Histone Tail Cleavage as a Mechanism for Epigenetic Regulation
Abstract
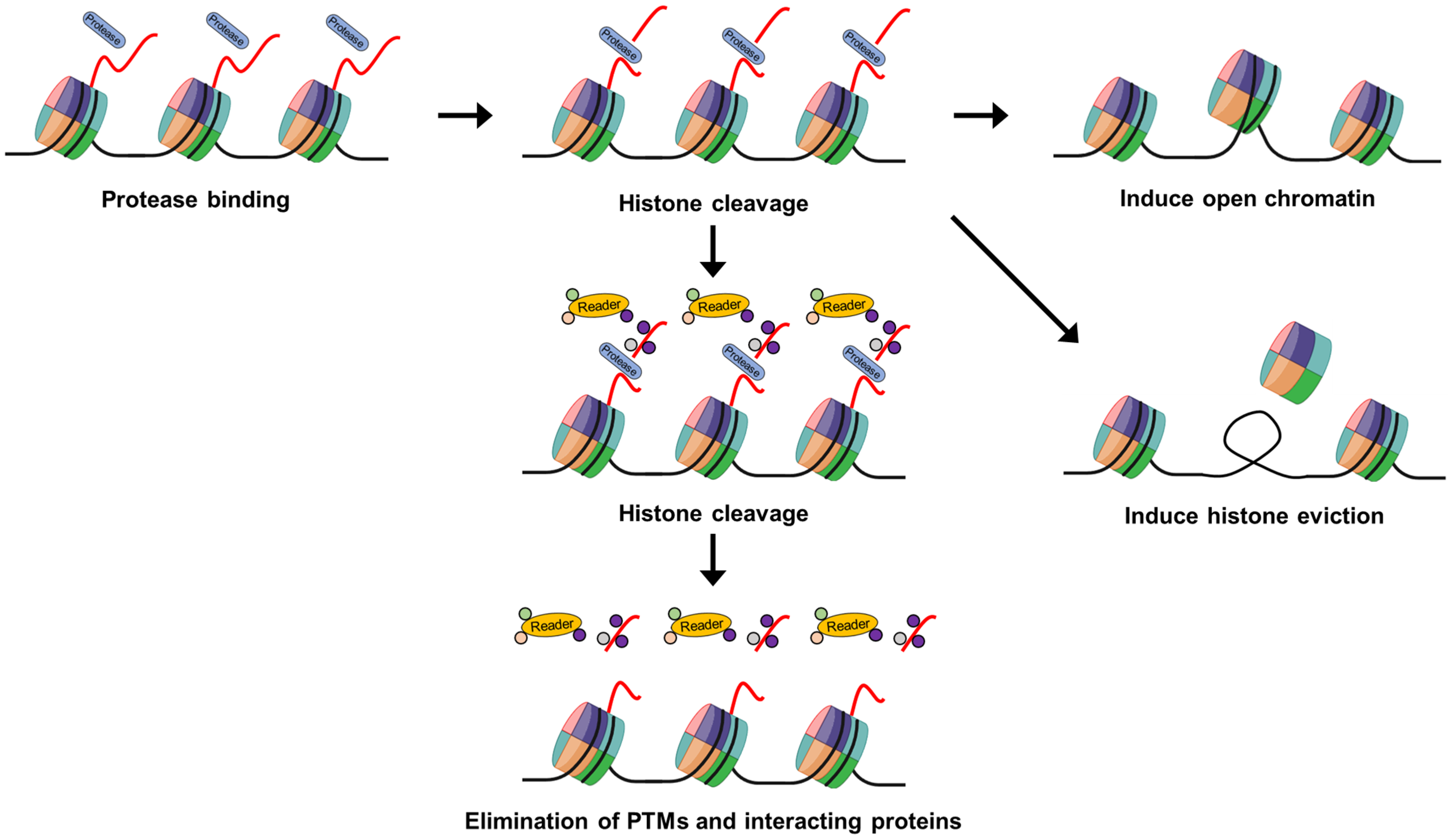
:1. Introduction
2. Cleavage of Histones H2A
| Histone | Protease | Cleavage Site(s) | Biological Significance of Activity | Model | Reference |
|---|---|---|---|---|---|
| H2A | H2A-specific protease | Val114-Leu115 | Unknown | Calf thymus | [25] |
| Neutrophil elastase | Val114-Leu115 | Neutrophil extracellular trap (NET) formation | Neutrophil | [31] | |
| Histone H2A specific protease (H2Asp) | Asn90-Asp91 | Unknown | Chicken liver extract | [33] | |
| Cathepsin L | Leu23-Gln24 | Embryonic stem cells (ESCs) differentiation | Mouse embryonic stem cells (mESCs) | [38] | |
| H2B | Tryptase | Unknown | Mast cell differentiation | Mouse mast cells | [35,36] |
| H3 | Tryptase | Unknown | Mast cell differentiation | Mouse mast cells | [35,36] |
| Cathepsin L | Ala21-Thr22, Arg26-Lys27, Ala31-Thr32 | Embryonic stem cells (ESCs) differentiation | Human embryonic stem cells (hESCs) | [37] | |
| Ala21-Thr22, Thr22-Lys23, Lys23-Ala24, | Embryonic stem cells (ESCs) differentiation | Mouse embryonic stem cells (mESCs) | [38] | ||
| Ala24-Ala25, Arg26-Lys27, Lys27-Ser28 | |||||
| Yeast endopeptidase | Ala21-Thr22 | Induced under nutrient deprivation and sporulation | Saccharomyces cerevisiae | [38] | |
| JMJD5 | Lys9-Ser10 | Induced under DNA damage | Human lung cancer cells | [39] | |
| Glutamate dehydrogenase | Lys23-Ala24, Lys27-Ser28 | Unknown | Chicken liver extracts | [40] | |
| Unknown | Unknown | Unknown | Tetrahymena micronuclei | [41] | |
| FMDV 3C protease | Leu20-Ala21 | Host cell transcription shutoff | Hamster kidney fibroblast cells | [42] | |
| MMP-9 | Lys18-Gln19 | Osteclastogenesis | Bone marrow macrophages | [43] | |
| Melanomagenesis | Human melanoma | [44] | |||
| Colonic carcinogenesis | Human colon cancer cells | [45] | |||
| Cathepsin D | Lys23-Ala24 | Involution mammary gland | Mouse mammary gland | [46] | |
| Vacuolor protease B (PrB) | Lys23-Ala24 | Unknown | Saccharomyces cerevisiae | [47] | |
| Granzyme A | Unknown | Staurosporine-induced cell death | Human B lymphoblastoid cell | [48] | |
| H4 | Granzyme A | Unknown | Staurosporine-induced cell death | Human B lymphoblastoid cell | [49] |
| Trypsin and Chymotrypsin | Arg17-Arg19 | Intestinal cell differentiation | Human colon cancer cells | [50] |
3. Cleavage of Histones H2B
4. Cleavage of Histones H3
5. Cleavage of Histones H4
6. Epigenetic Regulation through Histone Cleavage Mechanism
