CXCR4 Is a Potential Target for Anti-HIV Gene Therapy
Abstract
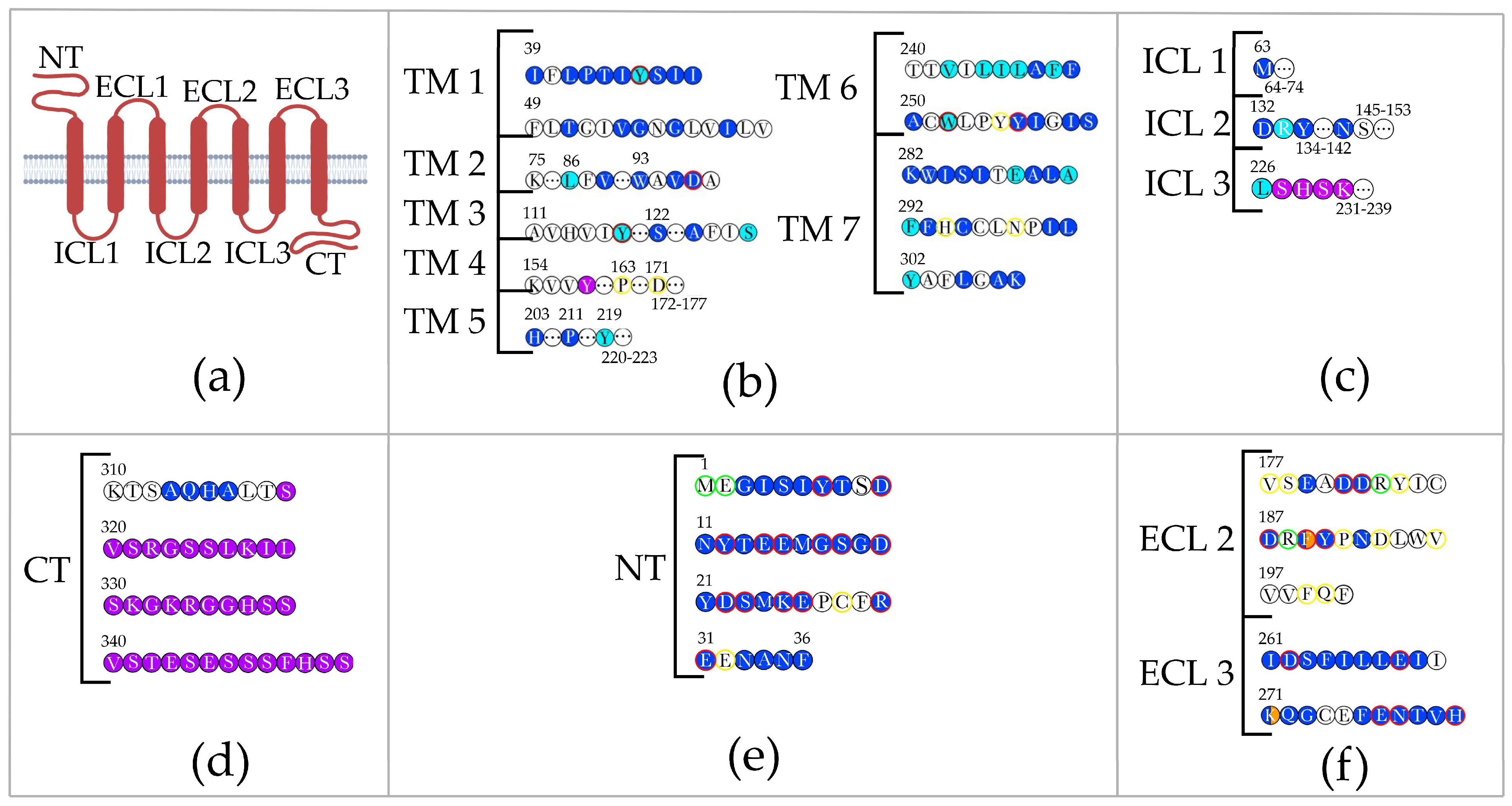
:1. Introduction
2. Interaction of CXCR4 with Ligands
3. Interaction of CXCR4 with the V3 Loop in HIV gp120
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- HIV and AIDS. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 10 October 2023).
