Thyroid Hormone Upregulates Cav1.2 Channels in Cardiac Cells via the Downregulation of the Channels’ β4 Subunit
Abstract
1. Introduction
2. Results
2.1. T3 Increased the Expression of α1c at the mRNA and Protein Levels in H9c2 Cells
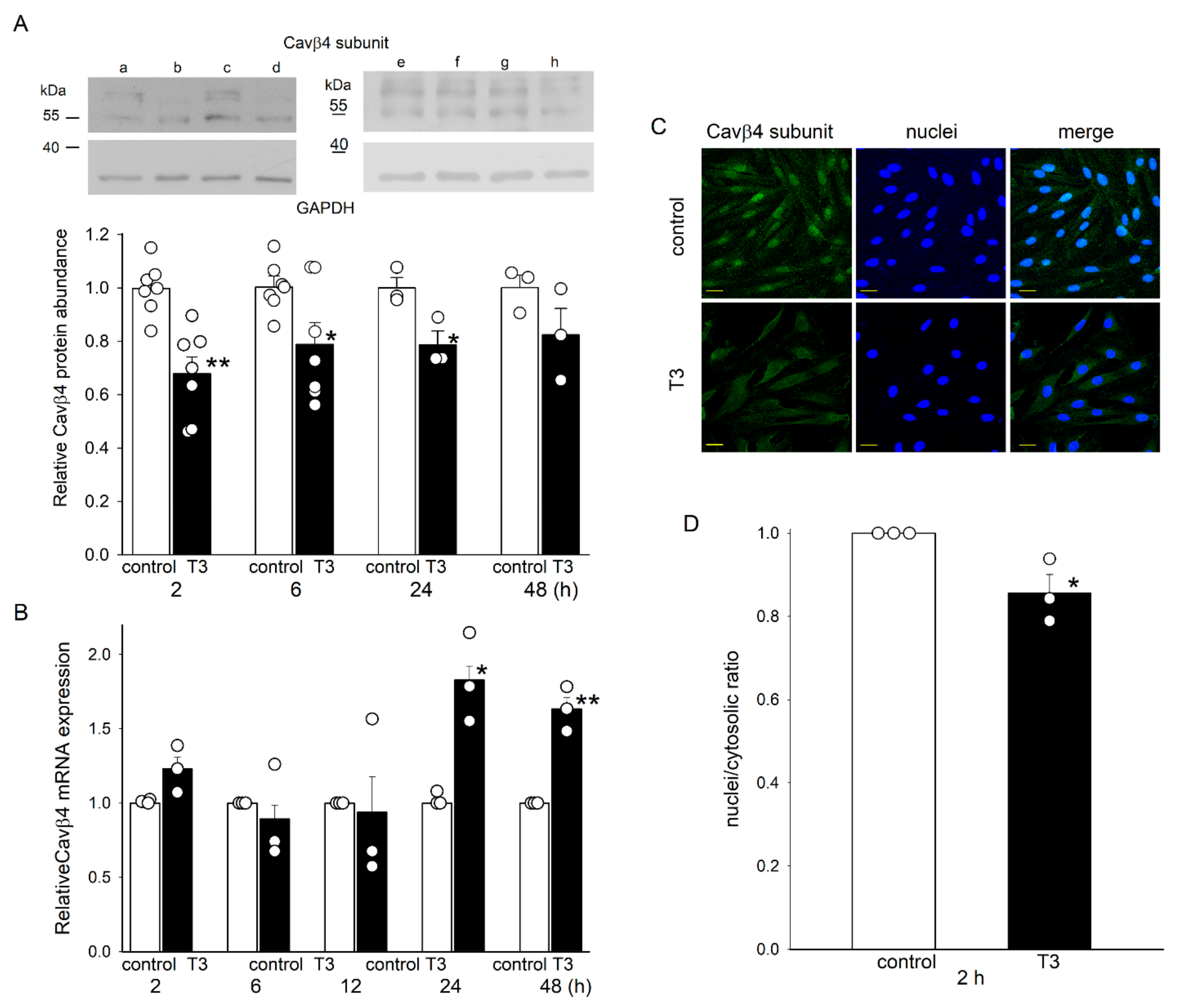
2.2. T3 Decreased the Expression of Cavβ4 at the Protein Level and Its Nuclear Translocation in H9c2 Cells
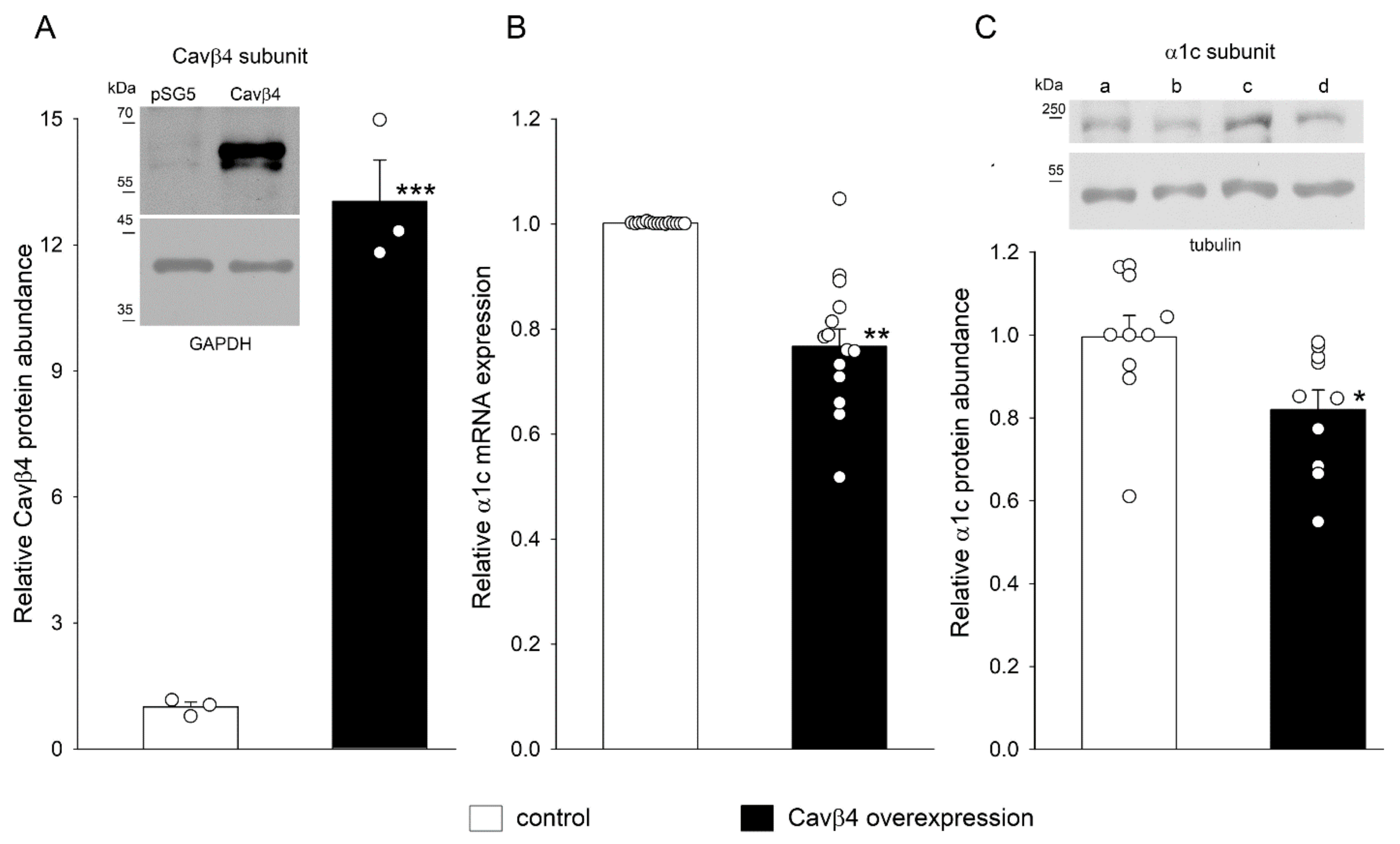
2.3. Cavβ4 Overexpression Decreased α1c mRNA Expression and Protein Abundance in H9c2 Cells
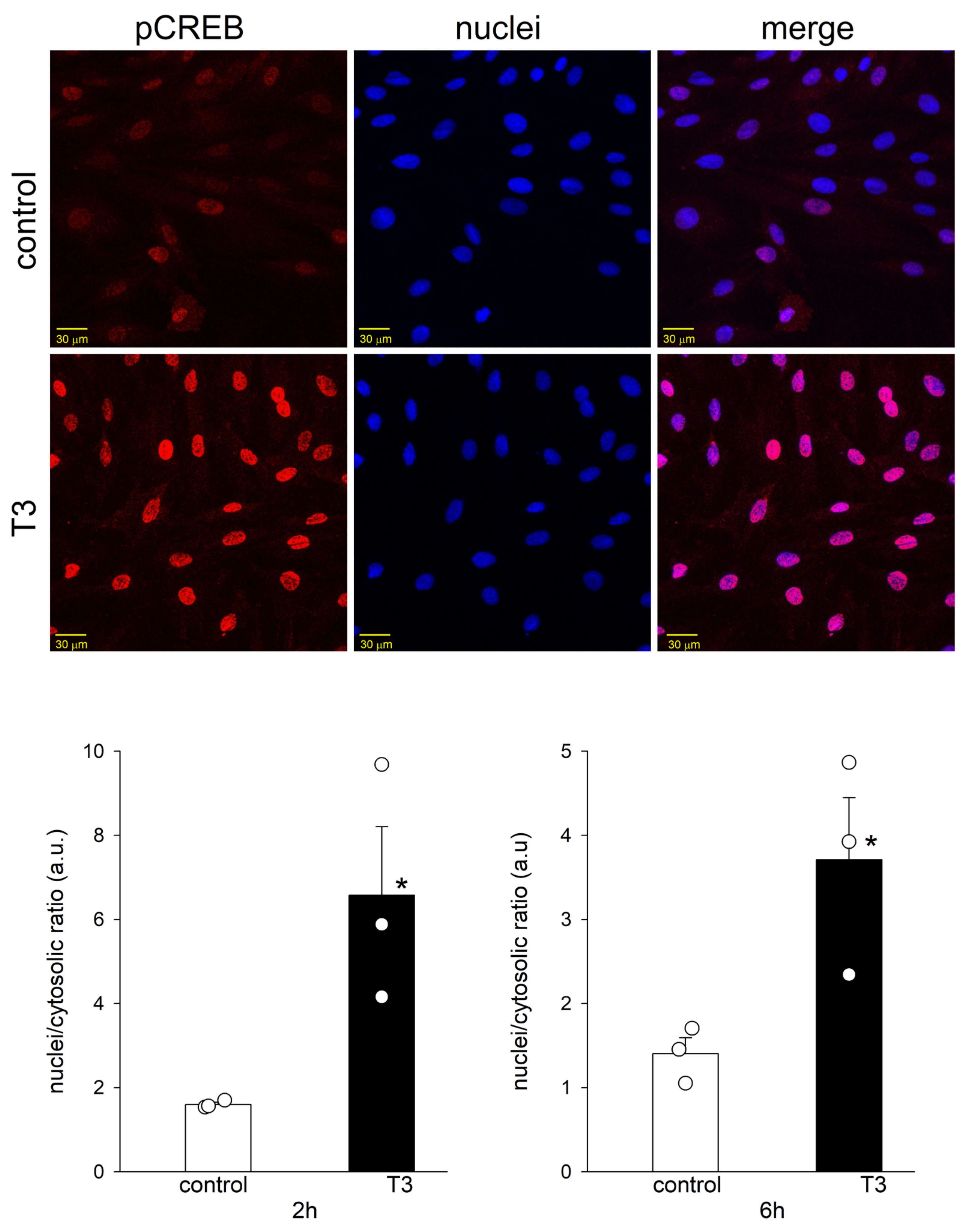
2.4. T3 Altered the Nuclear Translocation, but Not the Protein Abundance of pCREB
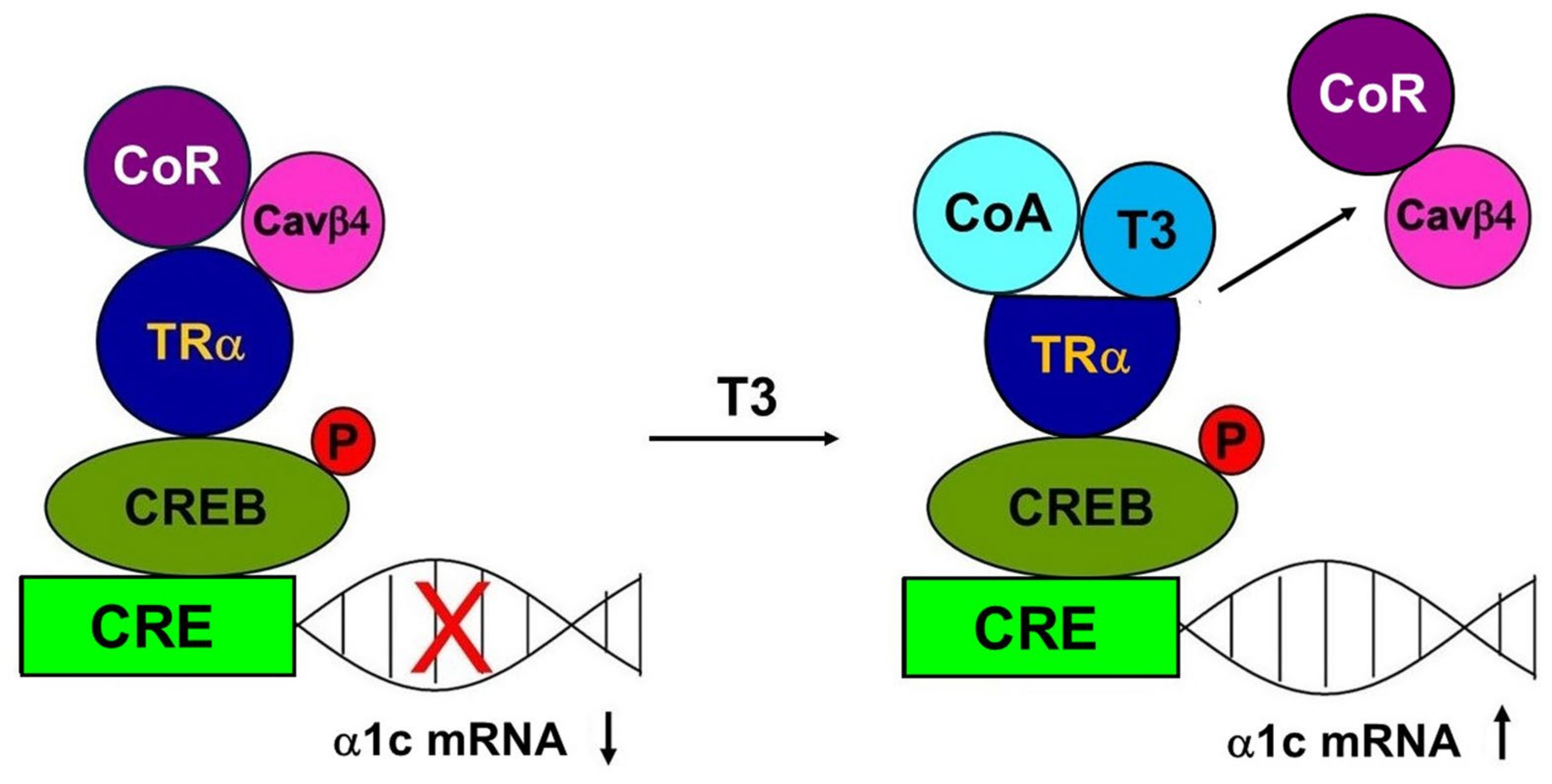
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Heart and Ventricular Myocyte Isolation
4.3. Cell Culture and Transfection
4.4. qRT-PCR Assays
4.5. Western Blotting
4.6. Immunofluorescence
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grais, I.M.; Sowers, J.R. Thyroid and the Heart. Am. J. Med. 2014, 127, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Bassett, J.H.D.; Harvey, C.B.; Williams, G.R. Mechanisms of Thyroid Hormone Receptor-Specific Nuclear and Extra Nuclear Actions. Mol. Cell. Endocrinol. 2003, 213, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Ma, M.; Washizuka, T.; Komura, S.; Yoshida, T.; Hosaka, Y.; Hatada, K.; Chinushi, M.; Yamamoto, T.; Watanabe, K.; et al. Thyroid Hormone Regulates mRNA Expression and Currents of Ion Channels in Rat Atrium. Biochem. Biophys. Res. Commun. 2003, 308, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Bers, D.M. Cardiac Excitation-Contraction Coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-J.; Yeh, Y.-H.; Lin, K.-H.; Chang, G.-J.; Kuo, C.-T. Molecular Characterization of Thyroid Hormone-Inhibited Atrial L-Type Calcium Channel Expression: Implication for Atrial Fibrillation in Hyperthyroidism. Basic Res. Cardiol. 2011, 106, 163–174. [Google Scholar] [CrossRef]
