Leonurine Inhibits Hepatic Lipid Synthesis to Ameliorate NAFLD via the ADRA1a/AMPK/SCD1 Axis
Abstract
1. Introduction
2. Results
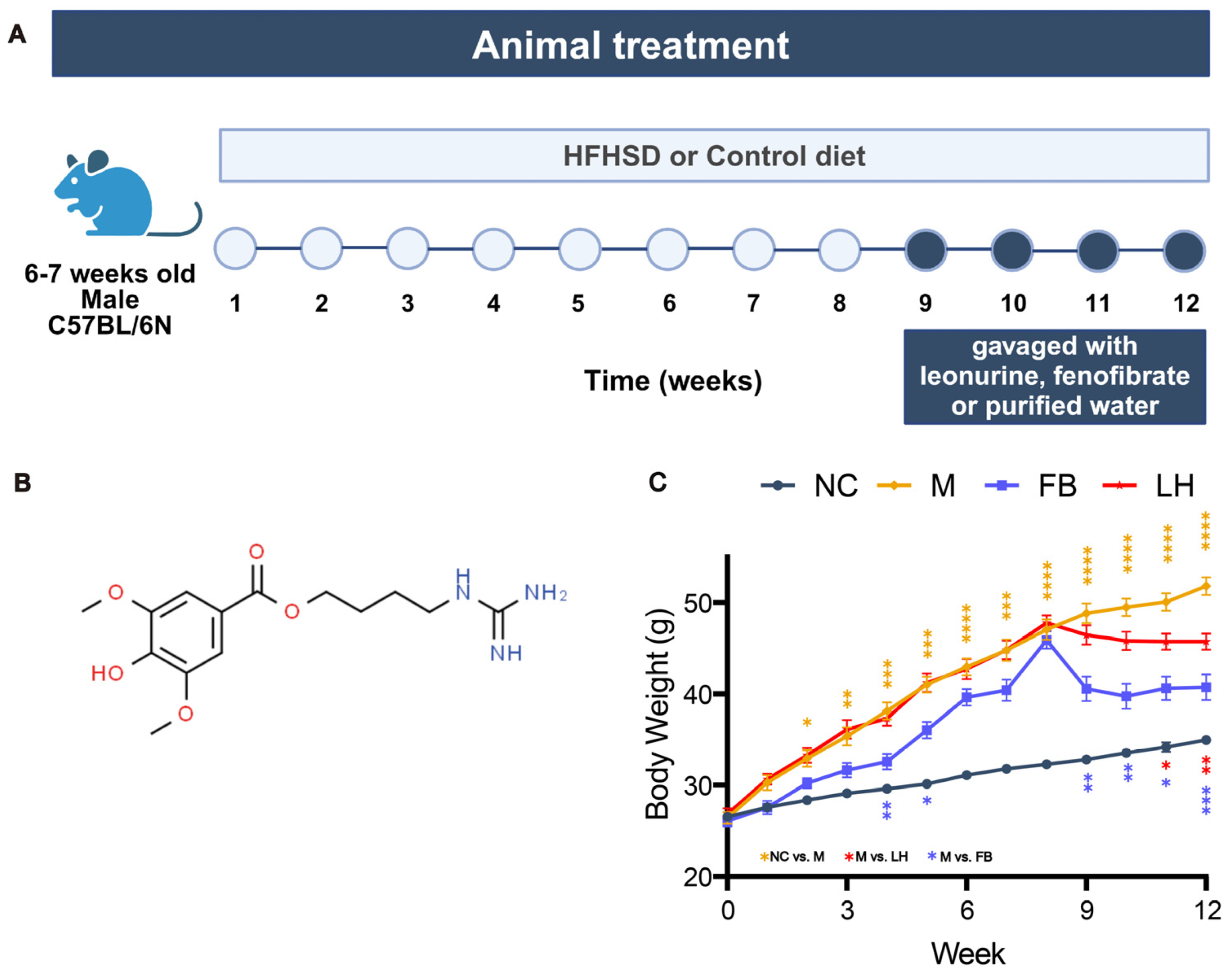
2.1. Four Weeks of Leonurine Treatment Resulted in Significant Weight Loss in HFHSD-Induced Mice
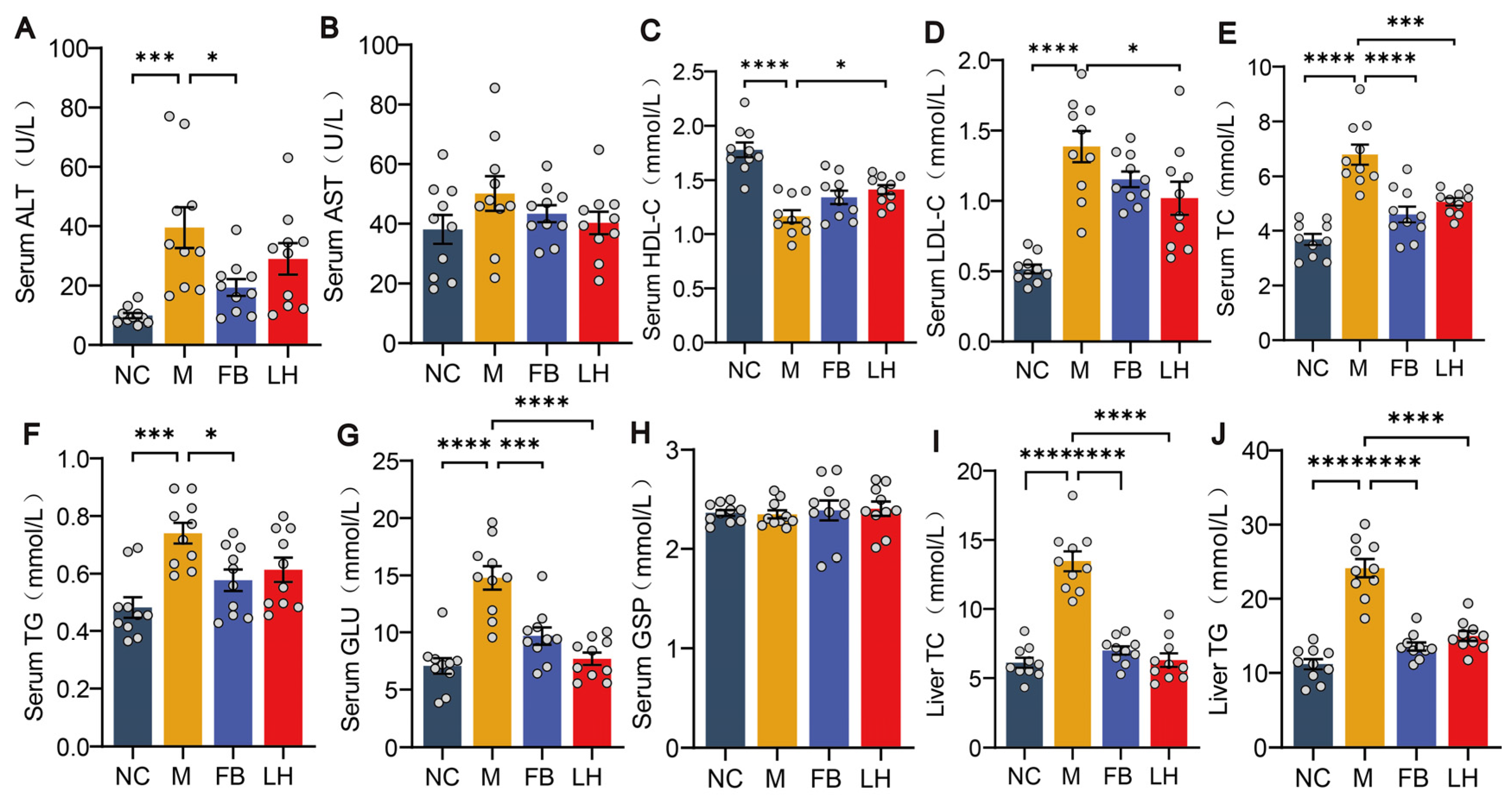
2.2. Leonurine Improved Serum and Hepatic Biochemical Parameters in NAFLD Mice
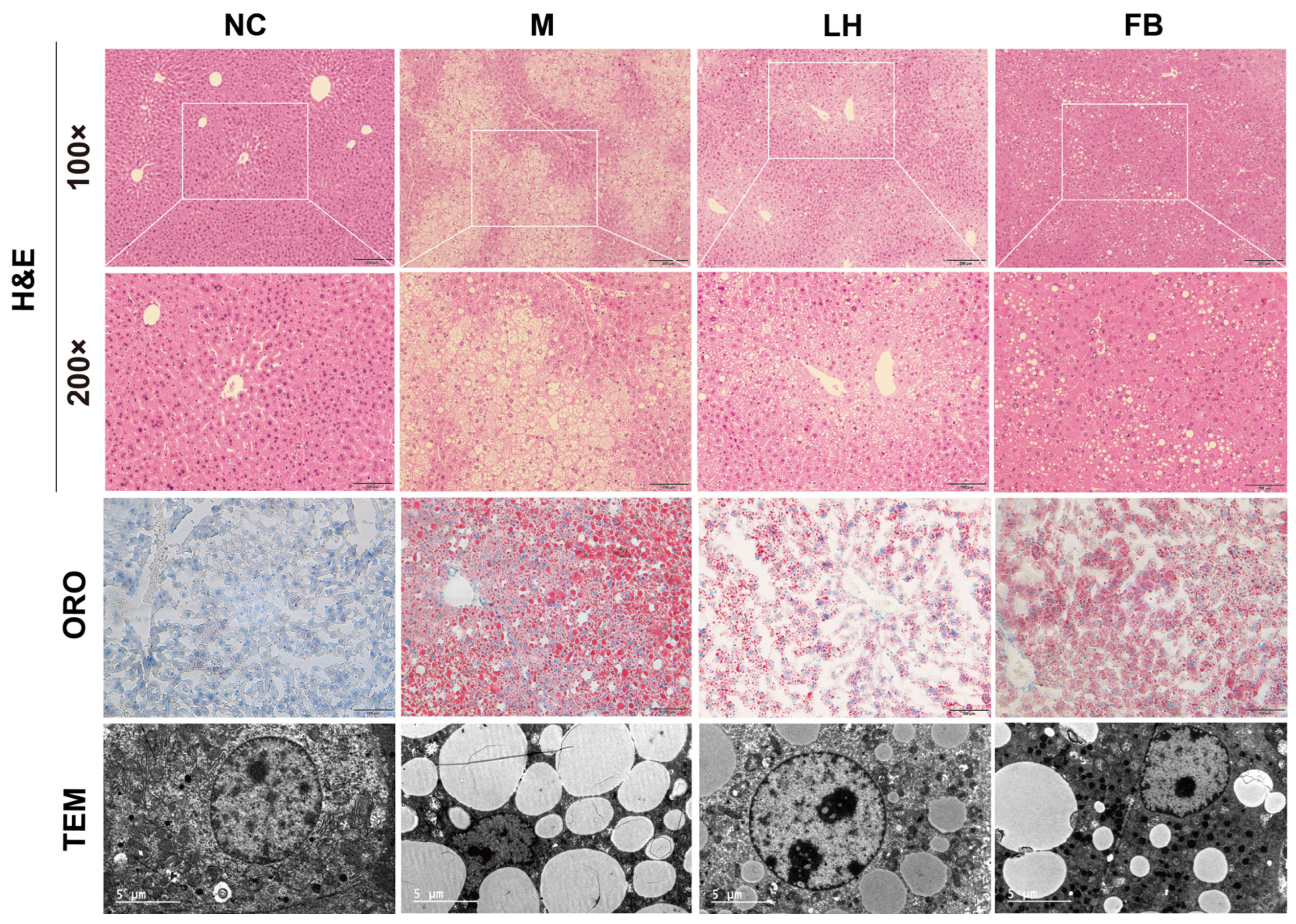
2.3. Leonurine Reversed Hepatic Histopathologic Changes in Mice with NAFLD
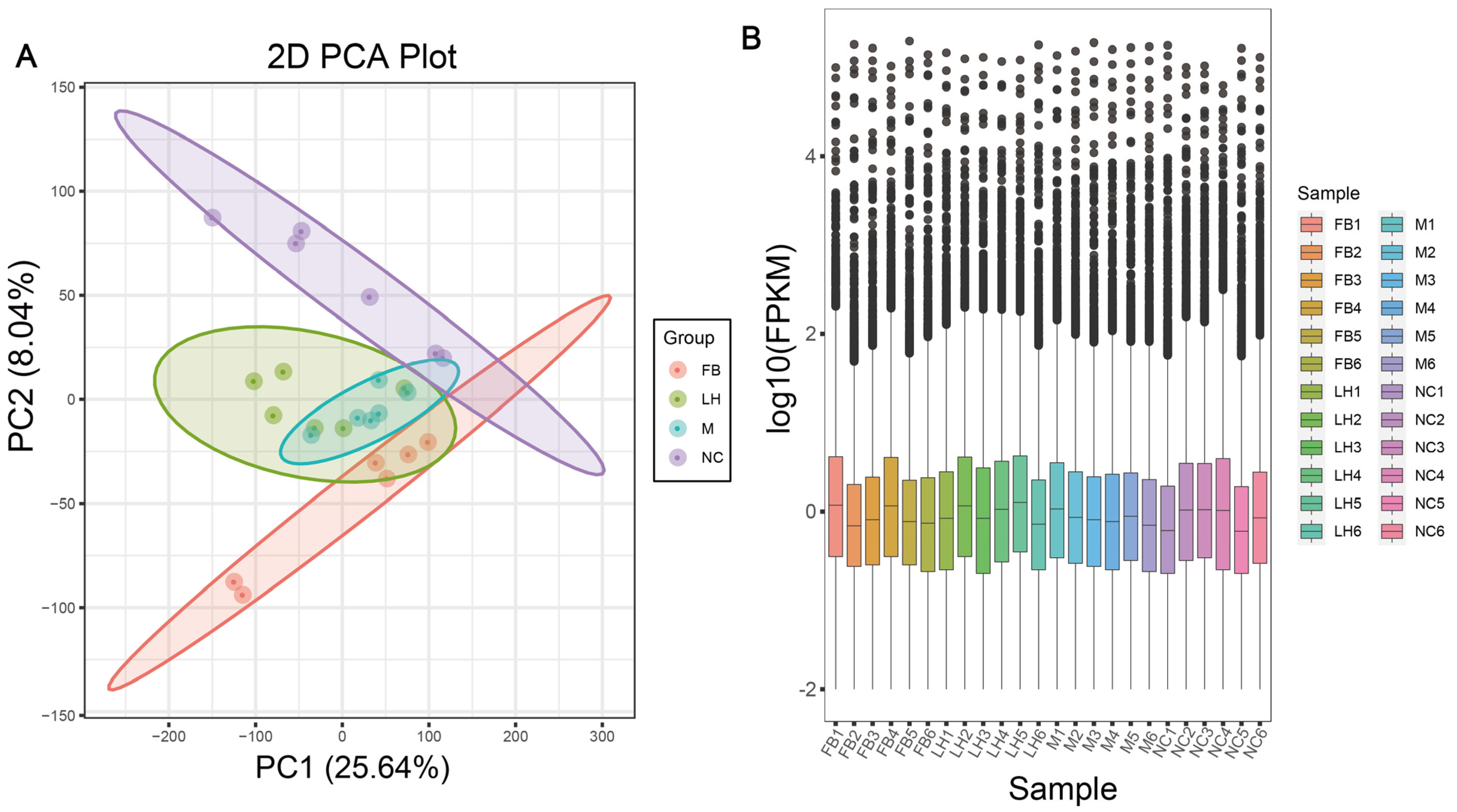
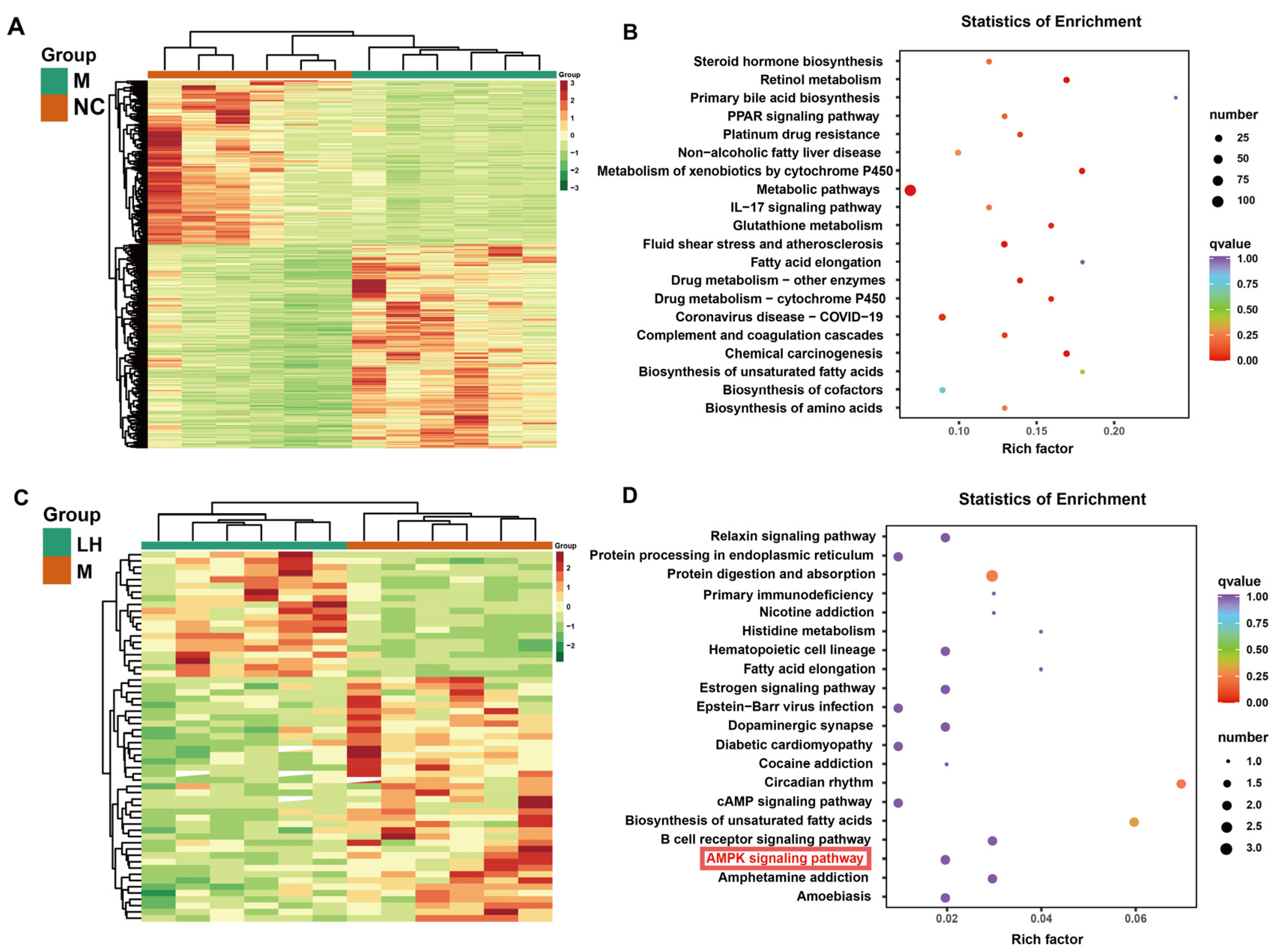
2.4. Leonurine Regulated Hepatic RNA Expression Profile in NAFLD Mice
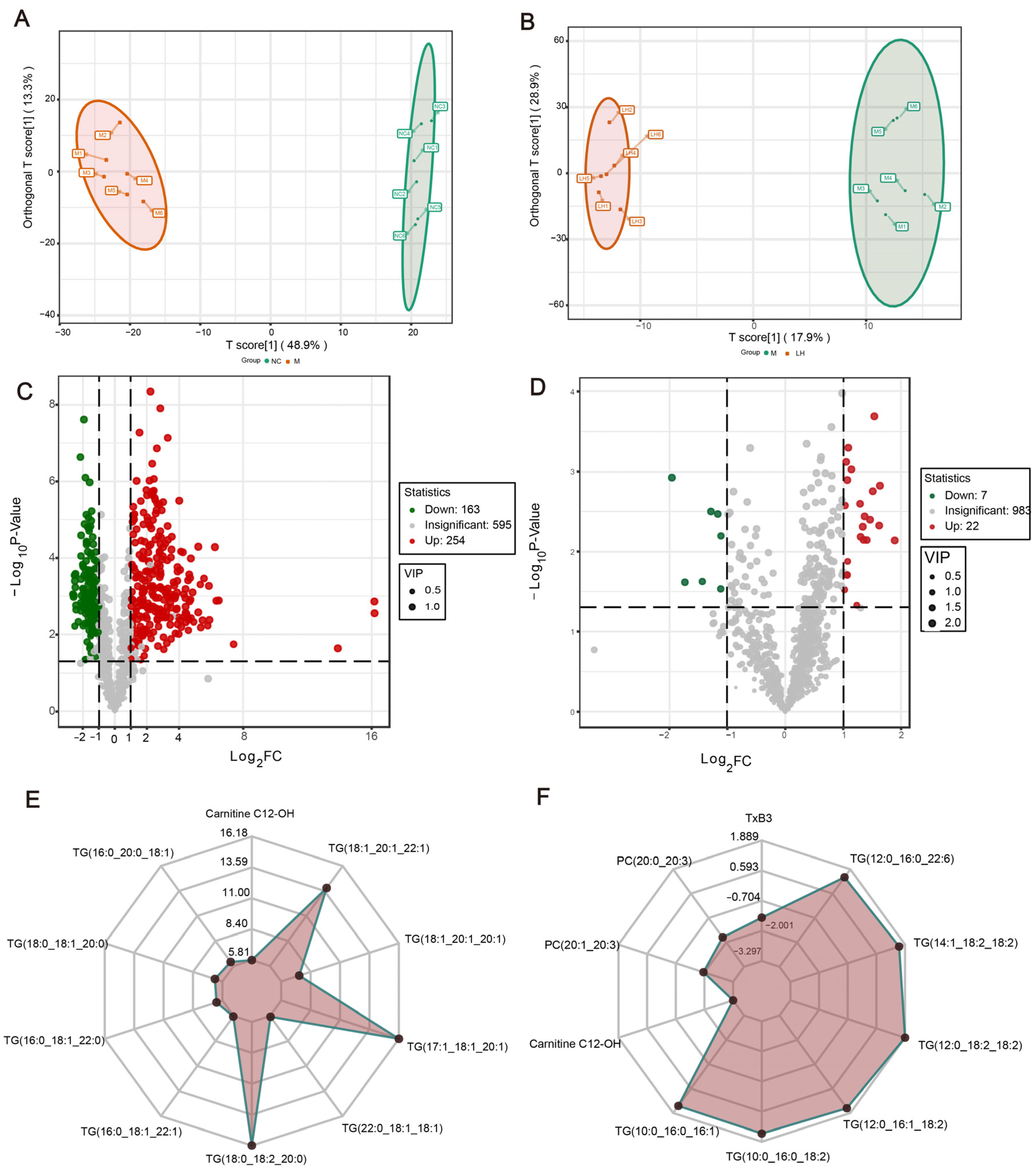
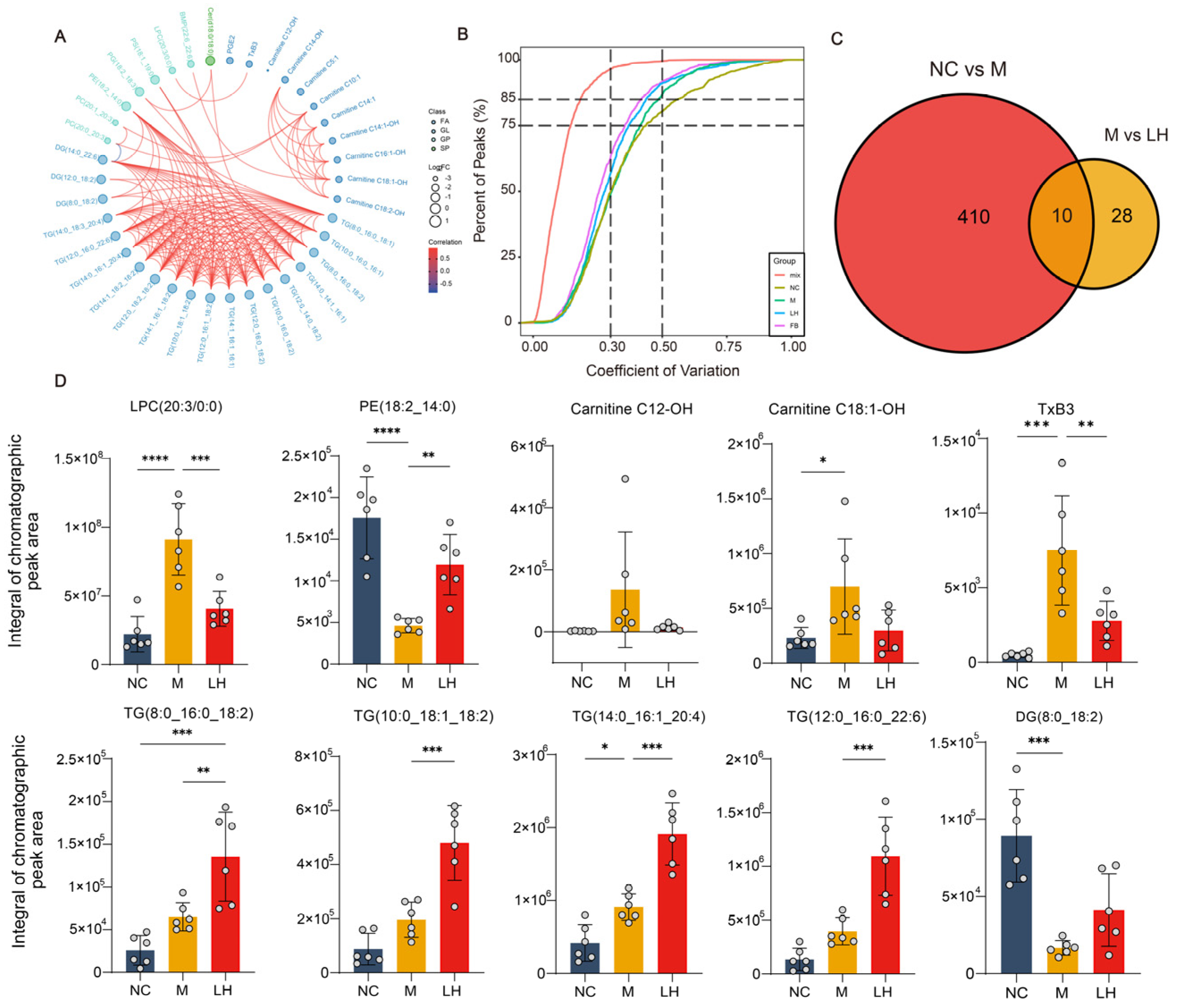
2.5. Leonurine Modulates Hepatic Lipid Profiles in NAFLD Mice
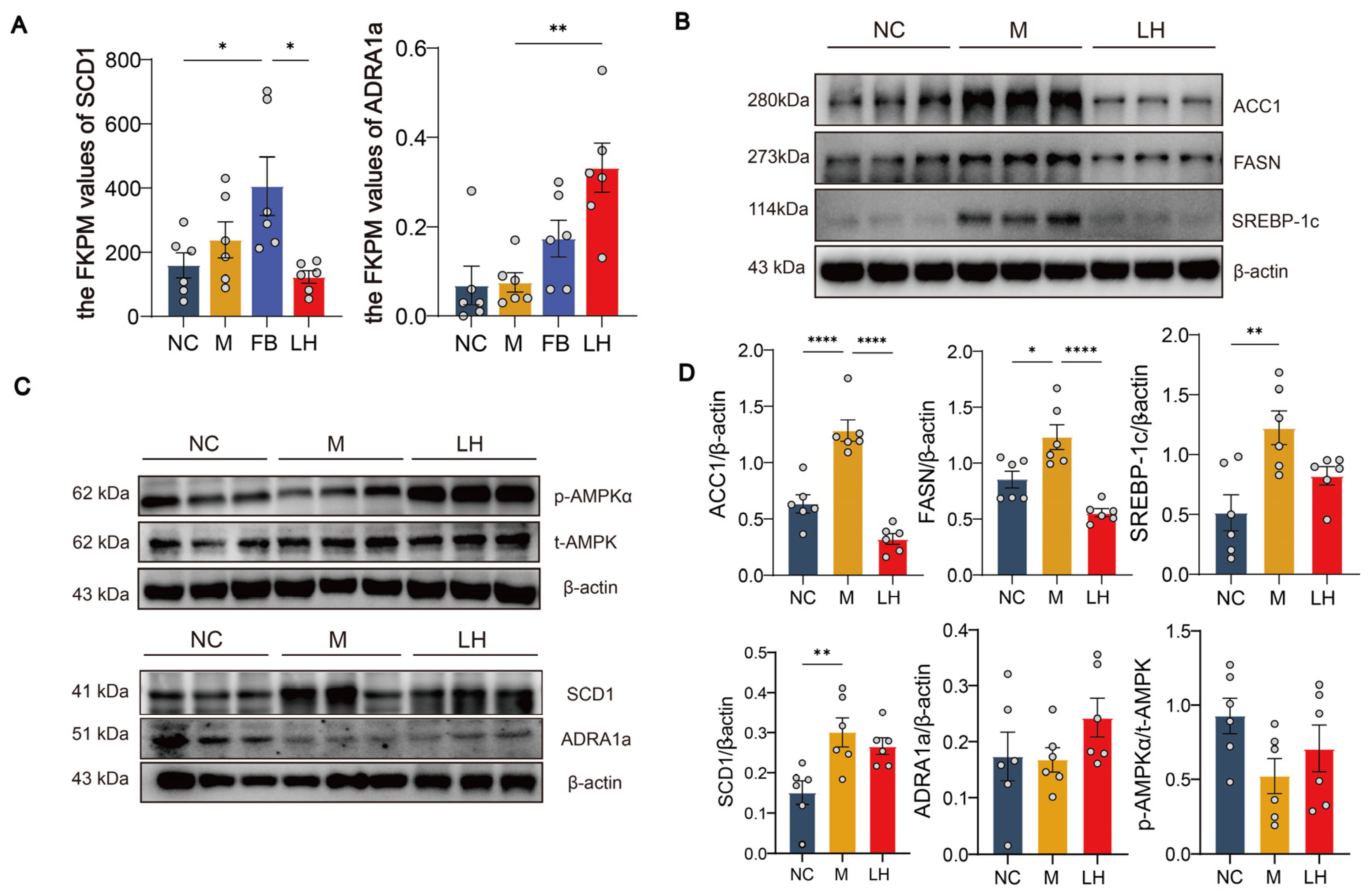
2.6. Leonurine Regulated Hepatic ADRA1a/AMPK/SCD1 Axis in HFHSD-Induced NAFLD Mice
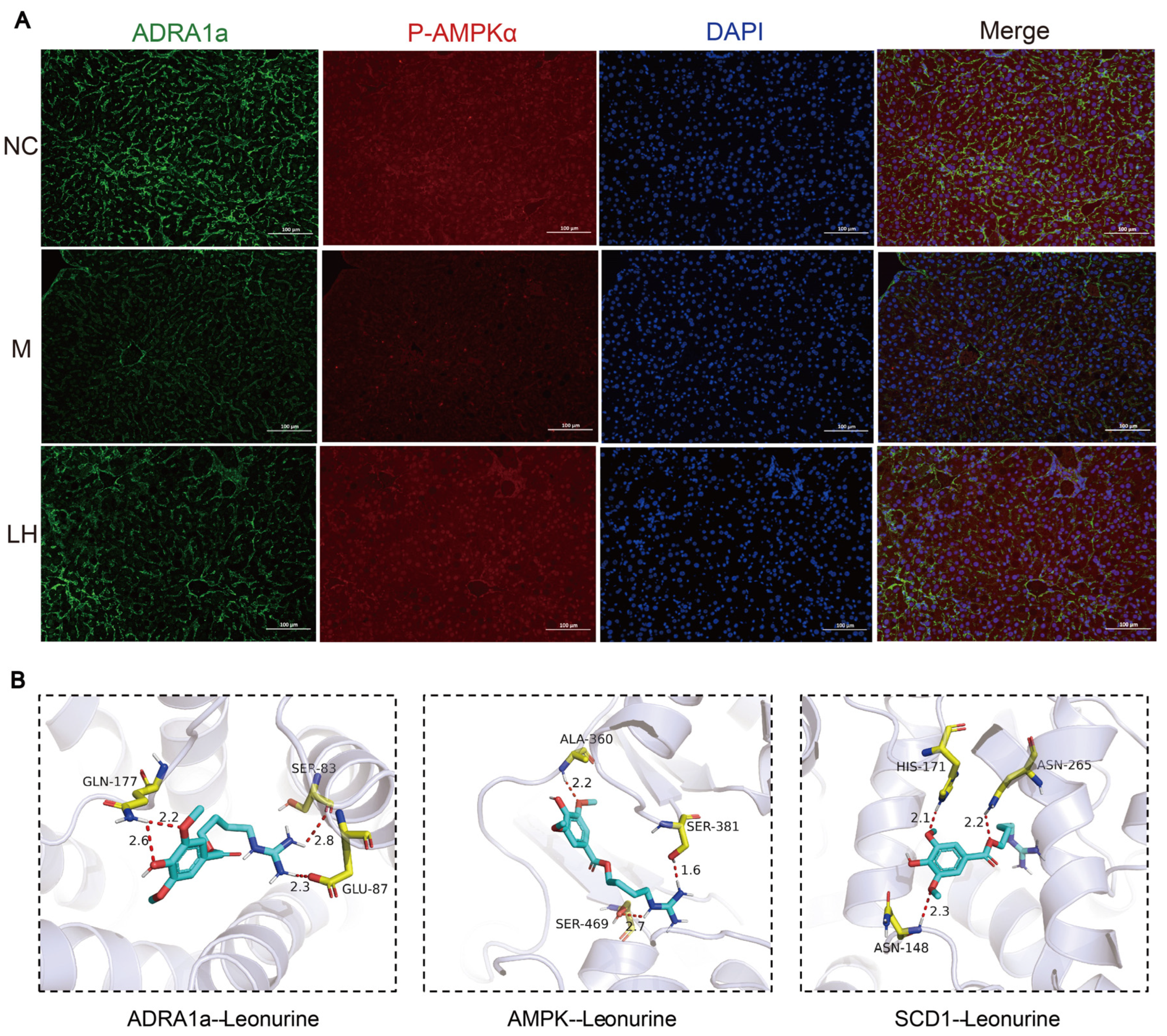
2.7. Effects on Immunofluorescence
2.8. Molecular Docking of Leonurine with ADRA1a, AMPK, and SCD1 Proteins
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Animal Experiment
4.3. Histological Analysis
4.4. Transmission Electron Microscopy Observations
4.5. Biochemical Analysis
4.6. Transcriptomic Analysis
4.7. Lipidomic Analysis
4.8. Molecular Docking
4.9. Western Blot
4.10. Immunofluorescence Analyses
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Powell, E.E.; Wong, V.W.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef]
- Guo, X.; Yin, X.; Liu, Z.; Wang, J. Non-Alcoholic Fatty Liver Disease (NAFLD) Pathogenesis and Natural Products for Prevention and Treatment. Int. J. Mol. Sci. 2022, 23, 15489. [Google Scholar] [CrossRef] [PubMed]
- Sheka, A.C.; Adeyi, O.; Thompson, J.; Hameed, B.; Crawford, P.A.; Kramuddin, S. Nonalcoholic Steatohepatitis: A Review. JAMA 2020, 323, 1175–1183. [Google Scholar] [CrossRef]
