylquinic and 4-caffeoylquinic acids. LWT 2021, 152, 112343. [Google Scholar] [CrossRef]
- Zhai, H.; Dong, W.; Tang, Y.; Hu, R.; Yu, X.; Chen, X. Characterization of the volatile flavour compounds in Yunnan Arabica coffee prepared by different primary processing methods using HS-SPME/GC-MS and HS-GC-IMS. LWT 2024, 192, 115717. [Google Scholar] [CrossRef]
- Bi, X.; Yu, H.; Hu, F.; Fu, X.; Li, Y.; Li, Y.; Yang, Y.; Liu, D.; Li, G.; Shi, R. A Systematic Analysis of the Correlation between Flavor Active Differential Metabolites and Multiple Bean Ripening Stages of Coffea arabica L. Molecules 2023, 29, 180. [Google Scholar] [CrossRef]
- Wintgens, J.N. Coffee-Growing, Processing, Sustainable Production: A Guidebook for Growers, Processors, Traders and Researchers; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Yu, H.; Bi, X.; Li, Z.; Fu, X.; Li, Y.; Li, Y.; Yang, Y.; Liu, D.; Li, G.; Dong, W. Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L. Genes 2024, 15, 459. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, L.; Simkin, A.J.; George Priya Doss, C.; Siva, R. Fruit ripening: Dynamics and integrated analysis of carotenoids and anthocyanins. BMC Plant Biol. 2022, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Kazama, E.H.; da Silva, R.P.; Tavares, T.d.O.; Correa, L.N.; de Lima Estevam, F.N.; Nicolau, F.E.d.A.; Maldonado Júnior, W. Methodology for selective coffee harvesting in management zones of yield and maturation. Precis. Agric. 2021, 22, 711–733. [Google Scholar] [CrossRef]
- Osorio Pérez, V.; Matallana Pérez, L.G.; Fernandez-Alduenda, M.R.; Alvarez Barreto, C.I.; Gallego Agudelo, C.P.; Montoya Restrepo, E.C. Chemical composition and sensory quality of coffee fruits at different stages of maturity. Agronomy 2023, 13, 341. [Google Scholar] [CrossRef]
- Li, X.; Jiang, J.; Chen, Z.; Jackson, A. Transcriptomic, Proteomic and Metabolomic Analysis of Flavonoid Biosynthesis During Fruit Maturation in Rubus chingii Hu. Front. Plant Sci. 2021, 12, 706667. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; van Velzen, E.J.; Hoefsloot, H.C.; Smilde, A.K. Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metabolomics 2010, 6, 119–128. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Chen, C. Pigments in Fruits and Vegetables: Genomics and Dietetics; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Baby, B.; Antony, P.; Vijayan, R. Antioxidant and anticancer properties of berries. Crit. Rev. Food Sci. Nutr. 2018, 58, 2491–2507. [Google Scholar] [CrossRef]
- Young, A.J.; Lowe, G.L. Carotenoids—Antioxidant properties. Antioxidants 2018, 7, 28. [Google Scholar] [CrossRef]
- Esquivel, P.; Viñas, M.; Steingass, C.B.; Gruschwitz, M.; Guevara, E.; Carle, R.; Schweiggert, R.M.; Jiménez, V.M. Coffee (Coffea arabica L.) by-products as a source of carotenoids and phenolic compounds—Evaluation of varieties with different peel color. Front. Sustain. Food Syst. 2020, 4, 590597. [Google Scholar] [CrossRef]
- Viñas, M.; Gruschwitz, M.; Schweiggert, R.; Guevara, E.; Carle, R.; Esquivel, P.; Jiménez, V. Identification of phenolic and carotenoid compounds in coffee (Coffea arabica) pulp, peels and mucilage by HPLC electrospray ionization mass spectrometry. In Proceedings of the 24th Internatonal Conference on Coffee Science ASIC, San Jose, CA, USA, 12–16 November 2012; pp. 12–16. [Google Scholar]
- Kay, C.D.; Pereira-Caro, G.; Ludwig, I.A.; Clifford, M.N.; Crozier, A. Anthocyanins and flavanones are more bioavailable than previously perceived: A review of recent evidence. Annu. Rev. Food Sci. Technol. 2017, 8, 155–180. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Luo, M.; Xu, J.; Wang, M.; Huang, B.; Miao, Y.; Liu, D. Integrative Analysis of Metabolomic and Transcriptomic Data Reveals the Mechanism of Color Formation in Corms of Pinellia ternata. Int. J. Mol. Sci. 2023, 24, 7990. [Google Scholar] [CrossRef]
- Liang, Z.; Liang, H.; Guo, Y.; Yang, D. Cyanidin 3-O-galactoside: A natural compound with multiple health benefits. Int. J. Mol. Sci. 2021, 22, 2261. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Deng, B.; Tian, S.; Guo, M.; Liu, H.; Zhao, X. Metabolic and transcriptomic analyses reveal different metabolite biosynthesis profiles between leaf buds and mature leaves in Ziziphus jujuba mill. Food Chem. 2021, 347, 129005. [Google Scholar] [CrossRef]
- Yang, H.; Li, H.; Li, Q. Biosynthetic regulatory network of flavonoid metabolites in stems and leaves of Salvia miltiorrhiza. Sci. Rep. 2022, 12, 18212. [Google Scholar] [CrossRef]
