Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure
Abstract
1. Introduction
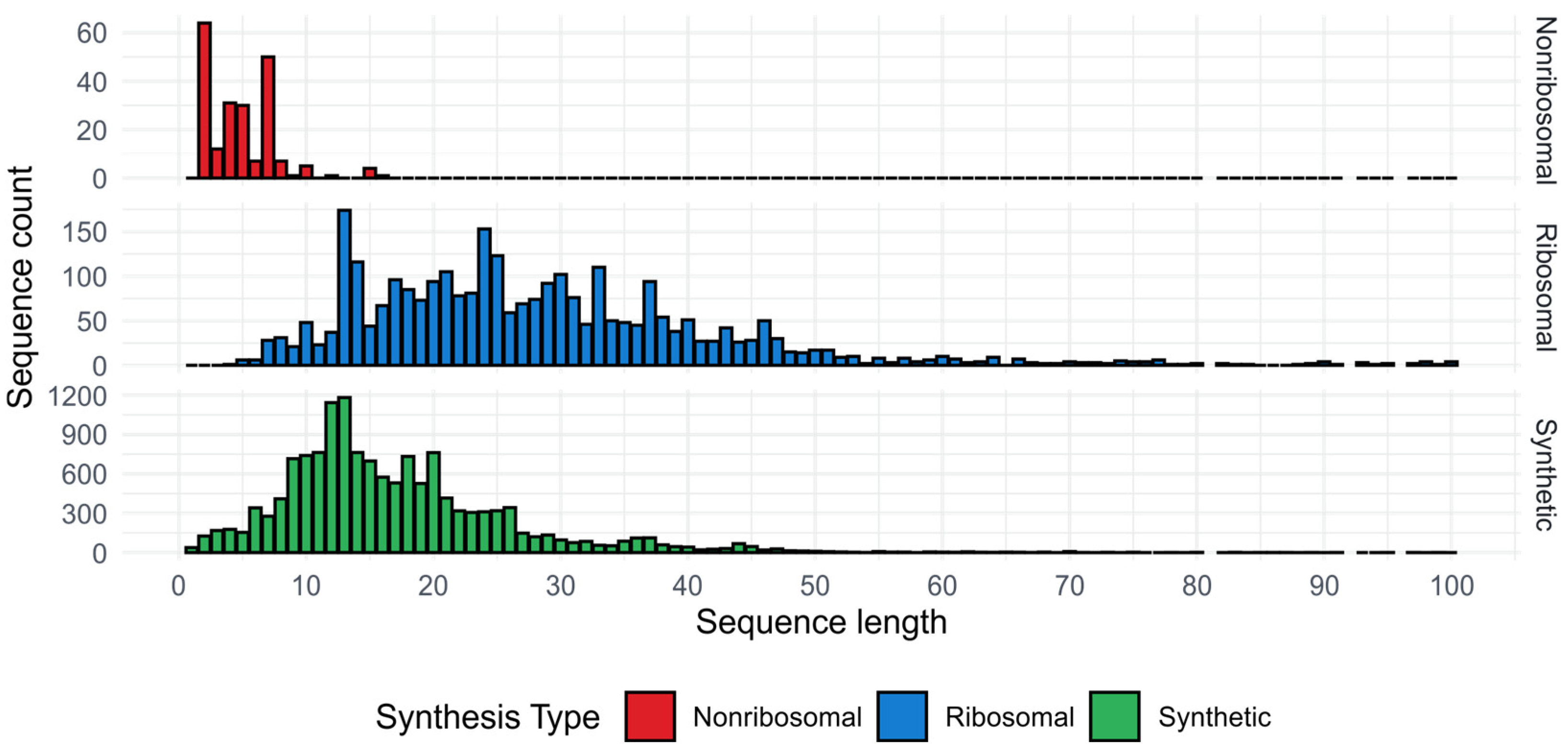
2. Optimization of AMPs
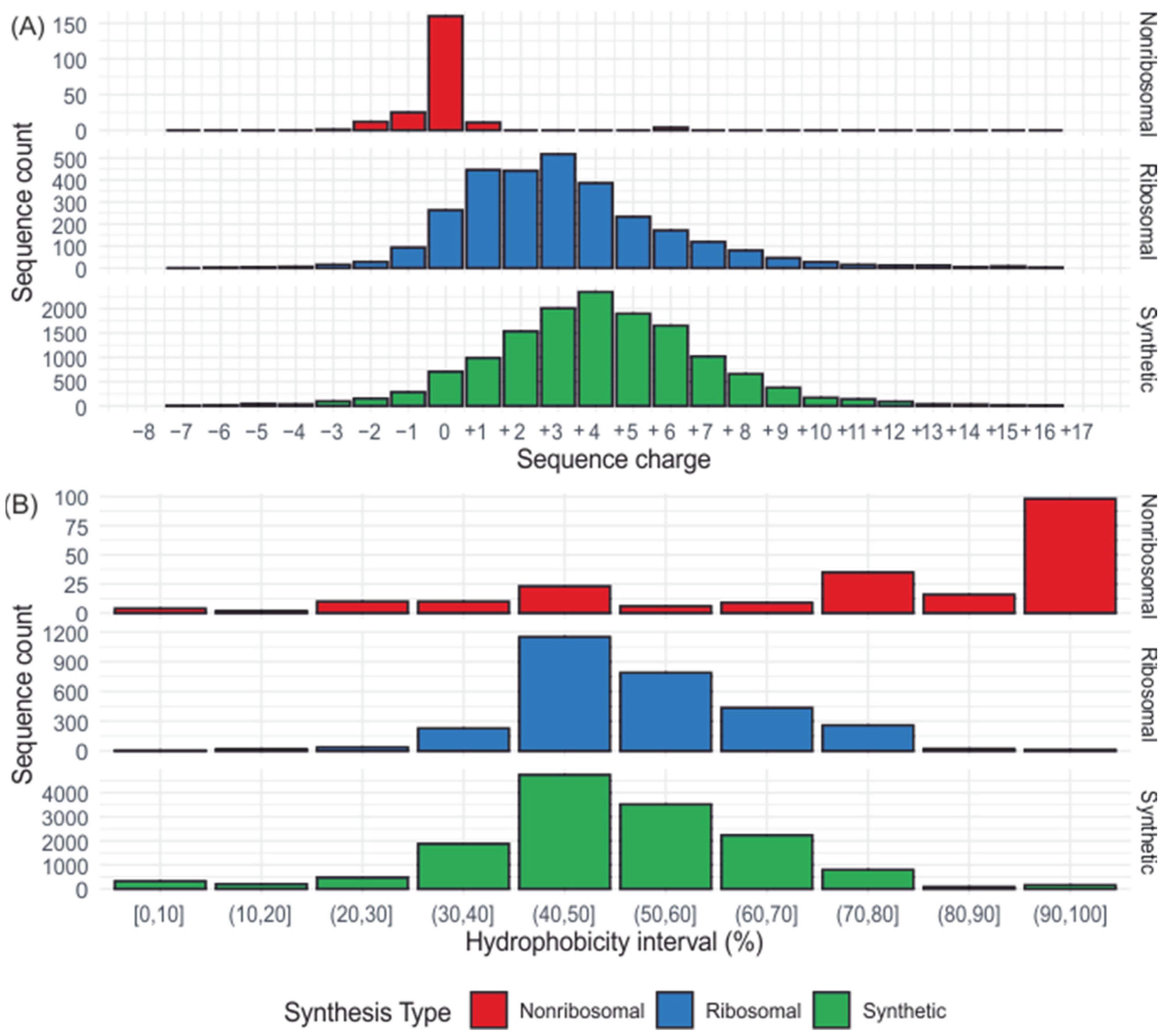
2.1. Charge
2.2. Hydrophobicity
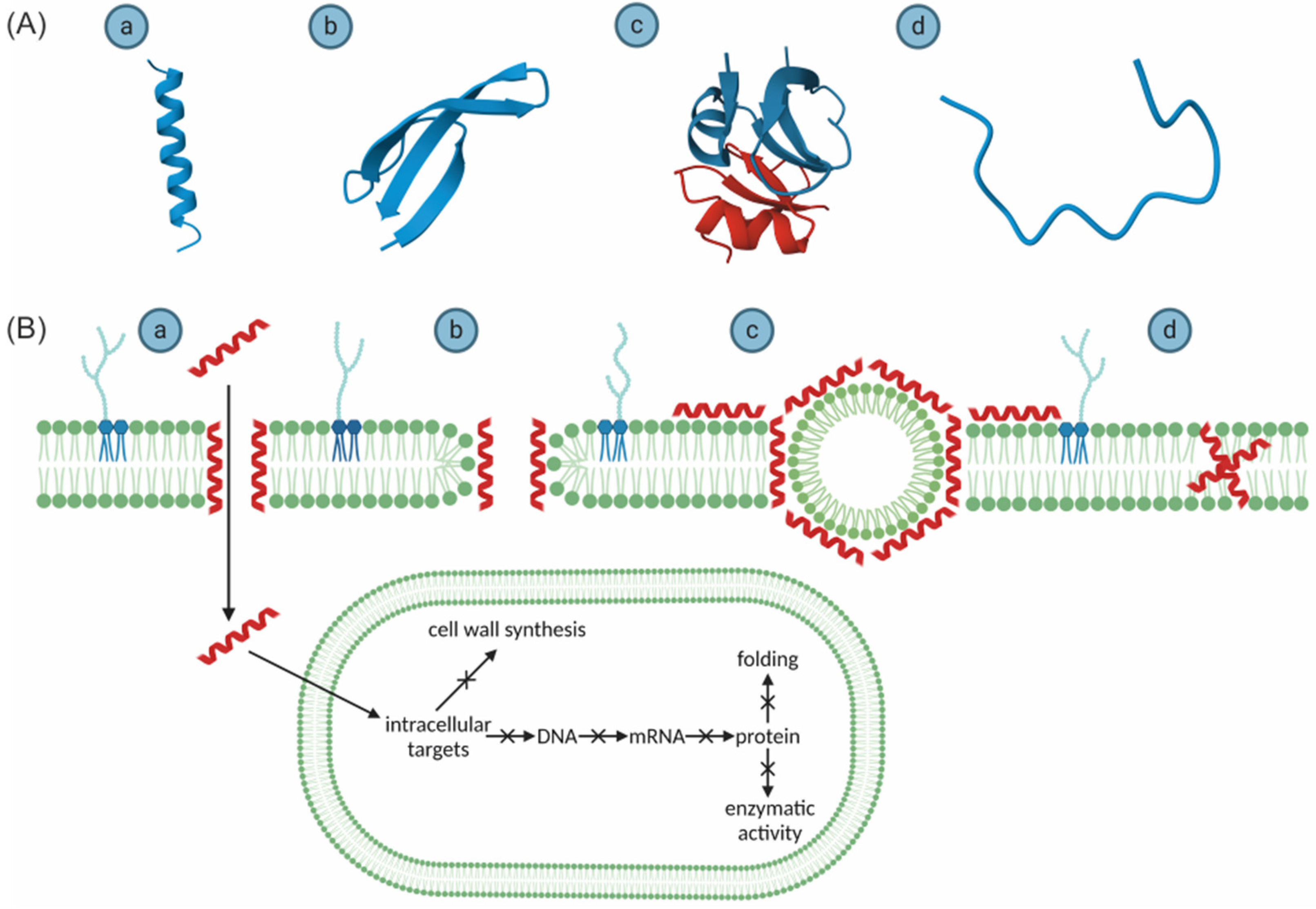
2.3. Structure
2.4. De Novo Design
3. Application of AMPs
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mendelsohn, J.A. “Like All That Lives”: Biology, Medicine and Bacteria in the Age of Pasteur and Koch. Hist. Philos. Life Sci. 2002, 24, 3–36. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic Discovery: History, Methods and Perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.d.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic Resistance in Microbes: History, Mechanisms, Therapeutic Strategies and Future Prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- MacGowan, A.; Macnaughton, E. Antibiotic Resistance. Medicine 2017, 45, 622–628. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic Resistance—The Need for Global Solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. BioNanoScience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Kwon, J.H.; Powderly, W.G. The Post-Antibiotic Era Is Here. Science 2021, 373, 471. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Lucero-Prisno, D.E., III; Shomuyiwa, D.O.; Kouwenhoven, M.B.N.; Dorji, T.; Odey, G.O.; Miranda, A.V.; Ogunkola, I.O.; Adebisi, Y.A.; Huang, J.; Xu, L.; et al. Top 10 Public Health Challenges to Track in 2023: Shifting Focus beyond a Global Pandemic. Public Health Chall. 2023, 2, e86. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 18 January 2024).
