Differential Protein Expression in Extracellular Vesicles Defines Treatment Responders and Non-Responders in Multiple Sclerosis
Abstract
1. Introduction
2. Results
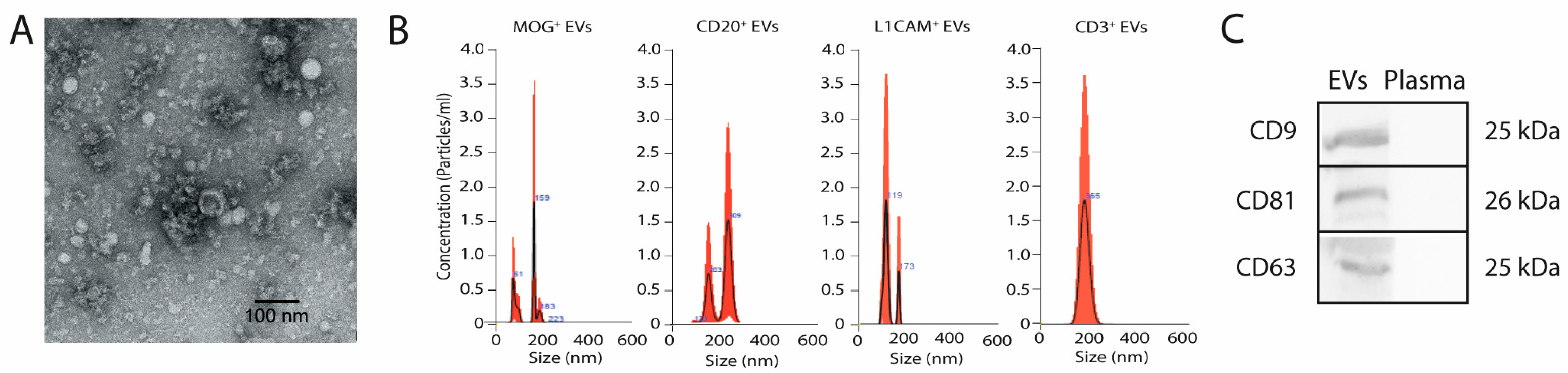
2.1. Characterisation of Brain- and Immune System-Derived Extracellular Vesicles: Size, Morphology, and Marker Profiling
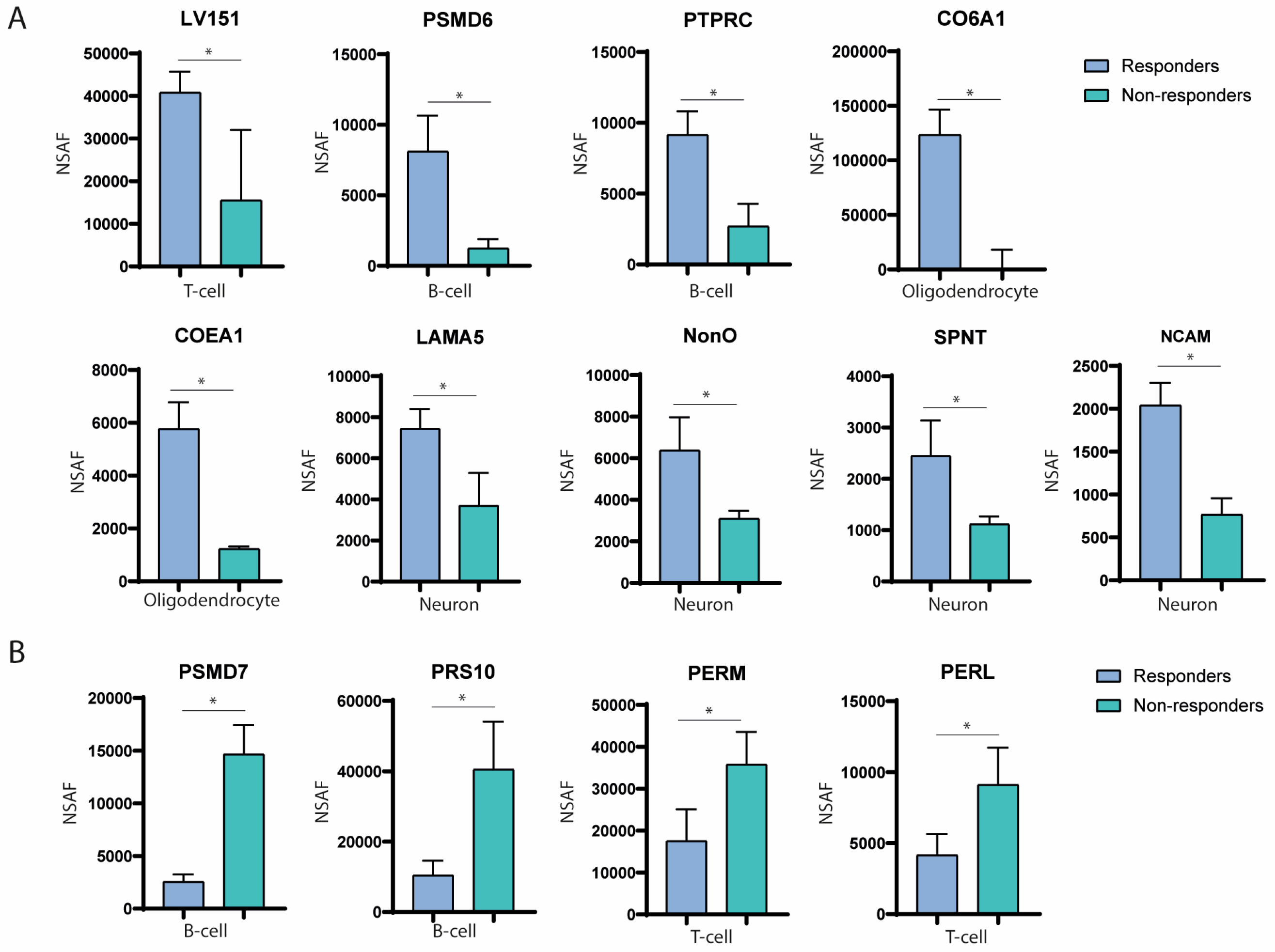
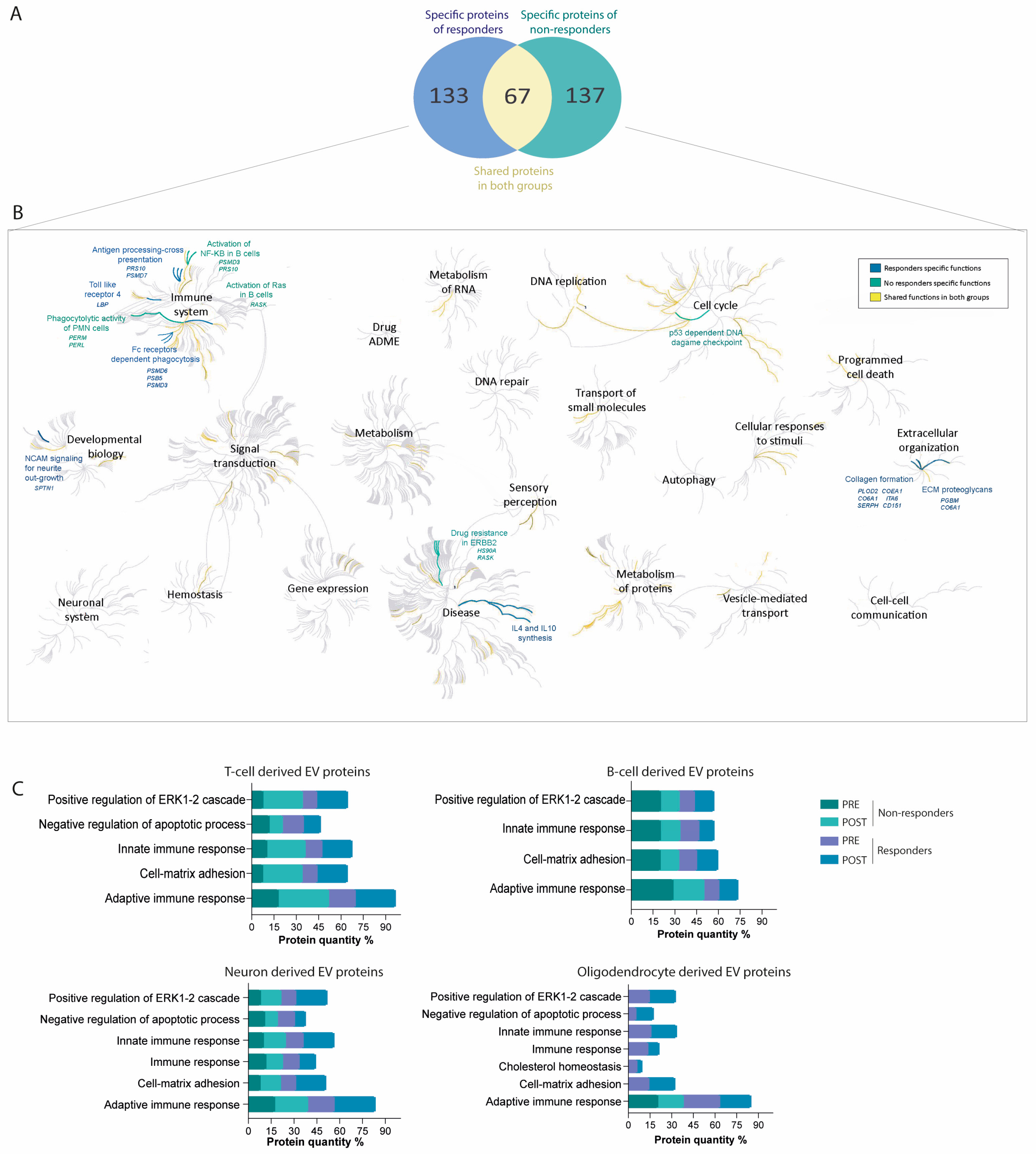
2.2. Immune System-Derived Extracellular Vesicles Are Involved in Immune Modulation in Responder Patients
2.3. Nervous System-Derived Extracellular Vesicle Subpopulation Has a Role in the Extracellular Matrix Reorganisation and White Matter Repair in Responder Patients
2.4. Proteins Associated with Failure of Treatment Indicate DNA Damage, Nuclear Factor Kappa B Pathway Activation, and Drug Resistance
2.5. Differential Protein Cargo in EVs before and after Treatment Confirms Immune Response Modulation and Cell-Matrix Adhesion Pathways in Treatment Responders
2.6. Protein–Protein Interaction Networks Predominate among Differential Proteins in Responders and Non-Responders, Highlighting Key Functional Associations
2.7. Most Abundant Differentially Expressed Proteins in Extracellular Vesicles among Responders and Non-Responders
3. Discussion
4. Materials and Methods
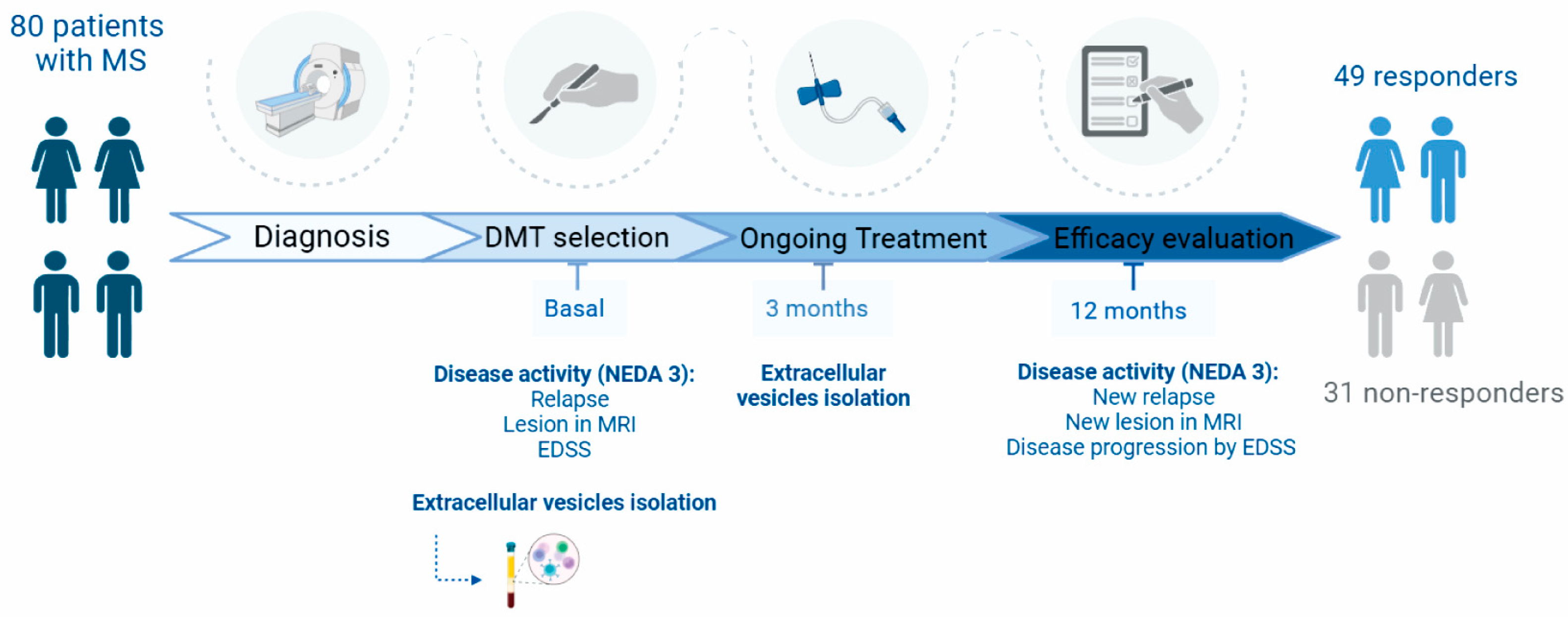
4.1. Study Design and Participants
4.2. Clinical Data
4.3. Outcome Measures
- Relapses: New or recurrent neurological symptoms not associated with fever, lasting for at least 24 h, followed by 30 days of stability or improvement.
- MRI Activity: Presence of one or more new or enlarged lesions on a T2-weighted scan at 12 months.
- Disease Progression: An increase of 1.5 points in the EDSS score if the baseline EDSS score was 0; an increase of 1.0 point for baseline EDSS scores between 1 and 5.5; an increase of 0.5 points from baseline EDSS ≥6; or a 20% increase in the 9-Hole Peg Test (9-HPT) at 12 months compared to baseline.
4.4. Treatment Response
4.5. Extracellular Vesicle Isolation
4.6. Extracellular Vesicle Characterisation