- Silvestro, S.; Gugliandolo, A.; Chiricosta, L.; Diomede, F.; Trubiani, O.; Bramanti, P.; Pizzicannella, J.; Mazzon, E. MicroRNA Profiling of HL-1 Cardiac Cells-Derived Extracellular Vesicles. Cells 2021, 10, 273. [Google Scholar] [CrossRef]
- Tadmouri, A.; Kiyonaka, S.; Barbado, M.; Rousset, M.; Fablet, K.; Sawamura, S.; Bahembera, E.; Pernet-Gallay, K.; Arnoult, C.; Miki, T.; et al. Cacnb4 Directly Couples Electrical Activity to Gene Expression, a Process Defective in Juvenile Epilepsy. EMBO J. 2012, 31, 3730–3744. [Google Scholar] [CrossRef]
- Tammineni, E.R.; Carrillo, E.D.; Soto-Acosta, R.; Angel-Ambrocio, A.H.; García, M.C.; Bautista-Carbajal, P.; Del Angel, R.M.; Sánchez, J.A. The Β4 Subunit of Cav1.2 Channels Is Required for an Optimal Interferon Response in Cardiac Muscle Cells. Sci. Signal 2018, 11, eaaj1676. [Google Scholar] [CrossRef]
- Hescheler, J.; Meyer, R.; Plant, S.; Krautwurst, D.; Rosenthal, W.; Schultz, G. Morphological, Biochemical, and Electrophysiological Characterization of a Clonal Cell (H9c2) Line from Rat Heart. Circ. Res. 1991, 69, 1476–1486. [Google Scholar] [CrossRef]
- Jump, D.B.; Narayan, P.; Towle, H.; Oppenheimer, J.H. Rapid Effects of Triiodothyronine on Hepatic Gene Expression. Hybridization Analysis of Tissue-Specific Triiodothyronine Regulation of mRNAS14. J. Biol. Chem. 1984, 259, 2789–2797. [Google Scholar] [CrossRef]