- Chen, C.-Y.; Yang, K.-Y.; Peng, C.-K.; Sheu, C.-C.; Chan, M.-C.; Feng, J.-Y.; Wang, S.-H.; Chen, C.-M.; Zheng, Z.-R.; Liang, S.-J.; et al. Clinical Outcome of Nosocomial Pneumonia Caused by Carbapenem-Resistant Gram-Negative Bacteria in Critically Ill Patients: A Multicenter Retrospective Observational Study. Sci. Rep. 2022, 12, 7501. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Hooper, D.C. Hospital-Acquired Infections Due to Gram-Negative Bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef]
- Temkin, E.; Carmeli, Y. Zero or More: Methodological Challenges of Counting and Estimating Deaths Related to Antibiotic-Resistant Infections. Clin. Infect. Dis. 2019, 69, 2029–2034. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016. [Google Scholar]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States, 2019; Centres for Disease Control and Prevention, US Department of Health and Human Services: Washington, DC, USA, 2019. [Google Scholar]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.W.; Harper, D.; et al. Alternatives to Antibiotics-a Pipeline Portfolio Review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Ryu, M.; Park, J.; Yeom, J.-H.; Joo, M.; Lee, K. Rediscovery of Antimicrobial Peptides as Therapeutic Agents. J. Microbiol. 2021, 59, 113–123. [Google Scholar] [CrossRef]
- Hassan, M.; Flanagan, T.W.; Kharouf, N.; Bertsch, C.; Mancino, D.; Haikel, Y. Antimicrobial Proteins: Structure, Molecular Action, and Therapeutic Potential. Pharmaceutics 2023, 15, 72. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human Antimicrobial Peptides as Therapeutics for Viral Infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef]
- Maróti, G.; Kereszt, A.; Kondorosi, É.; Mergaert, P. Natural Roles of Antimicrobial Peptides in Microbes, Plants and Animals. Res. Microbiol. 2011, 162, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The Immunology of Host Defence Peptides: Beyond Antimicrobial Activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 235805. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Silva, O.N.; Lu, T.K.; Franco, O.L. Antimicrobial Peptides: Role in Human Disease and Potential as Immunotherapies. Pharmacol. Ther. 2017, 178, 132–140. [Google Scholar] [CrossRef]
- Raffatellu, M. Learning from Bacterial Competition in the Host to Develop Antimicrobials. Nat. Med. 2018, 24, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Suneja, G.; Nain, S.; Sharma, R. Microbiome: A Source of Novel Bioactive Compounds and Antimicrobial Peptides. In Microbial Diversity in Ecosystem Sustainability and Biotechnological Applications: Volume 1. Microbial Diversity in Normal & Extreme Environments; Satyanarayana, T., Johri, B.N., Das, S.K., Eds.; Springer: Singapore, 2019; pp. 615–630. ISBN 9789811383151. [Google Scholar]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The Interaction of Antimicrobial Peptides with Membranes. Adv. Colloid. Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Ahmed, T.A.E.; Hammami, R. Recent Insights into Structure–Function Relationships of Antimicrobial Peptides. J. Food Biochem. 2019, 43, e12546. [Google Scholar] [CrossRef]
- Abraham, D.J.; Leo, A.J. Extension of the Fragment Method to Calculate Amino Acid Zwitterion and Side Chain Partition Coefficients. Proteins Struct. Funct. Bioinform. 1987, 2, 130–152. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and Consequences of Bacterial Resistance to Antimicrobial Peptides. Drug Resist. Updates 2016, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B.; et al. Antibiotic-Resistant Bacteria Show Widespread Collateral Sensitivity to Antimicrobial Peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K.; et al. Integrated Evolutionary Analysis Reveals Antimicrobial Peptides with Limited Resistance. Nat. Commun. 2019, 10, 4538. [Google Scholar] [CrossRef]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef]
- Costa, F.; Teixeira, C.; Gomes, P.; Martins, M.C.L. Clinical Application of AMPs. In Antimicrobial Peptides: Basics for Clinical Application; Matsuzaki, K., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; pp. 281–298. ISBN 9789811335884. [Google Scholar]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial Peptides as Therapeutic Agents: Opportunities and Challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial Peptides: Promising Alternatives in the Post Feeding Antibiotic Era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, N.; Wang, X.; Hao, Y.; Mao, R.; Li, Z.; Wang, Z.; Teng, D.; Wang, J. An Enhanced Variant Designed from DLP4 Cationic Peptide Against Staphylococcus Aureus CVCC 546. Front. Microbiol. 2020, 11, 1057. [Google Scholar] [CrossRef]