- Specific EV markers: Western blot analysis was performed using antibodies against CD9 (Thermo Fisher Scientific, Waltham, MA, USA), CD81 (1:250, Abcam, Cambridge, UK), and CD63 (1:250, Abcam, Cambridge, UK), followed by goat anti-mouse or anti-rabbit HRP antibodies (1:750, Invitrogen, Waltham, MA, USA). Blots were visualised using Pierce ECL chemiluminescence (Thermo Fisher Scientific, Waltham, MA, USA) and a UVITEC–Cambridge imaging system, as previously described [16].
- EV morphology: Transmission electron microscopy (JEOL JEM1010) was used to analyse EV morphology as previously described [16].
- EV size and concentration: NTA was performed using the NanoSight NS500 nanoparticle analyser (Malvern Instruments, Worcestershire, UK), equipped with fast video capture and particle-tracking software. Measurements were conducted in triplicate, following the manufacturer’s instructions, as previously described [16].
4.7. Proteomics Analysis
4.7.1. Protein Tryptic Digestion
4.7.2. Mass Spectrometric Analysis
4.7.3. Protein Quantification by SWATH-MS (Sequential Window Acquisition for All Theoretical Mass Spectra)
4.7.4. Data Analysis
4.8. Biological Functions and Pathway Study
4.9. Functional Enrichment Analysis
4.10. Protein–Protein Interaction Network Analysis
4.11. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bernard, C.C.; de Rosbo, N. Multiple sclerosis: An autoimmune disease of multifactorial etiology. Curr. Opin. Immunol. 1992, 4, 760–765. [Google Scholar] [CrossRef]
- Wingerchuk, D.M.; Carter, J.L. Multiple sclerosis: Current and emerging disease-modifying therapies and treatment strategies. Mayo Clin. Proc. 2014, 89, 225–240. [Google Scholar] [CrossRef]
- Montalban, X.; Gold, R.; Thompson, A.J.; Otero-Romero, S.; Amato, M.P.; Chandraratna, D.; Clanet, M.; Comi, G.; Derfuss, T.; Fazekas, F.; et al. ECTRIMS/EAN Guideline on the pharmacological treatment of people with multiple sclerosis. Mult. Scler. J. 2018, 24, 96–120. [Google Scholar] [CrossRef] [PubMed]
- Aronson, J.K.; Ferner, R.E. Biomarkers-A General Review. Curr. Protoc. Pharmacol. 2017, 76, 9.23.1–9.23.17. [Google Scholar] [CrossRef] [PubMed]
- Sandi, D.; Kokas, Z.; Biernacki, T.; Bencsik, K.; Klivényi, P.; Vécsei, L. Proteomics in Multiple Sclerosis: The Perspective of the Clinician. Int. J. Mol. Sci. 2022, 23, 5162. [Google Scholar] [CrossRef]
- Barro, C.; Chitnis, T.; Weiner, H.L. Blood neurofilament light: A critical review of its application to neurologic disease. Ann. Clin. Transl. Neurol. 2020, 7, 2508–2523. [Google Scholar] [CrossRef]
