Targeting of Specialized Metabolites Biosynthetic Enzymes to Membranes and Vesicles by Posttranslational Palmitoylation: A Mechanism of Non-Conventional Traffic and Secretion of Fungal Metabolites
Abstract
:1. Introduction
2. Diverse Localization of Secondary Metabolites Biosynthetic Enzymes
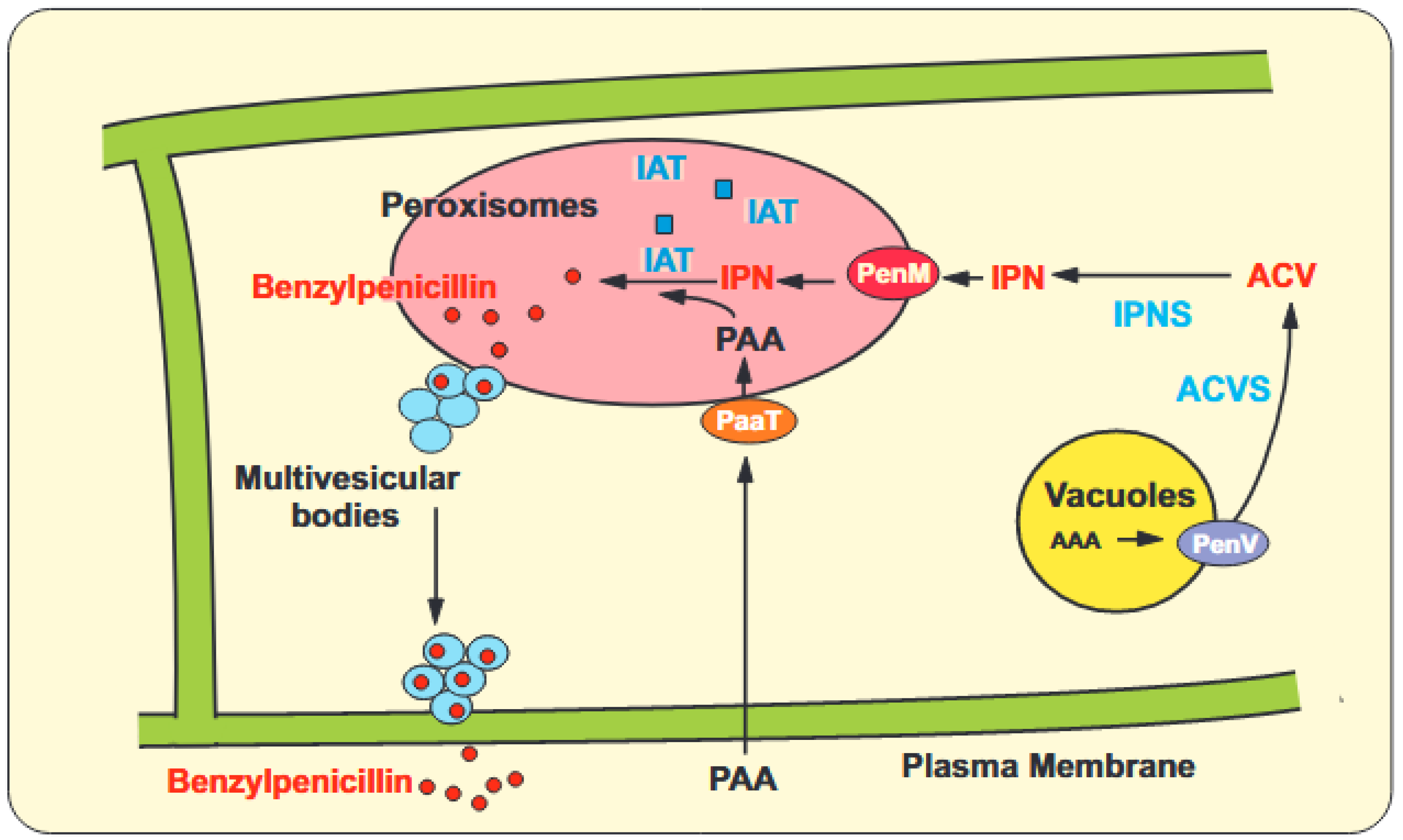
2.1. Penicillins, Cephalosporin C and Cyclosporins
2.2. Aflatoxins: Integration in Endosomes of Enzymes from Distinct Subcellular Compartments
2.3. The Alternaria Alternata AK Toxin Family: A Model of Polyketide Biosynthesis in Peroxisomes
2.4. Peroxisomal Enzymes Involved in the Biosynthesis of Fusarinine Siderophores
2.5. Trichothecenes: Localization in Toxisomes, a Novel Concept Involving Vesicles Fusions
2.6. Mycophenolic Acid: Diverse Localization of the Biosynthetic Enzymes
2.7. Peptidyl Alkaloids: An Extracellular Biosynthetic Enzyme
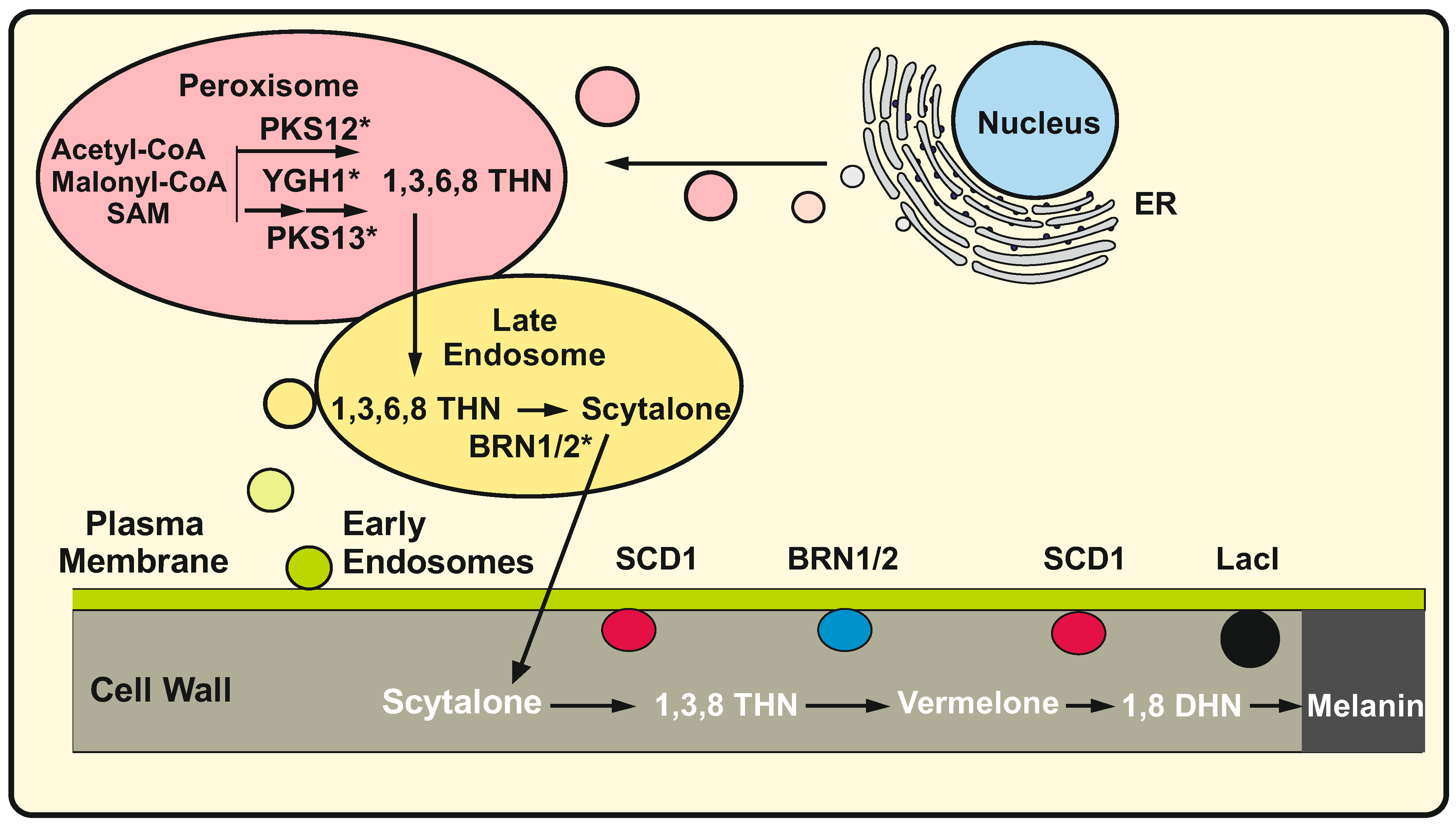
2.8. Melanin: The Second Half of the Pathway in Botrytis Cinerea Is Extracellular
2.9. Characteristic Features of Enzymes Involved in the Biosynthesis of Other Less Known Specialized Metabolites
3. Cytosolic Traffic of Peroxisomes, Vesicles and Endosomes: Peroxisomes Move to the Hyphal Tips by Hitchhiking on Early Endosomes
4. S-Acylation of Proteins: Palmitoylation
4.1. Palmitoylation of Proteins in Yeasts
4.2. Palmitoyl Transferases in Filamentous Fungi
4.2.1. Palmitoylation of the RAS GTPase in Basidiomycetes and Ascomycetes
4.2.2. Protein Palmitoylation in Plant Pathogenic Fungi: The Cargo Adaptor Protein Complex
5. Targeting of Specialized Metabolites Biosynthetic Enzymes to Vesicles/Endosomes by Posttranslational Palmitoylation
6. Connection between Protein Palmitoylation and the Calcium/Calcineurin Regulatory Cascade
Palmitoylation of Calcineurin Targets This Phosphatase to the Golgi and Plasma Membrane