- Hwang, P.M.; Vogel, H.J. Structure–Function Relationships of Antimicrobial Peptides. Biochem. Cell Biol. 1998, 76, 235–246. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The structure-mechanism relationship and mode of actions of antimicrobial peptides: A review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]
- Tan, P.; Fu, H.; Ma, X. Design, Optimization, and Nanotechnology of Antimicrobial Peptides: From Exploration to Applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Chilimoniuk, J.; Rödiger, S.; Gagat, P. Proteomic Screening for Prediction and Design of Antimicrobial Peptides with AmpGram. Int. J. Mol. Sci. 2020, 21, 4310. [Google Scholar] [CrossRef]
- Sidorczuk, K.; Gagat, P.; Pietluch, F.; Kała, J.; Rafacz, D.; Bąkała, L.; Słowik, J.; Kolenda, R.; Rödiger, S.; Fingerhut, L.C. Benchmarks in Antimicrobial Peptide Prediction Are Biased Due to the Selection of Negative Data. Brief. Bioinform. 2022, 23, bbac343. [Google Scholar] [CrossRef]
- Santos-Júnior, C.D.; Pan, S.; Zhao, X.-M.; Coelho, L.P. Macrel: Antimicrobial Peptide Screening in Genomes and Metagenomes. PeerJ 2020, 8, e10555. [Google Scholar] [CrossRef] [PubMed]
- Fingerhut, L.C.H.W.; Miller, D.J.; Strugnell, J.M.; Daly, N.L.; Cooke, I.R. Ampir: An R Package for Fast Genome-Wide Prediction of Antimicrobial Peptides. Bioinformatics 2021, 36, 5262–5263. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Deep Learning Improves Antimicrobial Peptide Recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef]
- Lawrence, T.J.; Carper, D.L.; Spangler, M.K.; Carrell, A.A.; Rush, T.A.; Minter, S.J.; Weston, D.J.; Labbé, J.L. amPEPpy 1.0: A Portable and Accurate Antimicrobial Peptide Prediction Tool. Bioinformatics 2021, 37, 2058–2060. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific α-Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E.W.; Zhang, G.; Zhang, Y. I-TASSER-MTD: A Deep-Learning-Based Platform for Multi-Domain Protein Structure and Function Prediction. Nat. Protoc. 2022, 17, 2326–2353. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de Novo Structure Prediction for Linear Peptides in Solution and in Complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein Secondary Structure Prediction Based on Position-Specific Scoring Matrices 1 1Edited by G. Von Heijne. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ramos-Llorens, M.; Bello-Madruga, R.; Valle, J.; Andreu, D.; Torrent, M. PyAMPA: A High-Throughput Prediction and Optimization Tool for Antimicrobial Peptides. mSystems 2024, 9, e01358-23. [Google Scholar] [CrossRef]
- Bessalle, R.; Haas, H.; Goria, A.; Shalit, I.; Fridkin, M. Augmentation of the Antibacterial Activity of Magainin by Positive-Charge Chain Extension. Antimicrob. Agents Chemother. 1992, 36, 313–317. [Google Scholar] [CrossRef]
- Higgs, R.; Lynn, D.J.; Cahalane, S.; Alaña, I.; Hewage, C.M.; James, T.; Lloyd, A.T.; O’Farrelly, C. Modification of Chicken Avian β-Defensin-8 at Positively Selected Amino Acid Sites Enhances Specific Antimicrobial Activity. Immunogenetics 2007, 59, 573–580. [Google Scholar] [CrossRef]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Effects of Net Charge and the Number of Positively Charged Residues on the Biological Activity of Amphipathic A-helical Cationic Antimicrobial Peptides. Biopolymers 2008, 90, 369–383. [Google Scholar] [CrossRef]
- Yount, N.Y.; Bayer, A.S.; Xiong, Y.Q.; Yeaman, M.R. Advances in Antimicrobial Peptide Immunobiology. Biopolymers 2006, 84, 435–458. [Google Scholar] [CrossRef]
- Frederiksen, N.; Hansen, P.R.; Zabicka, D.; Tomczak, M.; Urbas, M.; Domraceva, I.; Björkling, F.; Franzyk, H. Alternating Cationic-Hydrophobic Peptide/Peptoid Hybrids: Influence of Hydrophobicity on Antibacterial Activity and Cell Selectivity. ChemMedChem 2020, 15, 2544–2561. [Google Scholar] [CrossRef]
- Shang, D.; Zhang, Q.; Dong, W.; Liang, H.; Bi, X. The Effects of LPS on the Activity of Trp-Containing Antimicrobial Peptides against Gram-Negative Bacteria and Endotoxin Neutralization. Acta Biomater. 2016, 33, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Yount, N.; Yeaman, M. Immunocontinuum: Perspectives in Antimicrobial Peptide Mechanisms of Action and Resistance. Protein Pept. Lett. 2005, 12, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Dennison, S.; Harris, F.; Phoenix, D. Are Oblique Orientated α-Helices Used by Antimicrobial Peptides for Membrane Invasion? Protein Pept. Lett. 2005, 12, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Glukhov, E.; Stark, M.; Burrows, L.L.; Deber, C.M. Basis for Selectivity of Cationic Antimicrobial Peptides for Bacterial Versus Mammalian Membranes. J. Biol. Chem. 2005, 280, 33960–33967. [Google Scholar] [CrossRef]
- Haney, E.F.; Lau, F.; Vogel, H.J. Solution Structures and Model Membrane Interactions of Lactoferrampin, an Antimicrobial Peptide Derived from Bovine Lactoferrin. Biochim. Biophys. Acta (BBA) Biomembr. 2007, 1768, 2355–2364. [Google Scholar] [CrossRef]