- Otero-Ortega, L.; Fernández-Fournier, M.; Irizar, H.; Otaegui, D.; Díez-Tejedor, E. Editorial: Innovative OMICs-technology applications to reach a diagnosis and bring new therapies to immune-mediated neurological diseases. Front. Immunol. 2023, 14, 1348986. [Google Scholar] [CrossRef] [PubMed]
- Gobena, S.; Admassu, B.; Kinde, M.Z.; Gessese, A.T. Proteomics and Its Current Application in Biomedical Area: Concise Review. Sci. World J. 2024, 2024, 4454744. [Google Scholar] [CrossRef]
- Drabik, A.; Bierczynska-Krzysik, A.; Bodzon-Kulakowska, A.; Suder, P.; Kotlinska, J.; Silberring, J. Proteomics in neurosciences. Mass Spectrom Rev. 2007, 26, 432–450. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef]
- Sáenz-Cuesta, M.; Osorio-Querejeta, I.; Otaegui, D. Extracellular vesicles in multiple sclerosis: What are they telling us? Front. Cell. Neurosci. 2014, 8, 100. [Google Scholar] [CrossRef]
- Geraci, F.; Ragonese, P.; Barreca, M.M.; Aliotta, E.; Mazzola, M.A.; Realmuto, S.; Vazzoler, G.; Savettieri, G.; Sconzo, G.; Salemi, G. Differences in Intercellular Communication During Clinical Relapse and Gadolinium-Enhanced MRI in Patients With Relapsing Remitting Multiple Sclerosis: A Study of the Composition of Extracellular Vesicles in Cerebrospinal Fluid. Front. Cell. Neurosci. 2018, 12, 418. [Google Scholar] [CrossRef] [PubMed]
- Sáenz-Cuesta, M.; Alberro, A.; Muñoz-Culla, M.; Osorio-Querejeta, I.; Fernandez-Mercado, M.; Lopetegui, I.; Tainta, M.; Prada, Á.; Castillo-Triviño, T.; Falcón-Pérez, J.M.; et al. The First Dose of Fingolimod Affects Circulating Extracellular Vesicles in Multiple Sclerosis Patients. Int. J. Mol. Sci. 2018, 19, 2448. [Google Scholar] [CrossRef]
- Dalla Costa, G.; Finardi, A.; Garzetti, L.; Carandini, T.; Comi, G.; Martinelli, V.; Furlan, R. Disease-modifying treatments modulate myeloid cells in multiple sclerosis patients. Neurol. Sci. 2018, 39, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Torres Iglesias, G.; Fernández-Fournier, M.; Botella, L.; Piniella, D.; Laso-García, F.; Frutos, M.C.G.-D.; Chamorro, B.; Puertas, I.; Barranco, A.T.; Fuentes, B.; et al. Brain and immune system-derived extracellular vesicles mediate regulation of complement system, extracellular matrix remodeling, brain repair and antigen tolerance in Multiple Sclerosis. Brain Behav. Immun. 