- Shang, X.; Pan, H.; Wang, X.; He, H.; Li, M. Leonurus japonicus Houtt.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2014, 152, 14–32. [Google Scholar] [CrossRef]
- Fierascu, R.C.; Fierascu, I.; Ortan, A.; Fierascu, I.C.; Anuta, V.; Velescu, B.S.; Pituru, S.M.; Dinu-Pirvu, C.E. Leonurus cardiaca L. as a Source of Bioactive Compounds: An Update of the European Medicines Agency Assessment Report (2010). Biomed. Res. Int. 2019, 2019, 4303215. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Lin, Y.-K.; Liu, X.-H.; Wang, L.; Yu, M.; Li, D.-J.; Zhu, Y.-Z.; Du, M.-R. Leonurine: From Gynecologic Medicine to Pleiotropic Agent. Chin. J. Integr. Med. 2020, 26, 152–160. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, W.; Wen, Y.; Xiong, Q.; Liu, H.; Wu, J.; Zou, Y.; Zhu, Y. SCM-198 attenuates early atherosclerotic lesions in hypercholesterolemic rabbits via modulation of the inflammatory and oxidative stress pathways. Atherosclerosis 2012, 224, 43–50. [Google Scholar] [CrossRef]
- Liu, X.H.; Pan, L.L.; Gong, Q.H.; Zhu, Y.Z. Antiapoptotic Effect of Novel Compound from Herba leonuri-Leonurine (SCM-198): A Mechanism Through Inhibition of Mitochondria Dysfunction in H9c2 Cells. Curr. Pharm. Biotechnol. 2010, 11, 895–905. [Google Scholar] [CrossRef]
- Liu, X.-H.; Pan, L.-L.; Deng, H.-Y.; Xiong, Q.-H.; Wu, D.; Huang, G.-Y.; Gong, Q.-H.; Zhu, Y.-Z. Leonurine (SCM-198) attenuates myocardial fibrotic response via inhibition of NADPH oxidase 4. Free. Radic. Biol. Med. 2013, 54, 93–104. [Google Scholar] [CrossRef]
- Lee, M.-R.; Park, K.I.; Ma, J.Y. Leonurus japonicus Houtt Attenuates Nonalcoholic Fatty Liver Disease in Free Fatty Acid-Induced HepG2 Cells and Mice Fed a High-Fat Diet. Nutrients 2017, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Suguro, R.; Chen, S.; Yang, D.; Yang, Z.; Miao, L.; Wu, W.; Zeng, W.; Liu, X.; Zhu, Y.Z. Anti-hypercholesterolemic Effects and a Good Safety Profile of SCM-198 in Animals: From ApoE Knockout Mice to Rhesus Monkeys. Front. Pharmacol. 2018, 9, 1468. [Google Scholar] [CrossRef] [PubMed]