- Sparks, K.A.; Gleason, N.J.; Gist, R.; Langston, R.; Greathouse, D.V.; Koeppe, R.E.I. Comparisons of Interfacial Phe, Tyr, and Trp Residues as Determinants of Orientation and Dynamics for GWALP Transmembrane Peptides. Biochemistry 2014, 53, 3637–3645. [Google Scholar] [CrossRef]
- Shahmiri, M.; Cornell, B.; Mechler, A. Phenylalanine Residues Act as Membrane Anchors in the Antimicrobial Action of Aurein 1.2. Biointerphases 2017, 12, 05G605. [Google Scholar] [CrossRef]
- Lander, A.J.; Mercado, L.D.; Li, X.; Taily, I.M.; Findlay, B.L.; Jin, Y.; Luk, L.Y.P. Roles of Inter- and Intramolecular Tryptophan Interactions in Membrane-Active Proteins Revealed by Racemic Protein Crystallography. Commun. Chem. 2023, 6, 154. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Pasupuleti, M.; Mörgelin, M.; Davoudi, M.; Alenfall, J.; Chalupka, A.; Malmsten, M. Boosting Antimicrobial Peptides by Hydrophobic Oligopeptide End Tags. J. Biol. Chem. 2009, 284, 17584–17594. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Ringstad, L.; Kasetty, G.; Mizuno, H.; Rutland, M.W.; Malmsten, M. Membrane Selectivity by W-Tagging of Antimicrobial Peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 1081–1091. [Google Scholar] [CrossRef]
- Malmsten, M.; Kasetty, G.; Pasupuleti, M.; Alenfall, J.; Schmidtchen, A. Highly Selective End-Tagged Antimicrobial Peptides Derived from PRELP. PLoS ONE 2011, 6, e16400. [Google Scholar] [CrossRef] [PubMed]
- Molhoek, E.M.; Van Dijk, A.; Veldhuizen, E.J.A.; Dijk-Knijnenburg, H.; Mars-Groenendijk, R.H.; Boele, L.C.L.; Kaman-van Zanten, W.E.; Haagsman, H.P.; Bikker, F.J. Chicken Cathelicidin-2-Derived Peptides with Enhanced Immunomodulatory and Antibacterial Activities against Biological Warfare Agents. Int. J. Antimicrob. Agents 2010, 36, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, A.; Molhoek, E.M.; Veldhuizen, E.J.A.; Bokhoven, J.L.M.T.; Wagendorp, E.; Bikker, F.; Haagsman, H.P. Identification of Chicken Cathelicidin-2 Core Elements Involved in Antibacterial and Immunomodulatory Activities. Mol. Immunol. 2009, 46, 2465–2473. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shen, T.; Liu, Y.; Zhou, J.; Shi, S.; Wang, Y.; Zhao, Z.; Yan, Z.; Liao, C.; Wang, C. Enhancing the Antibacterial Activity of Antimicrobial Peptide PMAP-37(F34-R) by Cholesterol Modification. BMC Vet. Res. 2020, 16, 419. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, L.; Liu, Y.; Shen, T.; Zhang, C.; Liu, Z.; Feng, X.; Wang, C. Antimicrobial Peptide PMAP-37 Analogs: Increasing the Positive Charge to Enhance the Antibacterial Activity of PMAP-37. J. Pept. Sci. 2019, 25, e3220. [Google Scholar] [CrossRef]
- Liu, T.; Zhu, N.; Zhong, C.; Zhu, Y.; Gou, S.; Chang, L.; Bao, H.; Liu, H.; Zhang, Y.; Ni, J. Effect of N-Methylated and Fatty Acid Conjugation on Analogs of Antimicrobial Peptide Anoplin. Eur. J. Pharm. Sci. 2020, 152, 105453. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Zhu, N.; Zhu, Y.; Liu, T.; Gou, S.; Xie, J.; Yao, J.; Ni, J. Antimicrobial Peptides Conjugated with Fatty Acids on the Side Chain of D-Amino Acid Promises Antimicrobial Potency against Multidrug-Resistant Bacteria. Eur. J. Pharm. Sci. 2020, 141, 105123. [Google Scholar] [CrossRef]
- Liu, H.; Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Ma, X.; Wang, X.; Wang, J. Fatty Acid Modified-Antimicrobial Peptide Analogues with Potent Antimicrobial Activity and Topical Therapeutic Efficacy against Staphylococcus Hyicus. Appl. Microbiol. Biotechnol. 2021, 105, 5845–5859. [Google Scholar] [CrossRef]
- Chatterjee, J.; Rechenmacher, F.; Kessler, H. N-Methylation of Peptides and Proteins: An Important Element for Modulating Biological Functions. Angew. Chem. Int. Ed. 2013, 52, 254–269. [Google Scholar] [CrossRef]
- Smirnova, M.P.; Kolodkin, N.I.; Kolobov, A.A.; Afonin, V.G.; Afonina, I.V.; Stefanenko, L.I.; Shpen’, V.M.; Shamova, O.V.; Kolobov, A.A. Indolicidin Analogs with Broad-Spectrum Antimicrobial Activity and Low Hemolytic Activity. Peptides 2020, 132, 170356. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T. Structural Features of Helical Antimicrobial Peptides: Their Potential to Modulate Activity on Model Membranes and Biological Cells. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1462, 71–87. [Google Scholar] [CrossRef]
- Dempsey, C.E. The Actions of Melittin on Membranes. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 1990, 1031, 143–161. [Google Scholar] [CrossRef]
- Asthana, N.; Yadav, S.P.; Ghosh, J.K. Dissection of Antibacterial and Toxic Activity of Melittin. J. Biol. Chem. 2004, 279, 55042–55050. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational Design of α-Helical Antimicrobial Peptides with Enhanced Activities and Specificity/Therapeutic Index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [PubMed]