- Krane, I.M.; Spindel, E.R.; Chin, W.W. Thyroid Hormone Decreases the Stability and the Poly(A) Tract Length of Rat Thyrotropin Beta-Subunit Messenger RNA. Mol. Endocrinol. 1991, 5, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Goulart-Silva, F.; de Souza, P.B.; Nunes, M.T. T3 Rapidly Modulates TSHβ mRNA Stability and Translational Rate in the Pituitary of Hypothyroid Rats. Mol. Cell Endocrinol. 2011, 332, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An Overview of microRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, E.D.; Escobar, Y.; González, G.; Hernández, A.; Galindo, J.M.; García, M.C.; Sánchez, J.A. Posttranscriptional Regulation of the Β2-Subunit of Cardiac L-Type Ca2+ Channels by MicroRNAs during Long-Term Exposure to Isoproterenol in Rats. J. Cardiovasc. Pharmacol. 2011, 58, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Dolphin, A.C. Voltage-gated Calcium Channels and Their Auxiliary Subunits: Physiology and Pathophysiology and Pharmacology. J. Physiol. 2016, 594, 5369–5390. [Google Scholar] [CrossRef]
- Vergnol, A.; Traoré, M.; Pietri-Rouxel, F.; Falcone, S. New Insights in CaVβ Subunits: Role in the Regulation of Gene Expression and Cellular Homeostasis. Front. Cell Dev. Biol. 2022, 10, 880441. [Google Scholar] [CrossRef]
- Foell, J.D.; Balijepalli, R.C.; Delisle, B.P.; Yunker, A.M.R.; Robia, S.L.; Walker, J.W.; McEnery, M.W.; January, C.T.; Kamp, T.J. Molecular Heterogeneity of Calcium Channel Beta-Subunits in Canine and Human Heart: Evidence for Differential Subcellular Localization. Physiol. Genomics 2004, 17, 183–200. [Google Scholar] [CrossRef]
- Ronjat, M.; Kiyonaka, S.; Barbado, M.; De Waard, M.; Mori, Y. Nuclear Life of the Voltage-Gated Cacnb4 Subunit and Its Role in Gene Transcription Regulation. Channels 2013, 7, 119–125. [Google Scholar] [CrossRef]