- Brocker, C.N.; Patel, D.P.; Velenosi, T.J.; Kim, D.; Yan, T.; Yue, J.; Li, G.; Krausz, K.W.; Gonzalez, F.J. Extrahepatic PPARalpha modulates fatty acid oxidation and attenuates fasting-induced hepatosteatosis in mice. J. Lipid Res. 2018, 59, 2140–2152. [Google Scholar] [CrossRef]
- Carling, D. AMPK signalling in health and disease. Curr. Opin. Cell Biol. 2017, 45, 31–37. [Google Scholar] [CrossRef]
- Szlasa, W.; Zendran, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid composition of the cancer cell membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.E.; Moore, J.N.; Carrick, J.B.; Barton, M.H. Effect of intravenous infusion of omega-3 and omega-6 lipid emulsions on equine monocyte fatty acid composition and inflammatory mediator production in vitro. Shock 2000, 14, 222–228. [Google Scholar] [CrossRef][Green Version]
- Sinha, R.A.; Bruinstroop, E.; Singh, B.K.; Yen, P.M. Nonalcoholic Fatty Liver Disease and Hypercholesterolemia: Roles of Thyroid Hormones, Metabolites, and Agonists. Thyroid. 2019, 29, 1173–1191. [Google Scholar] [CrossRef]
- Llovet, J.M.; Willoughby, C.E.; Singal, A.G.; Greten, T.F.; Heikenwälder, M.; El-Serag, H.B.; Finn, R.S.; Friedman, S.L. Nonalcoholic steatohepatitis-related hepatocellular carcinoma: Pathogenesis and treatment. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 487–503. [Google Scholar] [CrossRef]
- Wu, H.; Dai, A.; Chen, X.; Yang, X.; Li, X.; Huang, C.; Jiang, K.; Deng, G. Leonurine ameliorates the inflammatory responses in lipopolysaccharide-induced endometritis. Int. Immunopharmacol. 2018, 61, 156–161. [Google Scholar] [CrossRef]
- Rong, W.; Li, J.; Wang, L.; Luo, S.; Liang, T.; Qian, X.; Zhang, X.; Zhou, Q.; Zhu, Y.; Zhu, Q. Investigation of the protective mechanism of leonurine against acute myocardial ischemia by an integrated metabolomics and network pharmacology strategy. Front. Cardiovasc. Med. 2022, 9, 969553. [Google Scholar] [CrossRef]