- Hawrani, A.; Howe, R.A.; Walsh, T.R.; Dempsey, C.E. Origin of Low Mammalian Cell Toxicity in a Class of Highly Active Antimicrobial Amphipathic Helical Peptides. J. Biol. Chem. 2008, 283, 18636–18645. [Google Scholar] [CrossRef]
- Zhang, S.-K.; Song, J.; Gong, F.; Li, S.-B.; Chang, H.-Y.; Xie, H.-M.; Gao, H.-W.; Tan, Y.-X.; Ji, S.-P. Design of an α-Helical Antimicrobial Peptide with Improved Cell-Selective and Potent Anti-Biofilm Activity. Sci. Rep. 2016, 6, 27394. [Google Scholar] [CrossRef]
- Srivastava, S.; Kumar, A.; Tripathi, A.K.; Tandon, A.; Ghosh, J.K. Modulation of Anti-Endotoxin Property of Temporin L by Minor Amino Acid Substitution in Identified Phenylalanine Zipper Sequence. Biochem. J. 2016, 473, 4045–4062. [Google Scholar] [CrossRef]
- Song, J.; Wang, J.; Zhan, N.; Sun, T.; Yu, W.; Zhang, L.; Shan, A.; Zhang, A. Therapeutic Potential of Trp-Rich Engineered Amphiphiles by Single Hydrophobic Amino Acid End-Tagging. ACS Appl. Mater. Interfaces 2019, 11, 43820–43834. [Google Scholar] [CrossRef]
- Li, Y.; Bionda, N.; Yongye, A.; Geer, P.; Stawikowski, M.; Cudic, P.; Martinez, K.; Houghten, R.A. Dissociation of Antimicrobial and Hemolytic Activities of Gramicidin S through N–Methylation Modification. ChemMedChem 2013, 8, 1865–1872. [Google Scholar] [CrossRef]
- He, R.; Di Bonaventura, I.; Visini, R.; Gan, B.-H.; Fu, Y.; Probst, D.; Lüscher, A.; Köhler, T.; Van Delden, C.; Stocker, A.; et al. Design, Crystal Structure and Atomic Force Microscopy Study of Thioether Ligated D,L-Cyclic Antimicrobial Peptides against Multidrug Resistant Pseudomonas Aeruginosa. Chem. Sci. 2017, 8, 7464–7475. [Google Scholar] [CrossRef] [PubMed]
- Mani, R.; Tang, M.; Wu, X.; Buffy, J.J.; Waring, A.J.; Sherman, M.A.; Hong, M. Membrane-Bound Dimer Structure of a β-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR. Biochemistry 2006, 45, 8341–8349. [Google Scholar] [CrossRef]
- Harwig, S.S.L.; Waring, A.; Yang, H.J.; Cho, Y.; Tan, L.; Lehrer, R.I. Intramolecular Disulfide Bonds Enhance the Antimicrobial and Lytic Activities of Protegrins at Physiological Sodium Chloride Concentrations. Eur. J. Biochem. 1996, 240, 352–357. [Google Scholar] [CrossRef]
- Kokryakov, V.N.; Harwig, S.S.L.; Panyutich, E.A.; Shevchenko, A.A.; Aleshina, G.M.; Shamova, O.V.; Korneva, H.A.; Lehrer, R.I. Protegrins: Leukocyte Antimicrobial Peptides That Combine Features of Corticostatic Defensins and Tachyplesins. FEBS Lett. 1993, 327, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wu, C.; Yang, J. Membranolytic Selectivity of Cystine-stabilized Cyclic Protegrins. Eur. J. Biochem. 2000, 267, 3289–3300. [Google Scholar] [CrossRef]
- Wiradharma, N.; Khoe, U.; Hauser, C.A.E.; Seow, S.V.; Zhang, S.; Yang, Y.-Y. Synthetic Cationic Amphiphilic α-Helical Peptides as Antimicrobial Agents. Biomaterials 2011, 32, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, C.; Zhang, S.; Zhao, X.; Xu, H.; Zhao, X.; Lu, J.R. Designed Antimicrobial and Antitumor Peptides with High Selectivity. Biomacromolecules 2011, 12, 3839–3843. [Google Scholar] [CrossRef]
- Chen, C.; Hu, J.; Zeng, P.; Chen, Y.; Xu, H.; Lu, J.R. High Cell Selectivity and Low-Level Antibacterial Resistance of Designed Amphiphilic Peptide G(IIKK)3I-NH2. ACS Appl. Mater. Interfaces 2014, 6, 16529–16536. [Google Scholar] [CrossRef]
- Ahmad, A.; Asthana, N.; Azmi, S.; Srivastava, R.M.; Pandey, B.K.; Yadav, V.; Ghosh, J.K. Structure–Function Study of Cathelicidin-Derived Bovine Antimicrobial Peptide BMAP-28: Design of Its Cell-Selective Analogs by Amino Acid Substitutions in the Heptad Repeat Sequences. Biochim. Biophys. Acta (BBA) Biomembr. 2009, 1788, 2411–2420. [Google Scholar] [CrossRef]
- Ahmad, A.; Azmi, S.; Srivastava, R.M.; Srivastava, S.; Pandey, B.K.; Saxena, R.; Bajpai, V.K.; Ghosh, J.K. Design of Nontoxic Analogues of Cathelicidin-Derived Bovine Antimicrobial Peptide BMAP-27: The Role of Leucine as Well as Phenylalanine Zipper Sequences in Determining Its Toxicity. Biochemistry 2009, 48, 10905–10917. [Google Scholar] [CrossRef]
- Kumar, A.; Tripathi, A.K.; Kathuria, M.; Shree, S.; Tripathi, J.K.; Purshottam, R.K.; Ramachandran, R.; Mitra, K.; Ghosh, J.K. Single Amino Acid Substitutions at Specific Positions of the Heptad Repeat Sequence of Piscidin-1 Yielded Novel Analogs That Show Low Cytotoxicity and In Vitro and In Vivo Antiendotoxin Activity. Antimicrob. Agents Chemother. 2016, 60, 3687–3699. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.; Parry, D.A.D. α-Helical Coiled Coils and Bundles: How to Design an A-helical Protein. Proteins 1990, 7, 1–15. [Google Scholar] [CrossRef]