- Razvi, S.; Jabbar, A.; Pingitore, A.; Danzi, S.; Biondi, B.; Klein, I.; Peeters, R.; Zaman, A.; Iervasi, G. Thyroid Hormones and Cardiovascular Function and Diseases. J. Am. Coll. Cardiol. 2018, 71, 1781–1796. [Google Scholar] [CrossRef]
- Dillmann, W.H. Cellular Action of Thyroid Hormone on the Heart. Thyroid 2002, 12, 447–452. [Google Scholar] [CrossRef]
- Pantos, C.; Xinaris, C.; Mourouzis, I.; Perimenis, P.; Politi, E.; Spanou, D.; Cokkinos, D.V. Thyroid Hormone Receptor Alpha 1: A Switch to Cardiac Cell “Metamorphosis”? J. Physiol. Pharmacol. 2008, 59, 253–269. [Google Scholar] [PubMed]
- Hibino, H.; Pironkova, R.; Onwumere, O.; Rousset, M.; Charnet, P.; Hudspeth, A.J.; Lesage, F. Direct Interaction with a Nuclear Protein and Regulation of Gene Silencing by a Variant of the Ca2+-Channel Beta 4 Subunit. Proc. Natl. Acad. Sci. USA 2003, 100, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Mayr, B.; Montminy, M. Transcriptional Regulation by the Phosphorylation-Dependent Factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Morishima, M.; Wang, P.; Horii, K.; Horikawa, K.; Ono, K. Eicosapentaenoic Acid Rescues Cav1.2-L-Type Ca2+ Channel Decline Caused by Saturated Fatty Acids via Both Free Fatty Acid Receptor 4-Dependent and -Independent Pathways in Cardiomyocytes. Int. J. Mol. Sci. 2024, 25, 7570. [Google Scholar] [CrossRef]
- Servili, E.; Trus, M.; Sajman, J.; Sherman, E.; Atlas, D. Elevated Basal Transcription Can Underlie Timothy Channel Association with Autism Related Disorders. Prog. Neurobiol. 2020, 191, 101820. [Google Scholar] [CrossRef]