2023, 113, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Zaldívar, H.M.; Polakovicova, I.; Salas-Huenuleo, E.; Corvalán, A.H.; Kogan, M.J.; Yefi, C.P.; Andia, M.E. Extracellular vesicles through the blood-brain barrier: A review. Fluids Barriers CNS 2022, 19, 60. [Google Scholar] [CrossRef]
- Gutiérrez-Fernández, M.; de la Cuesta, F.; Tallón, A.; Cuesta, I.; Fernández-Fournier, M.; Laso-García, F.; Gómez-de Frutos, M.C.; Díez-Tejedor, E.; Otero-Ortega, L. Potential Roles of Extracellular Vesicles as Biomarkers and a Novel Treatment Approach in Multiple Sclerosis. Int. J. Mol. Sci. 2021, 22, 9011. [Google Scholar] [CrossRef] [PubMed]
- Riedhammer, C.; Weissert, R. Antigen Presentation, Autoantigens, and Immune Regulation in Multiple Sclerosis and Other Autoimmune Diseases. Front. Immunol. 2015, 6, 322. [Google Scholar] [CrossRef]
- Montecchi, T.; Shaba, E.; De Tommaso, D.; Di Giuseppe, F.; Angelucci, S.; Bini, L.; Landi, C.; Baldari, C.T.; Ulivieri, C. Differential Proteomic Analysis of Astrocytes and Astrocytes-Derived Extracellular Vesicles from Control and Rai Knockout Mice: Insights into the Mechanisms of Neuroprotection. Int. J. Mol. Sci. 2021, 22, 7933. [Google Scholar] [CrossRef]
- Vigneron, N.; Van den Eynde, B.J. Proteasome subtypes and regulators in the processing of antigenic peptides presented by class I molecules of the major histocompatibility complex. Biomolecules 2014, 4, 994–1025. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Bar-Or, A.; Piehl, F.; Preziosa, P.; Solari, A.; Vukusic, S.; Rocca, A.M. Multiple sclerosis. Nat. Rev. Dis. Primers. 2018, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Reske, D.; Thomas, A.V.; Petereit, H.F.; Fink, G.R.; Schroeter, M. Impact of immunomodulatory treatment on leukocyte cytokine production in multiple sclerosis patients and healthy donors. Neuroimmunomodulation 2009, 16, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Jang, S.H.; Tian, H.; Kim, S.J. NonO Is a Novel Co-factor of PRDM1 and Regulates Inflammatory Response in Monocyte Derived-Dendritic Cells. Front. Immunol. 2020, 11, 1436. [Google Scholar] [CrossRef]