- Song, Z.; Meng, S.; Tang, Z.; Yang, X.; He, Y.; Zheng, Y.; Guo, H.; Du, M.; Zhu, Y.; Wang, X. Injectable leonurine nanocrystal-loaded microspheres for long-term hyperlipidemia man-agement. Biomater Sci. 2023, 11, 4713–4726. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.-L.; Yuan, Q.-Q.; Xue, D.-J.; Yang, F.-M.; Zhu, Y.-Z.; Qian, H.-B. Effect and mechanism of leonurine on pressure overload-induced cardiac hypertrophy in rats. Zhongguo Zhong Yao Za Zhi 2022, 47, 461–468. [Google Scholar] [CrossRef]
- Qi, L.; Chen, X.; Pan, Y.; Zha, Z.; Tang, M.; Shi, C.; Yang, B.; Wang, H. Leonurine exerts a protective effect in dextran sodium sulfate-induced experimental inflammatory bowel disease mice model. Gen. Physiol. Biophys. 2022, 41, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Sanches, S.C.L.; Ramalho, L.N.Z.; Augusto, M.J.; Da Silva, D.M.; Ramalho, F.S. Nonalcoholic Steatohepatitis: A Search for Factual Animal Models. BioMed Res. Int. 2015, 2015, 574832. [Google Scholar] [CrossRef]
- Kuivaniemi, H.; Tromp, G. Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene 2019, 707, 151–171. [Google Scholar] [CrossRef]
- Bajaj, T.; Hausl, A.S.; Schmidt, M.V.; Gassen, N.C. FKBP5/FKBP51 on weight watch: Central FKBP5 links regulatory WIPI protein networks to autophagy and metabolic control. Autophagy 2022, 18, 2756–2758. [Google Scholar] [CrossRef]
- Pereira, M.J.; Palming, J.; Svensson, M.K.; Rizell, M.; Dalenbäck, J.; Hammar, M.; Fall, T.; Sidibeh, C.O.; Svensson, P.-A.; Eriksson, J.W. FKBP5 expression in human adipose tissue increases following dexamethasone exposure and is associated with insulin resistance. Metabolism 2014, 63, 1198–1208. [Google Scholar] [CrossRef]
- Morales, T.S.; Avis, E.C.; Paskowski, E.K.; Shabar, H.; Nowotarski, S.L.; DiAngelo, J.R. The Role of Spermidine Synthase (SpdS) and Spermine Synthase (Sms) in Regulating Triglyceride Storage in Drosophila. Med. Sci. 2021, 9, 27. [Google Scholar] [CrossRef]