7. Chromatin Dynamics and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bentley, G.A.; Lewit-Bentley, A.; Finch, J.T.; Podjarny, A.D.; Roth, M. Crystal structure of the nucleosome core particle at 16 A resolution. J. Mol. Biol. 1984, 176, 55–75. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D.; Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Reeve, J.N.; Sandman, K.; Daniels, C.J. Archaeal histones, nucleosomes, and transcription initiation. Cell 1997, 89, 999–1002. [Google Scholar] [CrossRef]
- Zhou, P.; Wu, E.; Alam, H.B.; Li, Y. Histone cleavage as a mechanism for epigenetic regulation: Current insights and perspectives. Curr. Mol. Med. 2014, 14, 1164–1172. [Google Scholar] [CrossRef]
- Stillman, B. Histone Modifications: Insights into Their Influence on Gene Expression. Cell 2018, 175, 6–9. [Google Scholar] [CrossRef]
- Arnaudo, A.M.; Garcia, B.A. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin 2013, 6, 24. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Beck, H.C.; Nielsen, E.C.; Matthiesen, R.; Jensen, L.H.; Sehested, M.; Finn, P.; Grauslund, M.; Hansen, A.M.; Jensen, O.N. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteom. 2006, 5, 1314–1325. [Google Scholar] [CrossRef]
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteom. 2007, 6, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Peng, C.; Montellier, E.; Lu, Z.; Chen, Y.; Ishii, H.; Debernardi, A.; Buchou, T.; Rousseaux, S.; Jin, F.; et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014, 10, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Fierz, B.; Muir, T.W. Chromatin as an expansive canvas for chemical biology. Nat. Chem. Biol. 2012, 8, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Xie, Z.; Dai, J.; Dai, L.; Tan, M.; Cheng, Z.; Wu, Y.; Boeke, J.D.; Zhao, Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom. 2012, 11, 100–107. [Google Scholar] [CrossRef]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Bohm, V.; Hieb, A.R.; Andrews, A.J.; Gansen, A.; Rocker, A.; Toth, K.; Luger, K.; Langowski, J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011, 39, 3093–3102. [Google Scholar] [CrossRef]
- Allis, C.D.; Bowen, J.K.; Abraham, G.N.; Glover, C.V.; Gorovsky, M.A. Proteolytic processing of histone H3 in chromatin: A physiologically regulated event in Tetrahymena micronuclei. Cell 1980, 20, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Gorovsky, M.A.; Keevert, J.B. Absence of histone F1 in a mitotically dividing, genetically inactive nucleus. Proc. Natl. Acad. Sci. USA 1975, 72, 2672–2676. [Google Scholar] [CrossRef] [PubMed]
- Eickbush, T.H.; Godfrey, J.E.; Elia, M.C.; Moudrianakis, E.N. H2a-specific proteolysis as a unique probe in the analysis of the histone octamer. J. Biol. Chem. 1988, 263, 18972–18978. [Google Scholar] [CrossRef]