7. Conclusions and Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Davies, J. Small molecules: The lexicon of biodiversity. J. Biotechnol. 2007, 129, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Kück, U.; Bloemendal, S.; Teichert, I. Putting fungi to work: Harvesting a cornucopia of drugs, toxins, and antibiotics. PLoS Pathog. 2014, 10, e1003950. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Cary, J.W. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 2015, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Bandara Herath, H.M.T.; Jacob, M.; Wilson, A.D.; Abbas, H.K.; Nanayakkara, N.P.D. New compounds and secondary metabolites from bioactive extracts of the fungus Armillaria tabescens. Nat. Prod. Res. 2013, 27, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.H.; Bovenberg, R.A.; Groothuis, M.H.; Kattevilder, F.; Smaal, E.B.; Van der Voort, L.H.; Verkleij, A.J. Involvement of microbodies in penicillin biosynthesis. Biochim. Biophys. Acta 1992, 1116, 210–213. [Google Scholar] [CrossRef]
- Brocard, C.; Harting, A. Peroxisome targeting signal 1: Is it really a simple tripeptide? Biochim. Biophys. Acta 2006, 1763, 1565–1573. [Google Scholar] [CrossRef]
- Kiel, J.A.K.W.; van den Berg, M.A.; Fusetti, F.; Poolman, B.; Bovenberg, R.A.L.; Veenhuis, M.; van der Klei, I.J. Matching the proteome to the genome: The microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genom. 2009, 9, 167–184. [Google Scholar] [CrossRef]
- Imazaki, A.; Tanaka, A.; Harimoto, Y.; Yamamoto, M.; Akimitsu, K.; Park, P.; Tsuge, T. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot. Cell 2010, 9, 682–694. [Google Scholar] [CrossRef]
- Meijer, W.H.; Gidijala, L.; Fekken, S.; Kiel, J.A.K.W.; van den Berg, M.A.; Lascaris, R.; Bovenberg, R.A.L.; van der Klei, I.J. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 2010, 76, 5702–5709. [Google Scholar] [CrossRef]