- Yang, J.; Peng, S.; Zhang, K. ARL4C depletion suppresses the resistance of ovarian cancer to carboplatin by disrupting cholesterol transport and autophagy via notch-RBP-Jkappa-H3K4Me3-OSBPL5. Hum Exp Toxicol. 2022, 41, 9603271221135064. [Google Scholar] [CrossRef]
- Docherty, J.R. Subtypes of functional alpha1-adrenoceptor. Cell Mol. Life Sci. 2010, 67, 405–417. [Google Scholar] [CrossRef]
- Ballou, L.M.; Tian, P.Y.; Lin, H.Y.; Jiang, Y.P.; Lin, R.Z. Dual regulation of glycogen synthase kinase-3beta by the alpha1A-adrenergic receptor. J. Biol. Chem. 2001, 276, 40910–40916. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, S.; Yoshihara, K.; Kawanabe, R.; Hatada, I.; Koga, K.; Tsuda, M. Stress-induced antinociception to noxious heat requires α1A-adrenaline receptors of spinal inhibitory neurons in mice. Mol. Brain 2022, 15, 6. [Google Scholar] [CrossRef]
- Oben, J.A.; Roskams, T.; Yang, S.; Lin, H.; Sinelli, N.; Li, Z.; Torbenson, M.; Huang, J.; Guarino, P.; Kafrouni, M.; et al. Sympathetic nervous system inhibition increases hepatic progenitors and reduces liver injury. Hepatology 2003, 38, 664–673. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Liu, X. MiR-3682 promotes the progression of hepatocellular carcinoma (HCC) via inactivating AMPK signaling by targeting ADRA1A. Ann. Hepatol. 2022, 27 (Suppl. S1), 100570. [Google Scholar] [CrossRef]
- Hutchinson, D.S.; Bengtsson, T. AMP-activated protein kinase activation by adrenoceptors in L6 skeletal muscle cells: Mediation by alpha1-adrenoceptors causing glucose uptake. Diabetes 2006, 55, 682–690. [Google Scholar] [CrossRef]