- Nyamweya, S.; Hegedus, A.; Jaye, A.; Rowland-Jones, S.; Flanagan, K.L.; Macallan, D.C. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 2013, 23, 221–240. [Google Scholar] [CrossRef] [PubMed]
- Barroso, H.; Borrego, P.; Bártolo, I.; Marcelino, J.M.; Família, C.; Quintas, A.; Taveira, N. Evolutionary and structural features of the C2, V3 and C3 envelope regions underlying the differences in HIV-1 and HIV-2 biology and infection. PLoS ONE 2011, 6, e14548. [Google Scholar]
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.F.; Swenson, L.C.; Bunnik, E.M.; Edo-Matas, D.; Schuitemaker, H.; van’t Wout, A.B.; Harrigan, P.R. Reconstructing the dynamics of HIV evolution within hosts from serial deep sequence data. PLoS Comput. Biol. 2012, 8, e1002753. [Google Scholar] [CrossRef] [PubMed]
- Bunnik, E.M.; Swenson, L.C.; Edo-Matas, D.; Huang, W.; Dong, W.; Frantzell, A.; Petropoulos, C.J.; Coakley, E.; Schuitemaker, H.; Harrigan, P.R.; et al. Detection of inferred CCR5-and CXCR4-using HIV-1 variants and evolutionary intermediates using ultra-deep pyrosequencing. PLoS Pathog. 2011, 7, e1002106. [Google Scholar] [CrossRef]
- Schuitemaker, H.; Koot, M.; Kootstra, N.A.; Dercksen, M.W.; De Goede, R.E.; Van Steenwijk, R.P.; Lange, J.M.; Schattenkerk, J.K.; Miedema, F.; Tersmette, M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 1992, 66, 1354–1360. [Google Scholar] [CrossRef]
- Maldini, C.R.; Claiborne, D.T.; Okawa, K.; Chen, T.; Dopkin, D.L.; Shan, X.; Power, K.A.; Trifonova, R.T.; Krupp, K.; Phelps, M.; et al. Dual CD4-based CAR T cells with distinct costimulatory domains mitigate HIV pathogenesis in vivo. Nat. Med. 2020, 26, 1776–1787. [Google Scholar] [CrossRef]
- Schriek, A.I.; van Haaren, M.M.; Poniman, M.; Dekkers, G.; Bentlage, A.E.; Grobben, M.; van Gils, M.J. Anti-HIV-1 Nanobody-IgG1 constructs with improved neutralization potency and the ability to mediate fc effector functions. Front. Immunol. 2022, 13, 893648. [Google Scholar] [CrossRef]
- Sheykhhasan, M.; Foroutan, A.; Manoochehri, H.; Khoei, S.G.; Poondla, N.; Saidijam, M. Could gene therapy cure HIV? Life Sci. 2021, 277, 119451. [Google Scholar] [CrossRef]
- Worgall, S.; Crystal, R.G. Gene Therapy. In Principles of Tissue Engineering, 5th ed.; Lanza, R., Langer, R., Vacanti, J.P., Atala, A., Eds.; Elsevier: London, UK, 2020; Volume 28, pp. 493–518. [Google Scholar]
- Wilkinson, A.C.; Igarashi, K.J.; Nakauchi, H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat. Rev. Genet. 2020, 21, 541–554. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Hütter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Müßig, A.; Allers, K.; Schneider, T.; Hofmann, J.; Kücherer, C.; Blau, O.; et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009, 360, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Abdul-Jawad, S.; McCoy, L.E.; Mok, H.P.; Peppa, D.; Salgado, M.; Martinez-Picado, J.; Nijhuis, M.; Wensing, A.M.J.; Lee, H.; et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 2019, 568, 244–248. [Google Scholar] [CrossRef]
- Yukl, S.A.; Boritz, E.; Busch, M.; Bentsen, C.; Chun, T.W.; Douek, D.; Eisele, E.; Haase, A.; Ho, Y.-C.; Hütter, G.; et al. Challenges in detecting HIV persistence during potentially curative interventions: A study of the Berlin patient. PLoS Pathog. 2013, 9, e1003347. [Google Scholar] [CrossRef]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [PubMed]
- Jiang, R.; Zhao, N.; He, X. 259. Parathyroid Hormone Gene Modified Mesenchymal Stem Cells for Hypoparathyroidism. Mol. Ther. 2015, 23, S102. [Google Scholar]