- Chen, S.; Wang, J.; Zhang, K.; Ma, B.; Li, X.; Wei, R.; Nian, H. LncRNA Neat1 targets NonO and miR-128-3p to promote antigen-specific Th17 cell responses and autoimmune inflammation. Cell Death Dis. 2023, 14, 610. [Google Scholar] [CrossRef]
- Zenobia, G.; Hajishengallis, G. Basic biology and role of interleukin-17 in immunity and inflammation. Periodontology 2015, 69, 142–159. [Google Scholar] [CrossRef]
- Tang, C.; Chen, S.; Qian, H.; Huang, W. Interleukin-23: As a drug target for autoimmune inflammatory diseases. Immunology 2012, 135, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Moser, T.; Akgün, K.; Proschmann, U.; Sellner, J.; Ziemssen, T. The role of TH17 cells in multiple sclerosis: Therapeutic implications. Autoimmun. Rev. 2020, 19, 102647. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Harder, K.W.; Huntington, N.D.; Hibbs, M.L.; Tarlinton, D.M. Lyn tyrosine kinase: Accentuating the positive and the negative. Immunity 2005, 22, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Cyster, J.G.; Healy, J.I.; Kishihara, K.; Mak, T.W.; Thomas, M.L.; Goodnow, C.C. Regulation of B-lymphocyte negative and positive selection by tyrosine phosphatase CD45. Nature 1996, 381, 325–328. [Google Scholar] [CrossRef]
- Hecker, M.; Rüge, A.; Putscher, E.; Boxberger, N.; Rommer, P.S.; Fitzner, B.; Zettl, U.K. Aberrant expression of alternative splicing variants in multiple sclerosis—A systematic review. Autoimmun. Rev. 2019, 18, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, S.; Yong, V.W. The extracellular matrix as a modifier of neuroinflammation and remyelination in multiple sclerosis. Brain 2021, 144, 1958–1973. [Google Scholar] [CrossRef]
- Fu, R.; Shen, Q.; Xu, P.; Luo, J.J.; Tang, Y. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 2014, 49, 1422–1434. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMPs and DAMPs: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Pots, H.; Gilsbach, B.K.; Parsons, D.; Veltman, D.M.; Ramachandra, S.G.; Li, H.; Kortholt, A.; Jin, T. C2GAP2 is a common regulator of Ras signaling for chemotaxis, phagocytosis, and macropinocytosis. Front. Immunol. 2022, 13, 1075386. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, H.; Takeuchi, S.; Kubota, K.; Kobayashi, Y.; Kozakai, S.; Ukai, I.; Shichiku, A.; Okubo, M.; Numasaki, M.; Kanemitsu, Y.; et al. Lipopolysaccharide (LPS)-binding protein stimulates CD14-dependent Toll-like receptor 4 internalization and LPS-induced TBK1-IKKϵ-IRF3 axis activation. J. Biol. Chem. 2018, 293, 10186–10201. [Google Scholar] [CrossRef] [PubMed]