- Watson, D.K.; Moudrianakis, E.N. Histone-dependent reconstitution and nucleosomal localization of a nonhistone chromosomal protein: The H2A-specific protease. Biochemistry 1982, 21, 248–256. [Google Scholar] [CrossRef]
- Okawa, Y.; Takada, K.; Minami, J.; Aoki, K.; Shibayama, H.; Ohkawa, K. Purification of N-terminally truncated histone H2A-monoubiquitin conjugates from leukemic cell nuclei: Probable proteolytic products of ubiquitinated H2A. Int. J. Biochem. Cell Biol. 2003, 35, 1588–1600. [Google Scholar] [CrossRef]
- Pantazis, P.; Sarin, P.S.; Gallo, R.C. Detection of the histone-2A related polypeptide in differentiated human myeloid cells (HL-60) and its distribution in human acute leukemia. Int. J. Cancer 1981, 27, 585–592. [Google Scholar] [CrossRef]
- Simpkins, H.; Mahon, K. The histone content of chromatin preparations from leukaemic cells. Br. J. Haematol. 1977, 37, 467–473. [Google Scholar] [CrossRef]
- Glibert, P.; Vossaert, L.; Van Steendam, K.; Lambrecht, S.; Van Nieuwerburgh, F.; Offner, F.; Kipps, T.; Dhaenens, M.; Deforce, D. Quantitative proteomics to characterize specific histone H2A proteolysis in chronic lymphocytic leukemia and the myeloid THP-1 cell line. Int. J. Mol. Sci. 2014, 15, 9407–9421. [Google Scholar] [CrossRef] [PubMed]
- Minami, J.; Takada, K.; Aoki, K.; Shimada, Y.; Okawa, Y.; Usui, N.; Ohkawa, K. Purification and characterization of C-terminal truncated forms of histone H2A in monocytic THP-1 cells. Int. J. Biochem. Cell Biol. 2007, 39, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Dhaenens, M.; Glibert, P.; Lambrecht, S.; Vossaert, L.; Van Steendam, K.; Elewaut, D.; Deforce, D. Neutrophil Elastase in the capacity of the “H2A-specific protease”. Int. J. Biochem. Cell Biol. 2014, 51, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Vogler, C.; Huber, C.; Waldmann, T.; Ettig, R.; Braun, L.; Izzo, A.; Daujat, S.; Chassignet, I.; Lopez-Contreras, A.J.; Fernandez-Capetillo, O.; et al. Histone H2A C-terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet. 2010, 6, e1001234. [Google Scholar] [CrossRef]
- Panda, P.; Chaturvedi, M.M.; Panda, A.K.; Suar, M.; Purohit, J.S. Purification and characterization of a novel histone H2A specific protease (H2Asp) from chicken liver nuclear extract. Gene 2013, 512, 47–54. [Google Scholar] [CrossRef]
- Coradin, M.; Cesare, J.; Lan, Y.; Zhu, Z.; Lund, P.J.; Sidoli, S.; Perez, Y.; Lu, C.; Porter, E.G.; Robert, C.W.M.; et al. Cleavage of histone H2A during embryonic stem cell differentiation destabilizes nucleosomes to counteract gene activation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Melo, F.R.; Vita, F.; Berent-Maoz, B.; Levi-Schaffer, F.; Zabucchi, G.; Pejler, G. Proteolytic histone modification by mast cell tryptase, a serglycin proteoglycan-dependent secretory granule protease. J. Biol. Chem. 2014, 289, 7682–7690. [Google Scholar] [CrossRef]