- Wang, A.; Ma, H.; Zhang, X.; Zhang, B.; Li, F. Transcriptomic analysis reveals the mechanism underlying the anthocyanin changes in Fragaria nilgerrensis Schlecht. and its interspecific hybrids. BMC Plant Biol. 2023, 23, 356. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Yu, S.; Li, M.; Dubcovsky, J.; Tian, L. Mutant combinations of lycopene ɛ-cyclase and β-carotene hydroxylase 2 homoeologs increased β-carotene accumulation in endosperm of tetraploid wheat (Triticum turgidum L.) grains. Plant Biotechnol. J. 2022, 20, 564–576. [Google Scholar] [CrossRef]
- Zhou, N.; Yan, Y.; Wen, Y.; Zhang, M.; Huang, Y. Integrated transcriptome and metabolome analysis unveils the mechanism of color-transition in Edgeworthia chrysantha tepals. BMC Plant Biol. 2023, 23, 567. [Google Scholar] [CrossRef]
- Chauffour, F.; Bailly, M.; Perreau, F.; Cueff, G.; Suzuki, H.; Collet, B.; Frey, A.; Clément, G.; Soubigou-Taconnat, L.; Balliau, T. Multi-omics analysis reveals sequential roles for ABA during seed maturation. Plant Physiol. 2019, 180, 1198–1218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, Y.; Wang, D. Transcriptomic and metabolomic analyses providing insights into the coloring mechanism of Docynia delavayi. Foods 2022, 11, 2899. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Song, M.; Wang, Z. Integrated metabolome and transcriptome analysis of fruit flavor and carotenoids biosynthesis differences between mature-green and tree-ripe of cv.“Golden phoenix” mangoes (Mangifera indica L.). Front. Plant Sci. 2022, 13, 816492. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bai, P.-P.; Gu, K.-J.; Yang, S.-Z.; Lin, H.-Y.; Shi, C.-G.; Zhao, Y.-P. Dynamic transcriptome and network-based analysis of yellow leaf mutant Ginkgo biloba. BMC Plant Biol. 2022, 22, 465. [Google Scholar] [CrossRef] [PubMed]
- Nwasike, C.; Ewert, S.; Jovanovic, S.; Haider, S.; Mujtaba, S. SET domain-mediated lysine methylation in lower organisms regulates growth and transcription in hosts. Ann. N. Y. Acad. Sci. 2016, 1376, 18–28. [Google Scholar] [CrossRef]
- Sun, X.; Feng, P.; Xu, X.; Guo, H.; Ma, J.; Chi, W.; Lin, R.; Lu, C.; Zhang, L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2011, 2, 477. [Google Scholar] [CrossRef]
- Koch, K. Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Biol. 1996, 47, 509–540. [Google Scholar] [CrossRef]
- Martínez-Noël, G.M.; Tognetti, J.A. Sugar signaling under abiotic stress in plants. In Plant Metabolites and Regulation under Environmental Stress; Elsevier: Amsterdam, The Netherlands, 2018; pp. 397–406. [Google Scholar]
- Zhong, S.; Fei, Z.; Chen, Y.-R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Rincon-Jimenez, A.; Tinoco, H.A.; Buitrago-Osorio, J.; Ocampo, O.; Berrio, L.V.; Rodriguez-Sotelo, J.L.; Arizmendi, C. Ripeness stage characterization of coffee fruits (Coffea arabica L. var. Castillo) applying chromaticity maps obtained from digital images. Mater. Today Proc. 2021, 44, 1271–1278. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Zhao, F.; Maren, N.A.; Kosentka, P.Z.; Liao, Y.Y.; Lu, H.; Duduit, J.R.; Huang, D.; Ashrafi, H.; Zhao, T.; Huerta, A.I.; et al. An optimized protocol for stepwise optimization of real-time RT-PCR analysis. Hortic. Res. 2021, 8, 179. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Xie, C.; Wu, Y.; Liu, H.; Zhang, X.; Du, H.; Li, X.; Zhang, C. Integrated Metabolomics and Transcriptomics Analyses Reveal the Regulatory Mechanisms of Anthocyanin and Carotenoid Accumulation in the Peel of Coffea arabica. Int. J. Mol. Sci. 2024, 25, 10754. https://doi.org/10.3390/ijms251910754
Wang Z, Xie C, Wu Y, Liu H, Zhang X, Du H, Li X, Zhang C. Integrated Metabolomics and Transcriptomics Analyses Reveal the Regulatory Mechanisms of Anthocyanin and Carotenoid Accumulation in the Peel of Coffea arabica. International Journal of Molecular Sciences. 2024; 25(19):10754. https://doi.org/10.3390/ijms251910754
Chicago/Turabian StyleWang, Zuquan, Chun Xie, Yihong Wu, Haobo Liu, Xuesong Zhang, Huabo Du, Xuejun Li, and Chuanli Zhang. 2024. "Integrated Metabolomics and Transcriptomics Analyses Reveal the Regulatory Mechanisms of Anthocyanin and Carotenoid Accumulation in the Peel of Coffea arabica" International Journal of Molecular Sciences 25, no. 19: 10754. https://doi.org/10.3390/ijms251910754
APA StyleWang, Z., Xie, C., Wu, Y., Liu, H., Zhang, X., Du, H., Li, X., & Zhang, C. (2024). Integrated Metabolomics and Transcriptomics Analyses Reveal the Regulatory Mechanisms of Anthocyanin and Carotenoid Accumulation in the Peel of Coffea arabica. International Journal of Molecular Sciences, 25(19), 10754. https://doi.org/10.3390/ijms251910754