- Garcia, D.; Hellberg, K.; Chaix, A.; Wallace, M.; Herzig, S.; Badur, M.G.; Lin, T.; Shokhirev, M.N.; Pinto, A.F.; Ross, D.S.; et al. Genetic Liver-Specific AMPK Activation Protects against Diet-Induced Obesity and NAFLD. Cell Rep. 2019, 26, 192–208.e6. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, S.; Zhai, A.; Zhang, B.; Tian, G. AMPK-Mediated Regulation of Lipid Metabolism by Phosphorylation. Biol. Pharm. Bull. 2018, 41, 985–993. [Google Scholar] [CrossRef]
- Mauvoisin, D.; Mounier, C. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie 2011, 93, 78–86. [Google Scholar] [CrossRef]
- Mori, H.; Peterson, S.K.; Simmermon, R.C.; Overmyer, K.A.; Nishii, A.; Paulsson, E.; Li, Z.; Jen, A.; Uranga, R.M.; Maung, J.N.; et al. Scd1 and monounsaturated lipids are required for autophagy and survival of adipocytes. Mol. Metab. 2024, 83, 101916. [Google Scholar] [CrossRef]
- Ralston, J.C.; Metherel, A.H.; Stark, K.D.; Mutch, D.M. SCD1 mediates the influence of exogenous saturated and monounsaturated fatty acids in adipocytes: Effects on cellular stress, inflammatory markers and fatty acid elongation. J Nutr Biochem. 2016, 27, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Ralston, J.C.; Badoud, F.; Cattrysse, B.; McNicholas, P.D.; Mutch, D.M. Inhibition of stearoyl-CoA desaturase-1 in differentiating 3T3-L1 preadipocytes upregulates elongase 6 and downregulates genes affecting triacylglycerol synthesis. Int. J. Obes. 2014, 38, 1449–1456. [Google Scholar] [CrossRef]
- Ran, H.; Zhu, Y.; Deng, R.; Zhang, Q.; Liu, X.; Feng, M.; Zhong, J.; Lin, S.; Tong, X.; Su, Q. Stearoyl-CoA desaturase-1 promotes colorectal cancer metastasis in response to glucose by sup-pressing PTEN. J. Exp. Clin. Cancer Res. 2018, 37, 54. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, J.; Zou, X.; Greggain, J.; Færgeman, N.J.; Liang, B.; Watts, J.L. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 2013, 54, 2504–2514. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.; Yao, X.; Fei, Y.; Lin, Z.; Li, Z.; Cai, K.; Zhao, Y.; Luo, Z. HCAR1/MCT1 Regulates Tumor Ferroptosis through the Lactate-Mediated AMPK-SCD1 Activity and Its Therapeutic Implications. Cell Rep. 2020, 33, 108487. [Google Scholar] [CrossRef] [PubMed]