- Xu, L.; Wang, J.; Liu, Y.; Xie, L.; Su, B.; Mou, D.; Wang, L.; Liu, T.; Wang, X.; Zhang, B.; et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019, 381, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Van Hout, A.; Klarenbeek, A.; Bobkov, V.; Doijen, J.; Arimont, M.; Zhao, C.; Heukers, R.; Rimkunas, R.; de Graaf, C.; Verrips, T.; et al. CXCR4-targeting nanobodies differentially inhibit CXCR4 function and HIV entry. Biochem. Pharmacol. 2018, 158, 402–412. [Google Scholar]
- Zhou, N.; Fang, J.; Mukhtar, M.; Acheampong, E.; Pomerantz, R.J. Inhibition of HIV-1 fusion with small interfering RNAs targeting the chemokine coreceptor CXCR4. Gene Ther. 2004, 11, 1703–1712. [Google Scholar] [CrossRef]
- Cunha-Santos, C.; Perdigao, P.R.L.; Martin, F.; Oliveira, J.G.; Cardoso, M.; Manuel, A.; Goncalves, J. Inhibition of HIV replication through siRNA carried by CXCR4-targeted chimeric nanobody. Cell. Mol. Life Sci. 2020, 77, 2859–2870. [Google Scholar] [CrossRef]
- Anderson, J.; Banerjea, A.; Planelles, V.; Akkina, R. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res. Hum. Retroviruses 2003, 19, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.S.M.; Snyder, E.Y.; Schooley, R.T. Strategies and progress in CXCR4-targeted anti-human immunodeficiency virus (HIV) therapeutic development. Clin. Infect. Dis. 2021, 73, 919–924. [Google Scholar] [PubMed]
- Leslie, G.J.; Wang, J.; Richardson, M.V.; Haggarty, B.S.; Hua, K.L.; Duong, J.; Secreto, A.J.; Jordon, A.P.O.; Romano, J.; Kumar, K.E.; et al. Potent and broad inhibition of HIV-1 by a peptide from the gp41 heptad repeat-2 domain conjugated to the CXCR4 amino terminus. PLoS Pathog. 2016, 12, e1005983. [Google Scholar]
- Zou, Y.R.; Kottmann, A.H.; Kuroda, M.; Taniuchi, I.; Littman, D.R. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 1998, 393, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Jones, D.; Borghesani, P.R.; Segal, R.A.; Nagasawa, T.; Kishimoto, T.; Bronson, R.T.; Springer, T.A. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4-and SDF-1-deficient mice. Proc. Natl. Acad. Sci. USA 1998, 95, 9448–9453. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, T.; Hirota, S.; Tachibana, K.; Takakura, N.; Nishikawa, S.I.; Kitamura, Y.; Yoshida, N.; Kikutani, H.; Kishimoto, T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 1996, 382, 635–638. [Google Scholar] [PubMed]
- Tachibana, K.; Hirota, S.; Iizasa, H.; Yoshida, H.; Kawabata, K.; Kataoka, Y.; Kitamura, Y.; Matsushima, K.; Yoshida, N.; Nishikawa, S.-I.; et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 1998, 393, 591–594. [Google Scholar] [CrossRef]
- Oberlin, E.; Amara, A.; Bachelerie, F.; Bessia, C.; Virelizier, J.-L.; Arenzana-Seisdedos, F.; Schwartz, O.; Heard, J.-M.; Clark-Lewis, I.; Legler, D.F.; et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 1996, 382, 833–835. [Google Scholar] [CrossRef]
- Pozzobon, T.; Goldoni, G.; Viola, A.; Molon, B. CXCR4 signaling in health and disease. Immunol. Lett. 2016, 177, 6–15. [Google Scholar] [CrossRef]
- Payne, D.; Drinkwater, S.; Baretto, R.; Duddridge, M.; Browning, M.J. Expression of chemokine receptors CXCR4, CXCR5 and CCR7 on B and T lymphocytes from patients with primary antibody deficiency. Clin. Exp. Immunol. 2009, 156, 254–262. [Google Scholar]
- Vila-Coro, A.J.; Rodríguez-Frade, J.M.; De Ana, A.M.; Moreno-Ortíz, M.C.; Martínez-A, C.; Mellado, M. The chemokine SDF-lα triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. 1999, 13, 1699–1710. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Zuba-Surma, E.; Kucia, M.; Reca, R.; Wojakowski, W.; Ratajczak, J. The pleiotropic effects of the SDF-1–CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia 2006, 20, 1915–1924. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Waite, J.; Brewer, F.; Sunshine, M.J.; Littman, D.R.; Zou, Y.R. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 2004, 200, 1145–1156. [Google Scholar]