- Martín, J.F.; Ullán, R.V.; García-Estrada, C. Role of peroxisomes in the biosynthesis and secretion of β-lactams and other secondary metabolites. J. Ind. Microbiol. Biotechnol. 2012, 39, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Lamas-Maceiras, M.; Vaca, I.; Rodríguez, E.; Casqueiro, J.; Martín, J.F. Amplification and disruption of the phenylacetyl-CoA ligase gene of Penicillium chrysogenum encoding an aryl-capping enzyme that supplies phenylacetic acid to the isopenicillin N-acyltransferase. Biochem. J. 2006, 395, 147–155. [Google Scholar] [CrossRef] [PubMed]
- García-Estrada, C.; Vaca, I.; Fierro, F.; Sjollema, K.; Veenhuis, M.; Martín, J.F. The unprocessed preprotein form IATC103S of the isopenicillin N acyltransferase is transported inside peroxisomes and regulates its self-processing. Fungal Genet. Biol. 2008, 45, 1043–1052. [Google Scholar] [CrossRef] [PubMed]
- Ullán, R.V.; Teijeira, F.; Guerra, S.M.; Vaca, I.; Martín, J.F. Characterization of a novel peroxisome membrane protein essential for conversion of isopenicillin N into cephalosporin C. Biochem. J. 2010, 432, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Meyers, D.M.; Obrian, G.; Du, W.L.; Bhatnagar, D.; Payne, G.A. Characterization of AFLJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl. Environ. Microbiol. 1998, 64, 3713–3717. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Mack, B.M.; Wei, Q.; Li, P.; Roze, L.V.; Dazzo, F.; Cary, J.W.; Bhatnagar, D.; Linz, J.E. Association with AflR in endosomes reveals new functions for AflJ in aflatoxin biosynthesis. Toxins 2012, 4, 1582–1600. [Google Scholar] [CrossRef]
- Tanaka, A.; Shiotani, H.; Yamamoto, M.; Tsuge, T. Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 1999, 12, 691–702. [Google Scholar] [CrossRef]
- Tanaka, A.; Tsuge, T. Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 2000, 13, 975–986. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Masunaka, A.; Tsuge, T.; Yamamoto, M.; Ohtani, K.; Fukumoto, T.; Gomi, K.; Peever, T.L.; Tada, Y.; Ichimura, K.; et al. ACTTS3 encoding a polyketide synthase is essential for the biosynthesis of ACT-toxin and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 2010, 23, 406–414. [Google Scholar] [CrossRef]