- Tsai, C.-T.; Wang, D.L.; Chen, W.-P.; Hwang, J.-J.; Hsieh, C.-S.; Hsu, K.-L.; Tseng, C.-D.; Lai, L.-P.; Tseng, Y.-Z.; Chiang, F.-T.; et al. Angiotensin II Increases Expression of alpha1C Subunit of L-Type Calcium Channel through a Reactive Oxygen Species and cAMP Response Element-Binding Protein-Dependent Pathway in HL-1 Myocytes. Circ. Res. 2007, 100, 1476–1485. [Google Scholar] [CrossRef]
- Satin, J.; Schroder, E.A.; Crump, S.M. L-Type Calcium Channel Auto-Regulation of Transcription. Cell Calcium 2011, 49, 306–313. [Google Scholar] [CrossRef]
- Schulte, J.S.; Seidl, M.D.; Nunes, F.; Freese, C.; Schneider, M.; Schmitz, W.; Müller, F.U. CREB Critically Regulates Action Potential Shape and Duration in the Adult Mouse Ventricle. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H1998–H2007. [Google Scholar] [CrossRef]
- Yang, X.; Mao, X.; Xu, G.; Xing, S.; Chattopadhyay, A.; Jin, S.; Salama, G. Estradiol Up-Regulates L-Type Ca2+ Channels via Membrane-Bound Estrogen Receptor/Phosphoinositide-3-Kinase/Akt/cAMP Response Element-Binding Protein Signaling Pathway. Heart Rhythm 2018, 15, 741–749. [Google Scholar] [CrossRef]
- Kim, D.; Smith, T.W.; Marsh, J.D. Effect of Thyroid Hormone on Slow Calcium Channel Function in Cultured Chick Ventricular Cells. J. Clin. Investig. 1987, 80, 88–94. [Google Scholar] [CrossRef]
- Rubinstein, I.; Binah, O. Thyroid Hormone Modulates Membrane Currents in Guinea-Pig Ventricular Myocytes. Naunyn Schmiedebergs Arch. Pharmacol. 1989, 340, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Leem, C.; So, I.; Kim, E.; Hong, S.; Ho, W.; Sung, H.; Earm, Y.E. Effects of Thyroid Hormone on the Calcium Current and Isoprenaline-Induced Background Current in Rabbit Ventricular Myocytes. J. Mol. Cell Cardiol. 