- Stein, J.V.; Nombela-Arrieta, C. Chemokine control of lymphocyte trafficking: A general overview. Immunology 2005, 116, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, D.; Gröning, S.; Schmitz, C.; Zierow, S.; Drucker, N.; Bakou, M.; Kohl, K.; Mertens, A.; Lue, H.; Weber, C.; et al. Macrophage migration inhibitory factor-CXCR4 receptor interactions. J. Biol. Chem. 2016, 291, 15881–15895. [Google Scholar] [PubMed]
- Bernhagen, J.; Krohn, R.; Lue, H.; Gregory, J.L.; Zernecke, A.; Koenen, R.R.; Dewor, M.; Georgiev, I.; Schober, A.; Leng, L.; et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 2007, 13, 587–596. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. The bicyclam AMD3100 story. Nat. Rev. Drug Discov. 2003, 2, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Carter, N.J.; Keating, G.M. Maraviroc. Drugs 2007, 67, 2277–2288. [Google Scholar] [CrossRef] [PubMed]
- Hatse, S.; Princen, K.; Gerlach, L.-O.; Bridger, G.; Henson, G.; De Clercq, E.; Schwartz, T.W.; Schols, D. Mutation of Asp171 and Asp262 of the chemokine receptor CXCR4 impairs its coreceptor function for human immunodeficiency virus-1 entry and abrogates the antagonistic activity of AMD3100. Mol. Pharmacol. 2001, 60, 164–173. [Google Scholar] [CrossRef]
- Kucia, M.; Jankowski, K.; Reca, R.; Wysoczynski, M.; Bandura, L.; Allendorf, D.J.; Zhang, J.; Ratajczak, J.; Ratajczak, M.Z. CXCR4–SDF-1 signalling, locomotion, chemotaxis and adhesion. J. Mol. Histol. 2004, 35, 233–245. [Google Scholar] [CrossRef]
- Wescott, M.P.; Kufareva, I.; Paes, C.; Goodman, J.R.; Thaker, Y.; Puffer, B.A.; Berdougo, E.; Rucker, J.B.; Handel, T.M.; Doranz, B.J. Signal transmission through the CXC chemokine receptor 4 (CXCR4) transmembrane helices. Proc. Natl. Acad. Sci. USA 2016, 113, 9928–9933. [Google Scholar] [CrossRef]
- Ahr, B.; Denizot, M.; Robert-Hebmann, V.; Brelot, A.; Biard-Piechaczyk, M. Identification of the cytoplasmic domains of CXCR4 involved in Jak2 and STAT3 phosphorylation. J. Biol. Chem. 2005, 280, 6692–6700. [Google Scholar] [CrossRef] [PubMed]
- Krupnick, J.G.; Benovic, J.L. The role of receptor kinases and arrestins in G protein–coupled receptor regulation. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 289–319. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.J.; Zhao, J.; Sun, Y.; Hu, W.; Wu, Y.L.; Cen, B.; Wu, G.; Pei, G. β-arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between β-arrestin and CXCR4. J. Biol. Chem. 2000, 275, 2479–2485. [Google Scholar] [PubMed]
- Lagane, B.; Chow, K.Y.C.; Balabanian, K.; Levoye, A.; Harriague, J.; Planchenault, T.; Baleux, F.; Gunera-Saad, N.; Arenzana-Seisdedos, F.; Bachelerie, F. CXCR4 dimerization and β-arrestin–mediated signaling account for the enhanced chemotaxis to CXCL12 in WHIM syndrome. Blood J. Am. Soc. Hematol. 2008, 112, 34–44. [Google Scholar]
- Duquenne, C.; Psomas, C.; Gimenez, S.; Guigues, A.; Carles, M.-J.; Barbuat, C.; Lavigne, J.-P.; Sotto, A.; Reynes, J.; Guglielmi, P.; et al. The two human CXCR4 isoforms display different HIV receptor activities: Consequences for the emergence of X4 strains. J. Immunol. 2014, 193, 4188–4194. [Google Scholar] [CrossRef] [PubMed]
- Roumen, L.; Scholten, D.J.; De Kruijf, P.; De Esch, I.J.P.; Leurs, R.; De Graaf, C. C (X) CR in silico: Computer-aided prediction of chemokine receptor–ligand interactions. Drug Discov. Today Technol. 2012, 9, e281–e291. [Google Scholar] [CrossRef] [PubMed]