- Messina, S. Small GTPase RAS in multiple sclerosis—Exploring the role of RAS GTPase in the etiology of multiple sclerosis. Small GTPases 2020, 11, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Nogueras-Ortiz, C.; Chawla, S.; Bæk, R.; Jørgensen, M.M.; Kapogiannis, D. Altered levels of toll-like receptors in circulating extracellular vesicles in multiple sclerosis. Cells 2019, 8, 1058. [Google Scholar] [CrossRef]
- Melrose, J.; Hayes, A.J.; Bix, G. The CNS/PNS extracellular matrix provides instructive guidance cues to neural cells and neuroregulatory proteins in neural development and repair. Int. J. Mol. Sci. 2021, 22, 5583. [Google Scholar] [CrossRef]
- Leshchyns’ka, I.; Sytnyk, V.; Morrow, J.S.; Schachner, M. Neural cell adhesion molecule (NCAM) association with PKCbeta2 via betaI spectrin is implicated in NCAM-mediated neurite outgrowth. J. Cell Biol. 2003, 161, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Ramser, E.M.; Buck, F.; Schachner, M.; Tilling, T. Binding of alphaII spectrin to 14-3-3beta is involved in NCAM-dependent neurite outgrowth. Mol. Cell. Neurosci. 2010, 45, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Kroksveen, A.C.; Guldbrandsen, A.; Vaudel, M.; Lereim, R.R.; Barsnes, H.; Myhr, K.M.; Torkildsen, Ø.; Berven, F.S. In-depth cerebrospinal fluid quantitative proteome and deglycoproteome analysis: Presenting a comprehensive picture of pathways and processes affected by multiple sclerosis. J. Proteome Res. 2017, 16, 179–194. [Google Scholar] [CrossRef]
- Lodge, P.A.; Allegretta, M.; Steinman, L.; Sriram, S. Myelin basic protein peptide specificity and T-cell receptor gene usage of HPRT mutant T-cell clones in patients with multiple sclerosis. Ann. Neurol. 1994, 36, 734–740. [Google Scholar] [CrossRef]
- Trotter, J.L.; Damico, C.A.; Cross, A.H.; Pelfrey, C.M.; Karr, R.W.; Fu, X.T.; McFarland, H.F. HPRT mutant T-cell lines from multiple sclerosis patients recognize myelin proteolipid protein peptides. J. Neuroimmunol. 1997, 75, 95–103. [Google Scholar] [CrossRef]