1994, 26, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Kosinski, C.; Gross, G.; Hanft, G. Effect of Hypo- and Hyperthyroidism on Binding of [3H]-Nitrendipine to Myocardial and Brain Membranes. Br. J. Clin. Pharmacol. 1990, 30 (Suppl. S1), 128S–130S. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, Z.; Wang, J.-W.; Yu, D.; Soon, J.L.; de Kleijn, D.P.V.; Foo, R.; Liao, P.; Colecraft, H.M.; Soong, T.W. Aberrant Splicing Promotes Proteasomal Degradation of L-Type CaV1.2 Calcium Channels by Competitive Binding for CaVβ Subunits in Cardiac Hypertrophy. Sci. Rep. 2016, 6, 35247. [Google Scholar] [CrossRef] [PubMed]
- Göttlicher, M.; Heck, S.; Herrlich, P. Transcriptional Cross-Talk, the Second Mode of Steroid Hormone Receptor Action. J. Mol. Med. 1998, 76, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Pertuz, M.; Sánchez-Pacheco, A.; Aranda, A. The Thyroid Hormone Receptor Antagonizes CREB-Mediated Transcription. EMBO J. 2003, 22, 3102–3112. [Google Scholar] [CrossRef]
- Chu, P.-J.; Larsen, J.K.; Chen, C.-C.; Best, P.M. Distribution and Relative Expression Levels of Calcium Channel Beta Subunits within the Chambers of the Rat Heart. J. Mol. Cell. Cardiol. 2004, 36, 423–434. [Google Scholar] [CrossRef]
- Wagner, E.J.; Tong, L.; Adelman, K. Integrator Is a Global Promoter-Proximal Termination Complex. Mol. Cell 2023, 83, 416–427. [Google Scholar] [CrossRef]
- Cheng, S.-Y.; Leonard, J.L.; Davis, P.J. Molecular Aspects of Thyroid Hormone Actions. Endocr. Rev. 2010, 31, 139–170. [Google Scholar] [CrossRef]
- Gavali, J.T.; Carrillo, E.D.; García, M.C.; Sánchez, J.A. The Mitochondrial K-ATP Channel Opener Diazoxide Upregulates STIM1 and Orai1 via ROS and the MAPK Pathway in Adult Rat Cardiomyocytes. Cell Biosci. 2020, 10, 96. [Google Scholar] [CrossRef]