- Stephens, B.S.; Ngo, T.; Kufareva, I.; Handel, T.M. Functional anatomy of the full-length CXCR4-CXCL12 complex systematically dissected by quantitative model-guided mutagenesis. Sci. Signal. 2020, 13, eaay5024. [Google Scholar] [CrossRef] [PubMed]
- Brelot, A.; Heveker, N.; Montes, M.; Alizon, M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 2000, 275, 23736–23744. [Google Scholar]
- Doranz, B.J.; Orsini, M.J.; Turner, J.D.; Hoffman, T.L.; Berson, J.F.; Hoxie, J.A.; Peiper, S.C.; Brass, L.F.; Doms, R.W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J. Virol. 1999, 73, 2752–2761. [Google Scholar]
- Zhou, N.; Luo, Z.; Luo, J.; Liu, D.; Hall, J.W.; Pomerantz, R.J.; Huang, Z. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J. Biol. Chem. 2001, 276, 42826–42833. [Google Scholar] [CrossRef]
- Saini, V.; Staren, D.M.; Ziarek, J.J.; Nashaat, Z.N.; Campbell, E.M.; Volkman, B.F.; Marchese, A.; Majetschak, M. The CXC chemokine receptor 4 ligands ubiquitin and stromal cell-derived factor-1α function through distinct receptor interactions. J. Biol. Chem. 2011, 286, 33466–33477. [Google Scholar] [CrossRef]
- Saini, V.; Marchese, A.; Tang, W.J.; Majetschak, M. Structural determinants of ubiquitin-CXC chemokine receptor 4 interaction. J. Biol. Chem. 2011, 286, 44145–44152. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, T.; Kimura, T.; Philpott, S.; Weiser, B.; Burger, H.; Zolla-Pazner, S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retroviruses 2007, 23, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Pollakis, G.; Kang, S.; Kliphuis, A.; Chalaby, M.I.; Goudsmit, J.; Paxton, W.A. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 2001, 276, 13433–13441. [Google Scholar] [PubMed]
- Choi, W.-T.; Tian, S.; Dong, C.-Z.; Kumar, S.; Liu, D.; Madani, N.; An, J.; Sodroski, J.G.; Huang, Z. Unique ligand binding sites on CXCR4 probed by a chemical biology approach: Implications for the design of selective human immunodeficiency virus type 1 inhibitors. J. Virol. 2005, 79, 15398–15404. [Google Scholar] [CrossRef]
- Tamamis, P.; Floudas, C.A. Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop. Biophys. J. 2013, 105, 1502–1514. [Google Scholar] [CrossRef]
- Lu, Z.-H.; Berson, J.F.; Chen, Y.-H.; Turner, J.D.; Zhang, T.-Y.; Sharron, M.; Jenks, M.H.; Wang, Z.-X.; Kim, J.; Rucker, J.; et al. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. USA 1997, 94, 6426–6431. [Google Scholar] [CrossRef]
- Chabot, D.J.; Zhang, P.F.; Quinnan, G.V.; Broder, C.C. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol. 1999, 73, 6598–6609. [Google Scholar]
- Brelot, A.; Heveker, N.; Adema, K.; Hosie, M.J.; Willett, B.; Alizon, M. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol. 1999, 73, 2576–2586. [Google Scholar]
- Tian, S.; Choi, W.-T.; Liu, D.; Pesavento, J.; Wang, Y.; An, J.; Sodroski, J.G.; Huang, Z. Distinct functional sites for human immunodeficiency virus type 1 and stromal cell-derived factor 1α on CXCR4 transmembrane helical domains. J. Virol. 2005, 79, 12667–12673. [Google Scholar] [CrossRef]
- He, S.; Wu, Y. Relationships between HIV-mediated chemokine coreceptor signaling, cofilin hyperactivation, viral tropism switch and HIV-mediated CD4 depletion. Curr. HIV Res. 2019, 17, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Yandrapally, S.; Mohareer, K.; Arekuti, G.; Vadankula, G.R.; Banerjee, S. HIV co-receptor-tropism: Cellular and molecular events behind the enigmatic co-receptor switching. Crit. Rev. Microbiol. 2021, 47, 499–516. [Google Scholar] [PubMed]
- Bunnik, E.M.; Quakkelaar, E.D.; Van Nuenen, A.C.; Boeser-Nunnink, B.; Schuitemaker, H. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J. Virol. 2007, 81, 525–531. [Google Scholar] [PubMed]