- Jelesarov, I.; Dürr, E.; Thomas, R.M.; Bosshard, H.R. Salt Effects on Hydrophobic Interaction and Charge Screening in the Folding of a Negatively Charged Peptide to a Coiled Coil (Leucine Zipper). Biochemistry 1998, 37, 7539–7550. [Google Scholar] [CrossRef]
- Dou, X.; Zhu, X.; Wang, J.; Dong, N.; Shan, A. Novel Design of Heptad Amphiphiles To Enhance Cell Selectivity, Salt Resistance, Antibiofilm Properties and Their Membrane-Disruptive Mechanism. J. Med. Chem. 2017, 60, 2257–2270. [Google Scholar] [CrossRef] [PubMed]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of Amphipathicity and Hydrophobicity to the Antimicrobial Activity and Cytotoxicity of β-Hairpin Peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef]
- Shao, C.; Zhu, Y.; Jian, Q.; Lai, Z.; Tan, P.; Li, G.; Shan, A. Cross-Strand Interaction, Central Bending, and Sequence Pattern Act as Biomodulators of Simplified β-Hairpin Antimicrobial Amphiphiles. Small 2021, 17, 2003899. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Orozco, R.Q.; Rezende, S.B.; Rodrigues, G.; Oshiro, K.G.N.; Cândido, E.S.; Franco, O.L. Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates? Front. Microbiol. 2020, 10, 3097. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Galpert-Cañizares, D.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Pérez-Machado, G.; Teijeira, M.; Antunes, A. Emerging Computational Approaches for Antimicrobial Peptide Discovery. Antibiotics 2022, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Zhu, Y.; Huang, P.; Gao, Q.; Li, X.; Chen, Z.; Liu, Y.; Jiang, J.; Gao, Y.; et al. Machine Learning and Genetic Algorithm-Guided Directed Evolution for the Development of Antimicrobial Peptides. J. Adv. Res. 2024, in press. [CrossRef]
- Yoshida, M.; Hinkley, T.; Tsuda, S.; Abul-Haija, Y.M.; McBurney, R.T.; Kulikov, V.; Mathieson, J.S.; Reyes, S.G.; Castro, M.D.; Cronin, L. Using Evolutionary Algorithms and Machine Learning to Explore Sequence Space for the Discovery of Antimicrobial Peptides. Chem 2018, 4, 533–543. [Google Scholar] [CrossRef]
- Maccari, G.; Luca, M.D.; Nifosí, R.; Cardarelli, F.; Signore, G.; Boccardi, C.; Bifone, A. Antimicrobial Peptides Design by Evolutionary Multiobjective Optimization. PLoS Comput. Biol. 2013, 9, e1003212. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, F.; Leier, A.; Xiang, D.; Shen, H.-H.; Marquez Lago, T.T.; Li, J.; Yu, D.-J.; Song, J. Comprehensive Assessment of Machine Learning-Based Methods for Predicting Antimicrobial Peptides. Brief. Bioinform. 2021, 22, bbab083. [Google Scholar] [CrossRef] [PubMed]
- Suda, S.; Field, D.; Barron, N. Antimicrobial Peptide Production and Purification. In Protein Chromatography: Methods and Protocols; Walls, D., Loughran, S.T., Eds.; Springer: New York, NY, USA, 2017; pp. 401–410. ISBN 978-1-4939-6412-3. [Google Scholar]
- Gaglione, R.; Pane, K.; Dell’Olmo, E.; Cafaro, V.; Pizzo, E.; Olivieri, G.; Notomista, E.; Arciello, A. Cost-Effective Production of Recombinant Peptides in Escherichia coli. New Biotechnol. 2019, 51, 39–48. [Google Scholar] [CrossRef]
- Tiwari, P.; Srivastava, Y.; Sharma, A.; Vinayagam, R. Antimicrobial Peptides: The Production of Novel Peptide-Based Therapeutics in Plant Systems. Life 2023, 13, 1875. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Oren, Z.; Pag, U.; Sahl, H.-G.; Shai, Y. The Consequence of Sequence Alteration of an Amphipathic α-Helical Antimicrobial Peptide and Its Diastereomers. J. Biol. Chem. 2002, 277, 33913–33921. [Google Scholar] [CrossRef]
- Li, Y.; Liu, T.; Liu, Y.; Tan, Z.; Ju, Y.; Yang, Y.; Dong, W. Antimicrobial Activity, Membrane Interaction and Stability of the D-Amino Acid Substituted Analogs of Antimicrobial Peptide W3R6. J. Photochem. Photobiol. B Biol. 2019, 200, 111645. [Google Scholar] [CrossRef]
- Zai, Y.; Ying, Y.; Ye, Z.; Zhou, M.; Ma, C.; Shi, Z.; Chen, X.; Xi, X.; Chen, T.; Wang, L. Broad-Spectrum Antimicrobial Activity and Improved Stability of a D-Amino Acid Enantiomer of DMPC-10A, the Designed Derivative of Dermaseptin Truncates. Antibiotics 2020, 9, 627. [Google Scholar] [CrossRef]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical Modifications Designed to Improve Peptide Stability: Incorporation of Non-Natural Amino Acids, Pseudo-Peptide Bonds, and Cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef]
- Rozek, A.; Powers, J.-P.S.; Friedrich, C.L.; Hancock, R.E.W. Structure-Based Design of an Indolicidin Peptide Analogue with Increased Protease Stability. Biochemistry 2003, 42, 14130–14138. [Google Scholar] [CrossRef] [PubMed]
- Schafmeister, C.E.; Po, J.; Verdine, G.L. An All-Hydrocarbon Cross-Linking System for Enhancing the Helicity and Metabolic Stability of Peptides. J. Am. Chem. Soc. 2000, 122, 5891–5892. [Google Scholar] [CrossRef]