- Exton, J.H. Hormonal regulation of phosphatidylcholine breakdown. Adv. Second Messenger Phosphoprot. Res. 1990, 24, 152–157. [Google Scholar]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Wendel, A.A.; Lewin, T.M.; Coleman, R.A. Glycerol-3-phosphate acyltransferases: Rate limiting enzymes of triacylglycerol bio-synthesis. Biochim. Biophys. Acta 2009, 1791, 501–506. [Google Scholar] [CrossRef]
- McLaren, D.G.; Han, S.; Murphy, B.A.; Wilsie, L.; Stout, S.J.; Zhou, H.; Roddy, T.P.; Gorski, J.N.; Metzger, D.E.; Shin, M.K.; et al. DGAT2 Inhibition Alters Aspects of Triglyceride Metabolism in Rodents but Not in Non-human Primates. Cell Metab. 2018, 27, 1236–1248.e6. [Google Scholar] [CrossRef]
- McFie, P.J.; Patel, A.; Stone, S.J. The monoacylglycerol acyltransferase pathway contributes to triacylglycerol synthesis in HepG2 cells. Sci. Rep. 2022, 12, 4943. [Google Scholar] [CrossRef]
- Lan, T.; Geng, X.J.; Zhang, S.J.; Zeng, X.X.; Ying, J.J.; Xu, Y.; Liu, S.Y.; Li, P.; Tong, Y.H.; Wang, W.; et al. Si-Ni-San inhibits hepatic Fasn expression and lipid accumulation in MAFLD mice through AMPK/p300/SREBP-1c axis. Phytomedicine 2024, 123, 155209. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, C.; Song, Y.; Chen, L.; Chen, X.; Zheng, G.; Yang, Y.; Cao, P.; Qiu, Z. Gallic acid impairs fructose-driven de novo lipogenesis and ameliorates hepatic steatosis via AMPK-dependent suppression of SREBP-1/ACC/FASN cascade. Eur. J. Pharmacol. 2023, 940, 175457. [Google Scholar] [CrossRef] [PubMed]
- Sheng, D.; Zhao, S.; Gao, L.; Zheng, H.; Liu, W.; Hou, J.; Jin, Y.; Ye, F.; Zhao, Q.; Li, R.; et al. BabaoDan attenuates high-fat diet-induced non-alcoholic fatty liver disease via activation of AMPK signaling. Cell Biosci. 