- Blewett, M.M.; Xie, J.; Zaro, B.W.; Backus, K.M.; Altman, A.; Teijaro, J.R.; Cravatt, B.F. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal. 2016, 9, rs10. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, J.A.; Boster, A.L.; Imitola, J.; O’Connell, C. Design of oral agents for the management of multiple sclerosis: Benefit and risk assessment for dimethyl fumarate. Drug Des. Dev. Ther. 2014, 8, 897–908. [Google Scholar] [CrossRef]
- Rossi, S.; Motta, C.; Studer, V.; Macchiarulo, G.; Volpe, E.; Barbieri, F.; Ruocco, G.; Finardi, A.; Mancino, R.; Weiss, S.; et al. Interleukin-1β causes excitotoxic neurodegeneration and multiple sclerosis disease progression by activating the apoptotic protein p53. Mol. Neurodegener. 2014, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yeung, T.K.; Wang, Z. Enhanced drug resistance in cells coexpressing ErbB2 with EGF receptor or ErbB3. Biochem. Biophys. Res. Commun. 2000, 277, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Al-Ani, M.; Elemam, N.M.; Hachim, I.Y.; Raju, T.K.; Muhammad, J.S.; Hachim, M.Y.; Bendardaf, R.; Maghazachi, A.A. Molecular examination of differentially expressed genes in the brains of experimental autoimmune encephalomyelitis mice post Herceptin treatment. J. Inflamm. Res. 2021, 14, 2601–2617. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 2, 162–173. [Google Scholar] [CrossRef]
- Río, J.; Peña, J.; Brieva, L.; García-Domínguez, J.; Rodríguez-Antigüedad, A.; Oreja-Guevara, C.; Costa-Frossard, L.; Arroyo, R. Monitoring response to disease-modifying treatment in multiple sclerosis. Neurol. Perspect. 2023, 1, 100119. [Google Scholar] [CrossRef]
- Casado-Fernández, L.; Laso-García, F.; Piniella, D.; de Frutos, M.-C.G.; Otero-Ortega, L.; Bravo, S.-B.; Fuentes-Gimeno, B.; Docando, F.; Alonso-López, E.; Ruiz-Ares, G.; et al. The proteomic signature of circulating extracellular vesicles following intracerebral hemorrhage: Novel insights into mechanisms underlying recovery. Neurobiol. Dis. 2024, 201, 106665. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, 638–646. [Google Scholar] [CrossRef]


| Responders (n = 49) | Non-Responders (n = 31) | p-Value | |
|---|---|---|---|
| Demographics | |||
| Women, n (%) | 25 (51%) | 18 (58.1%) | 0.34 |
| Age, years | 44.06 (9.03) | 43.82 (11.23) | 0.91 |