- Grigera, P.R.; Tisminetzky, S.G. Histone H3 modification in BHK cells infected with foot-and-mouth disease virus. Virology 1984, 136, 10–19. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Kirmizis, A.; Nelson, C.; Bartke, T.; Saksouk, N.; Cote, J.; Kouzarides, T. Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 2009, 16, 17–22. [Google Scholar] [CrossRef]
- Mandal, P.; Azad, G.K.; Tomar, R.S. Identification of a novel histone H3 specific protease activity in nuclei of chicken liver. Biochem. Biophys. Res. Commun. 2012, 421, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.; Verma, N.; Chauhan, S.; Tomar, R.S. Unexpected histone H3 tail-clipping activity of glutamate dehydrogenase. J. Biol. Chem. 2013, 288, 18743–18757. [Google Scholar] [CrossRef] [PubMed]
- Duarte, L.F.; Young, A.R.; Wang, Z.; Wu, H.A.; Panda, T.; Kou, Y.; Kapoor, A.; Hasson, D.; Mills, N.R.; Ma’ayan, A.; et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun. 2014, 5, 5210. [Google Scholar] [CrossRef]
- Lee, P.Y.; Park, B.C.; Chi, S.W.; Bae, K.H.; Kim, S.; Cho, S.; Kim, J.H.; Park, S.G. Histone H3 is Digested by Granzyme A During Compromised Cell Death in the Raji Cells. J. Microbiol. Biotechnol. 2015, 25, 1578–1582. [Google Scholar] [CrossRef]
- Kim, K.; Shin, Y.; Kim, J.; Ulmer, T.S.; An, W. H3K27me1 is essential for MMP-9-dependent H3N-terminal tail proteolysis during osteoclastogenesis. Epigenetics Chromatin 2018, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Goulet, B.; Baruch, A.; Moon, N.S.; Poirier, M.; Sansregret, L.L.; Erickson, A.; Bogyo, M.; Nepveu, A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 2004, 14, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Sudhan, D.R.; Siemann, D.W. Cathepsin L targeting in cancer treatment. Pharmacol. Ther. 2015, 155, 105–116. [Google Scholar] [CrossRef]
- Xue, Y.; Vashisht, A.A.; Tan, Y.; Su, T.; Wohlschlegel, J.A. PRB1 is required for clipping of the histone H3 N terminal tail in Saccharomyces cerevisiae. PLoS ONE 2014, 9, e90496. [Google Scholar] [CrossRef]
- Shin, Y.; Kim, S.; Ghate, N.B.; Rhie, S.K.; An, W. MMP-9 drives the melanomagenic transcription program through histone H3 tail proteolysis. Oncogene 2022, 41, 560–570. [Google Scholar] [CrossRef]
- Shin, Y.; Kim, S.; Liang, G.; An, W. MMP-9-dependent proteolysis of the histone H3 N-terminal tail: A critical epigenetic step in driving oncogenic transcription and colon tumorigenesis. Mol. Oncol. 2024, 18, 2001–2019. [Google Scholar] [CrossRef]