2019, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, Q.; Fu, J.; Ren, R. Polysaccharides derived from natural sources regulate triglyceride and cholesterol metabolism: A review of the mechanisms. Food Funct. 2019, 10, 2330–2339. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient op-timization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLOS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef]
| Target Name | Ligand Name | Docking Score (kcal/mol) | |||
|---|---|---|---|---|---|
| Round 1 | Round 2 | Round 3 | Mean | ||
| ADRA1a | Leonurine | −6.751 | −6.641 | −6.756 | −6.716 |
| AMPK | Leonurine | −6.309 | −5.613 | −6.295 | −6.072 |
| SCD1 | Leonurine | −8.585 | −8.416 | −8.473 | −8.491 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, W.; Pan, M.; Zheng, C.; Shen, H.; Pi, D.; Song, Q.; Liang, Z.; Zhen, J.; Pan, J.; Liu, L.; et al. Leonurine Inhibits Hepatic Lipid Synthesis to Ameliorate NAFLD via the ADRA1a/AMPK/SCD1 Axis. Int. J. Mol. Sci. 2024, 25, 10855. https://doi.org/10.3390/ijms251910855
Fan W, Pan M, Zheng C, Shen H, Pi D, Song Q, Liang Z, Zhen J, Pan J, Liu L, et al. Leonurine Inhibits Hepatic Lipid Synthesis to Ameliorate NAFLD via the ADRA1a/AMPK/SCD1 Axis. International Journal of Molecular Sciences. 2024; 25(19):10855. https://doi.org/10.3390/ijms251910855
Chicago/Turabian StyleFan, Wen, Maoxing Pan, Chuiyang Zheng, Haiyan Shen, Dajin Pi, Qingliang Song, Zheng Liang, Jianwei Zhen, Jinyue Pan, Lianghao Liu, and et al. 2024. "Leonurine Inhibits Hepatic Lipid Synthesis to Ameliorate NAFLD via the ADRA1a/AMPK/SCD1 Axis" International Journal of Molecular Sciences 25, no. 19: 10855. https://doi.org/10.3390/ijms251910855
APA StyleFan, W., Pan, M., Zheng, C., Shen, H., Pi, D., Song, Q., Liang, Z., Zhen, J., Pan, J., Liu, L., Yang, Q., & Zhang, Y. (2024). Leonurine Inhibits Hepatic Lipid Synthesis to Ameliorate NAFLD via the ADRA1a/AMPK/SCD1 Axis. International Journal of Molecular Sciences, 25(19), 10855. https://doi.org/10.3390/ijms251910855