| Clinical data | |||
| Time from diagnosis, months | 123.53 (120.48) | 153.3 (137.01) | 0.30 |
| Baseline EDSS | 1.89 (2.07) | 2 (2.29) | 0.88 |
| Treatments received | 0.59 | ||
| Natalizumab, n (%) | 12 (24.5) | 6 (19.4) | |
| Teriflunomide, n (%) | 2 (4.1) | 3 (9.7) | |
| Interferon, n (%) | 6 (12.2) | 0 (0) | |
| Dimethyl fumarate, n (%) | 7 (14.3) | 6 (19.4) | |
| Ocrelizumab, n (%) | 7 (14.2) | 3 (9.7) | |
| Siponimod, n (%) | 1 (2.0) | 0 (0) | |
| Cladribine, n (%) | 14 (28.6) | 11 (35.5) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres Iglesias, G.; López-Molina, M.; Botella, L.; Laso-García, F.; Chamorro, B.; Fernández-Fournier, M.; Puertas, I.; Bravo, S.B.; Alonso-López, E.; Díez-Tejedor, E.; et al. Differential Protein Expression in Extracellular Vesicles Defines Treatment Responders and Non-Responders in Multiple Sclerosis. Int. J. Mol. Sci. 2024, 25, 10761. https://doi.org/10.3390/ijms251910761
Torres Iglesias G, López-Molina M, Botella L, Laso-García F, Chamorro B, Fernández-Fournier M, Puertas I, Bravo SB, Alonso-López E, Díez-Tejedor E, et al. Differential Protein Expression in Extracellular Vesicles Defines Treatment Responders and Non-Responders in Multiple Sclerosis. International Journal of Molecular Sciences. 2024; 25(19):10761. https://doi.org/10.3390/ijms251910761
Chicago/Turabian StyleTorres Iglesias, Gabriel, MariPaz López-Molina, Lucía Botella, Fernando Laso-García, Beatriz Chamorro, Mireya Fernández-Fournier, Inmaculada Puertas, Susana B. Bravo, Elisa Alonso-López, Exuperio Díez-Tejedor, and et al. 2024. "Differential Protein Expression in Extracellular Vesicles Defines Treatment Responders and Non-Responders in Multiple Sclerosis" International Journal of Molecular Sciences 25, no. 19: 10761. https://doi.org/10.3390/ijms251910761
APA StyleTorres Iglesias, G., López-Molina, M., Botella, L., Laso-García, F., Chamorro, B., Fernández-Fournier, M., Puertas, I., Bravo, S. B., Alonso-López, E., Díez-Tejedor, E., Gutiérrez-Fernández, M., & Otero-Ortega, L. (2024). Differential Protein Expression in Extracellular Vesicles Defines Treatment Responders and Non-Responders in Multiple Sclerosis. International Journal of Molecular Sciences, 25(19), 10761. https://doi.org/10.3390/ijms251910761