- Castellano, A.; Wei, X.; Birnbaumer, L.; Perez-Reyes, E. Cloning and Expression of a Neuronal Calcium Channel Beta Subunit. J. Biol. Chem. 1993, 268, 12359–12366. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.J. ImageJ for Microscopy. Biotechniques 2007, 43, 25–30. [Google Scholar] [CrossRef]
- Kelley, J.B.; Paschal, B.M. Fluorescence-Based Quantification of Nucleocytoplasmic Transport. Methods 2019, 157, 106–114. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrillo, E.D.; Alvarado, J.A.; Hernández, A.; Lezama, I.; García, M.C.; Sánchez, J.A. Thyroid Hormone Upregulates Cav1.2 Channels in Cardiac Cells via the Downregulation of the Channels’ β4 Subunit. Int. J. Mol. Sci. 2024, 25, 10798. https://doi.org/10.3390/ijms251910798
Carrillo ED, Alvarado JA, Hernández A, Lezama I, García MC, Sánchez JA. Thyroid Hormone Upregulates Cav1.2 Channels in Cardiac Cells via the Downregulation of the Channels’ β4 Subunit. International Journal of Molecular Sciences. 2024; 25(19):10798. https://doi.org/10.3390/ijms251910798
Chicago/Turabian StyleCarrillo, Elba D., Juan A. Alvarado, Ascención Hernández, Ivonne Lezama, María C. García, and Jorge A. Sánchez. 2024. "Thyroid Hormone Upregulates Cav1.2 Channels in Cardiac Cells via the Downregulation of the Channels’ β4 Subunit" International Journal of Molecular Sciences 25, no. 19: 10798. https://doi.org/10.3390/ijms251910798
APA StyleCarrillo, E. D., Alvarado, J. A., Hernández, A., Lezama, I., García, M. C., & Sánchez, J. A. (2024). Thyroid Hormone Upregulates Cav1.2 Channels in Cardiac Cells via the Downregulation of the Channels’ β4 Subunit. International Journal of Molecular Sciences, 25(19), 10798. https://doi.org/10.3390/ijms251910798