- Melo, F.R.; Wallerman, O.; Paivandy, A.; Calounova, G.; Gustafson, A.M.; Sabari, B.R.; Zabucchi, G.; Allis, C.D.; Pejler, G. Tryptase-catalyzed core histone truncation: A novel epigenetic regulatory mechanism in mast cells. J. Allergy Clin. Immunol. 2017, 140, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Duncan, E.M.; Muratore-Schroeder, T.L.; Cook, R.G.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 2008, 135, 284–294. [Google Scholar] [CrossRef]
- Kim, K.; Punj, V.; Kim, J.M.; Lee, S.; Ulmer, T.S.; Lu, W.; Rice, J.C.; An, W. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016, 30, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Mahendra, G.; Kanungo, M.S. Age-related and steroid induced changes in the histones of the quail liver. Arch. Gerontol. Geriatr. 2000, 30, 109–114. [Google Scholar] [CrossRef]
- Shen, J.; Xiang, X.; Chen, L.; Wang, H.; Wu, L.; Sun, Y.; Ma, L.; Gu, X.; Liu, H.; Wang, L.; et al. JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep. 2017, 18, 2131–2143. [Google Scholar] [CrossRef]
- Vossaert, L.; Meert, P.; Scheerlinck, E.; Glibert, P.; Van Roy, N.; Heindryckx, B.; De Sutter, P.; Dhaenens, M.; Deforce, D. Identification of histone H3 clipping activity in human embryonic stem cells. Stem Cell Res. 2014, 13, 123–134. [Google Scholar] [CrossRef]
- Falk, M.M.; Grigera, P.R.; Bergmann, I.E.; Zibert, A.; Multhaup, G.; Beck, E. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J. Virol. 1990, 64, 748–756. [Google Scholar] [CrossRef]
- Tesar, M.; Marquardt, O. Foot-and-mouth disease virus protease 3C inhibits cellular transcription and mediates cleavage of histone H3. Virology 1990, 174, 364–374. [Google Scholar] [CrossRef]
- Goulet, B.; Sansregret, L.; Leduy, L.; Bogyo, M.; Weber, E.; Chauhan, S.S.; Nepveu, A. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol. Cancer Res. 2007, 5, 899–907. [Google Scholar] [CrossRef]
- Goulet, B.; Truscott, M.; Nepveu, A. A novel proteolytically processed CDP/Cux isoform of 90 kDa is generated by cathepsin L. Biol. Chem. 2006, 387, 1285–1293. [Google Scholar] [CrossRef]
- Grotsky, D.A.; Gonzalez-Suarez, I.; Novell, A.; Neumann, M.A.; Yaddanapudi, S.C.; Croke, M.; Martinez-Alonso, M.; Redwood, A.B.; Ortega-Martinez, S.; Feng, Z.; et al. BRCA1 loss activates cathepsin L-mediated degradation of 53BP1 in breast cancer cells. J. Cell Biol. 2013, 200, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Khalkhali-Ellis, Z.; Goossens, W.; Margaryan, N.V.; Hendrix, M.J. Cleavage of Histone 3 by Cathepsin D in the involuting mammary gland. PLoS ONE 2014, 9, e103230. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.Y.; Park, B.C.; Chi, S.W.; Bae, K.H.; Kim, S.; Cho, S.; Kang, S.; Kim, J.H.; Park, S.G. Histone H4 is cleaved by granzyme A during staurosporine-induced cell death in B-lymphoid Raji cells. BMB Rep. 2016, 49, 560–565. [Google Scholar] [CrossRef]