- Pham, T.K.; Kim, D.-H.; Lee, B.-J.; Kim, Y.-W. Truncated and Constrained Helical Analogs of Antimicrobial Esculentin-2EM. Bioorganic Med. Chem. Lett. 2013, 23, 6717–6720. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hu, Y.; Pu, Q.; He, T.; Zhang, Q.; Wu, W.; Xia, X.; Zhang, J. Novel Stapling by Lysine Tethering Provides Stable and Low Hemolytic Cationic Antimicrobial Peptides. J. Med. Chem. 2020, 63, 4081–4089. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Cho, J.H.; Kim, K.S.; Kim, Y.-B.; Kim, M.S.; Kim, S.C. Helix Stability Confers Salt Resistance upon Helical Antimicrobial Peptides *. J. Biol. Chem. 2004, 279, 13896–13901. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-Y.; Tu, C.-H.; Yip, B.-S.; Chen, H.-L.; Cheng, H.-T.; Huang, K.-C.; Lo, H.-J.; Cheng, J.-W. Easy Strategy To Increase Salt Resistance of Antimicrobial Peptides. Antimicrob. Agents Chemother. 2011, 55, 4918–4921. [Google Scholar] [CrossRef] [PubMed]
- Hayes, H.C.; Luk, L.Y.P.; Tsai, Y.-H. Approaches for Peptide and Protein Cyclisation. Org. Biomol. Chem. 2021, 19, 3983–4001. [Google Scholar] [CrossRef]
- Ye, Z.; Zhu, X.; Acosta, S.; Kumar, D.; Sang, T.; Aparicio, C. Self-Assembly Dynamics and Antimicrobial Activity of All L- and D-Amino Acid Enantiomers of a Designer Peptide. Nanoscale 2018, 11, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, X.; Wang, Q.; Meng, D. Unnatural Amino Acids: Promising Implications for the Development of New Antimicrobial Peptides. Crit. Rev. Microbiol. 2023, 49, 231–255. [Google Scholar] [CrossRef]
- Zhu, Y.; Hao, W.; Wang, X.; Ouyang, J.; Deng, X.; Yu, H.; Wang, Y. Antimicrobial Peptides, Conventional Antibiotics, and Their Synergistic Utility for the Treatment of Drug-Resistant Infections. Med. Res. Rev. 2022, 42, 1377–1422. [Google Scholar] [CrossRef]
- Kim, E.Y.; Rajasekaran, G.; Shin, S.Y. LL-37-Derived Short Antimicrobial Peptide KR-12-A5 and Its d-Amino Acid Substituted Analogs with Cell Selectivity, Anti-Biofilm Activity, Synergistic Effect with Conventional Antibiotics, and Anti-Inflammatory Activity. Eur. J. Med. Chem. 2017, 136, 428–441. [Google Scholar] [CrossRef]
- Reffuveille, F.; de la Fuente-Núñez, C.; Mansour, S.; Hancock, R.E.W. A Broad-Spectrum Antibiofilm Peptide Enhances Antibiotic Action against Bacterial Biofilms. Antimicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef]
- Song, M.; Liu, Y.; Huang, X.; Ding, S.; Wang, Y.; Shen, J.; Zhu, K. A Broad-Spectrum Antibiotic Adjuvant Reverses Multidrug-Resistant Gram-Negative Pathogens. Nat. Microbiol. 2020, 5, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial Peptides: Mechanism of Action, Activity and Clinical Potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.K.; Kumari, T.; Harioudh, M.K.; Yadav, P.K.; Kathuria, M.; Shukla, P.K.; Mitra, K.; Ghosh, J.K. Identification of GXXXXG Motif in Chrysophsin-1 and Its Implication in the Design of Analogs with Cell-Selective Antimicrobial and Anti-Endotoxin Activities. Sci. Rep. 2017, 7, 3384. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.K.; Kumari, T.; Tandon, A.; Sayeed, M.; Afshan, T.; Kathuria, M.; Shukla, P.K.; Mitra, K.; Ghosh, J.K. Selective Phenylalanine to Proline Substitution for Improved Antimicrobial and Anticancer Activities of Peptides Designed on Phenylalanine Heptad Repeat. Acta Biomater. 2017, 57, 170–186. [Google Scholar] [CrossRef]
- Porto, W.F.; Fensterseifer, I.C.M.; Ribeiro, S.M.; Franco, O.L. Joker: An Algorithm to Insert Patterns into Sequences for Designing Antimicrobial Peptides. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 2043–2052. [Google Scholar] [CrossRef]
- Porto, W.F.; Irazazabal, L.; Alves, E.S.F.; Ribeiro, S.M.; Matos, C.O.; Pires, Á.S.; Fensterseifer, I.C.M.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In Silico Optimization of a Guava Antimicrobial Peptide Enables Combinatorial Exploration for Peptide Design. Nat. Commun. 2018, 9, 1490. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gagat, P.; Ostrówka, M.; Duda-Madej, A.; Mackiewicz, P. Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure. Int. J. Mol. Sci. 2024, 25, 10821. https://doi.org/10.3390/ijms251910821
Gagat P, Ostrówka M, Duda-Madej A, Mackiewicz P. Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure. International Journal of Molecular Sciences. 2024; 25(19):10821. https://doi.org/10.3390/ijms251910821
Chicago/Turabian StyleGagat, Przemysław, Michał Ostrówka, Anna Duda-Madej, and Paweł Mackiewicz. 2024. "Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure" International Journal of Molecular Sciences 25, no. 19: 10821. https://doi.org/10.3390/ijms251910821
APA StyleGagat, P., Ostrówka, M., Duda-Madej, A., & Mackiewicz, P. (2024). Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure. International Journal of Molecular Sciences, 25(19), 10821. https://doi.org/10.3390/ijms251910821