- Tasca, S.; Ho, S.H.; Cheng-Mayer, C. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J. Virol. 2008, 82, 7089–7099. [Google Scholar] [CrossRef] [PubMed]
- Klasse, P.J. The molecular basis of HIV entry. Cell. Microbiol. 2012, 14, 1183–1192. [Google Scholar] [CrossRef]
- Choi, W.T.; An, J. Biology and clinical relevance of chemokines and chemokine receptors CXCR4 and CCR5 in human diseases. Exp. Biol. Med. 2011, 236, 637–647. [Google Scholar] [CrossRef]
- Trkola, A.; Dragic, T.; Arthos, J.; Binley, J.M.; Olson, W.C.; Allaway, G.P.; Moore, J.P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 1996, 384, 184–187. [Google Scholar] [CrossRef]
- Britton, C.; Poznansky, M.C.; Reeves, P. Polyfunctionality of the CXCR4/CXCL12 axis in health and disease: Implications for therapeutic interventions in cancer and immune-mediated diseases. FASEB J. 2021, 35, e21260. [Google Scholar]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.-S.; Domm, J.; Eustace, B.K.; Foell, J.; De La Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar]
- Ma, C.C.; Wang, Z.L.; Xu, T.; He, Z.Y.; Wei, Y.Q. The approved gene therapy drugs worldwide: From 1998 to 2019. Biotechnol. Adv. 2020, 40, 107502. [Google Scholar]
- Zimran, E.; Papa, L.; Djedaini, M.; Patel, A.; Iancu-Rubin, C.; Hoffman, R. Expansion and preservation of the functional activity of adult hematopoietic stem cells cultured ex vivo with a histone deacetylase inhibitor. Stem Cells Transl. Med. 2020, 9, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Ellison, S.M.; Liao, A.; Wood, S.; Taylor, J.; Youshani, A.S.; Rowlston, S.; Parker, H.; Armant, M.; Biffi, A.; Chan, L.; et al. Pre-clinical safety and efficacy of lentiviral vector-mediated ex vivo stem cell gene therapy for the treatment of mucopolysaccharidosis IIIA. Mol. Ther.-Methods Clin. Dev. 2019, 13, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Godbout, K.; Tremblay, J.P. Prime editing for human gene therapy: Where are we now? Cells 2023, 12, 536. [Google Scholar] [CrossRef] [PubMed]
- De Ravin, S.S.; Li, L.; Wu, X.; Choi, U.; Allen, C.; Koontz, S.; Lee, J.; Theobald-Whiting, N.; Chu, J.; Garofalo, M.; et al. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci. Transl. Med. 2017, 9, eaah3480. [Google Scholar] [CrossRef]
- Azhagiri, M.K.K.; Babu, P.; Venkatesan, V.; Thangavel, S. Homology-directed gene-editing approaches for hematopoietic stem and progenitor cell gene therapy. Stem Cell Res. Ther. 2021, 12, 500. [Google Scholar] [PubMed]
- Zhang, M.; Zhu, Z.; Xun, G.; Zhao, H. To cut or not to cut: Next-generation genome editors for precision genome engineering. Curr. Opin. Biomed. Eng. 2023, 28, 100489. [Google Scholar] [CrossRef]
- Wang, J.Y.; Doudna, J.A. CRISPR technology: A decade of genome editing is only the beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prokopovich, A.K.; Litvinova, I.S.; Zubkova, A.E.; Yudkin, D.V. CXCR4 Is a Potential Target for Anti-HIV Gene Therapy. Int. J. Mol. Sci. 2024, 25, 1187. https://doi.org/10.3390/ijms25021187
Prokopovich AK, Litvinova IS, Zubkova AE, Yudkin DV. CXCR4 Is a Potential Target for Anti-HIV Gene Therapy. International Journal of Molecular Sciences. 2024; 25(2):1187. https://doi.org/10.3390/ijms25021187
Chicago/Turabian StyleProkopovich, Appolinaria K., Irina S. Litvinova, Alexandra E. Zubkova, and Dmitry V. Yudkin. 2024. "CXCR4 Is a Potential Target for Anti-HIV Gene Therapy" International Journal of Molecular Sciences 25, no. 2: 1187. https://doi.org/10.3390/ijms25021187
APA StyleProkopovich, A. K., Litvinova, I. S., Zubkova, A. E., & Yudkin, D. V. (2024). CXCR4 Is a Potential Target for Anti-HIV Gene Therapy. International Journal of Molecular Sciences, 25(2), 1187. https://doi.org/10.3390/ijms25021187