- Marruecos, L.; Bertran, J.; Alvarez-Villanueva, D.; Mulero, M.C.; Guillen, Y.; Palma, L.G.; Floor, M.; Vert, A.; Arce-Gallego, S.; Pecharroman, I.; et al. Dynamic chromatin association of IkappaBalpha is regulated by acetylation and cleavage of histone H4. EMBO Rep. 2021, 22, e52649. [Google Scholar] [CrossRef]
- Stormberg, T.; Vemulapalli, S.; Filliaux, S.; Lyubchenko, Y.L. Effect of histone H4 tail on nucleosome stability and internucleosomal interactions. Sci. Rep. 2021, 11, 24086. [Google Scholar] [CrossRef]
- Allis, C.D.; Allen, R.L.; Wiggins, J.C.; Chicoine, L.G.; Richman, R. Proteolytic processing of h1-like histones in chromatin: A physiologically and developmentally regulated event in Tetrahymena micronuclei. J. Cell Biol. 1984, 99, 1669–1677. [Google Scholar] [CrossRef]
- Nurse, N.P.; Jimenez-Useche, I.; Smith, I.T.; Yuan, C. Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophys. J. 2013, 104, 1081–1088. [Google Scholar] [CrossRef]
- Polach, K.J.; Lowary, P.T.; Widom, J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 2000, 298, 211–223. [Google Scholar] [CrossRef]
- Das, C.; Tyler, J.K. Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta 2013, 1819, 332–342. [Google Scholar] [CrossRef]
- Lee, C.K.; Shibata, Y.; Rao, B.; Strahl, B.D.; Lieb, J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004, 36, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Patel, D.J. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. J. Biol. Chem. 2011, 286, 18363–18368. [Google Scholar] [CrossRef] [PubMed]
- Hadnagy, A.; Beaulieu, R.; Balicki, D. Histone tail modifications and noncanonical functions of histones: Perspectives in cancer epigenetics. Mol. Cancer Ther. 2008, 7, 740–748. [Google Scholar] [CrossRef]
- Azad, G.K.; Tomar, R.S. Proteolytic clipping of histone tails: The emerging role of histone proteases in regulation of various biological processes. Mol. Biol. Rep. 2014, 41, 2717–2730. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, Y. Histone Tail Cleavage as a Mechanism for Epigenetic Regulation. Int. J. Mol. Sci. 2024, 25, 10789. https://doi.org/10.3390/ijms251910789
Shin Y. Histone Tail Cleavage as a Mechanism for Epigenetic Regulation. International Journal of Molecular Sciences. 2024; 25(19):10789. https://doi.org/10.3390/ijms251910789
Chicago/Turabian StyleShin, Yonghwan. 2024. "Histone Tail Cleavage as a Mechanism for Epigenetic Regulation" International Journal of Molecular Sciences 25, no. 19: 10789. https://doi.org/10.3390/ijms251910789
APA StyleShin, Y. (2024). Histone Tail Cleavage as a Mechanism for Epigenetic Regulation. International Journal of Molecular Sciences, 25(19), 10789. https://doi.org/10.3390/ijms251910789