- Zhang, W.; Du, L.; Qu, Z.; Zhang, X.; Li, F.; Li, Z.; Qi, F.; Wang, X.; Jiang, Y.; Men, P.; et al. Compartmentalized biosynthesis of mycophenolic acid. Proc. Natl. Acad. Sci. USA 2019, 116, 13305–13310. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, C.; Na, Y.; Ren, D.; Zhang, C.; He, Y.; Wang, Y.; Xiang, S.; Ren, W.; Xu, L.; et al. Compartmentalization of Melanin Biosynthetic Enzymes Contributes to Self-Defense against Intermediate Compound Scytalone in Botrytis cinerea. MBio 2021, 12, e00007-21. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Xu, X.; Lowry, D.; Jackson, J.C.; Roberson, R.W.; Lin, X. Subcellular Compartmentalization and Trafficking of the Biosynthetic Machinery for Fungal Melanin. Cell Rep. 2016, 14, 2511–2518. [Google Scholar] [CrossRef]
- Upadhyay, S.; Xu, X.; Lin, X. Interactions between Melanin Enzymes and Their Atypical Recruitment to the Secretory Pathway by Palmitoylation. mBio 2016, 7, e01925-16. [Google Scholar] [CrossRef]
- Saikia, S.; Scott, B. Functional analysis and subcellular localization of two geranylgeranyl diphosphate synthases from Penicillium paxillin. Mol. Genet. Genom. 2009, 282, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Gründlinger, M.; Yasmin, S.; Lechner, B.E.; Geley, S.; Schrettl, M.; Hynes, M.; Haas, H. Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol. 2013, 88, 862–875. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Linz, J.E. Functional expression and subcellular localization of the aflatoxin pathway enzyme Ver-1 fused to enhanced green fluorescent protein. Appl. Environ. Microbiol. 2008, 74, 6385–6396. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-Y.; Linz, J.E. Functional expression and sub-cellular localization of the early aflatoxin pathway enzyme Nor-1 in Aspergillus parasiticus. Mycol. Res. 2009, 113, 591–601. [Google Scholar] [CrossRef]
- Lee, L.W.; Chiou, C.H.; Klomparens, K.L.; Cary, J.W.; Linz, J.E. Subcellular localization of aflatoxin biosynthetic enzymes Nor-1, Ver-1, and OmtA in time-dependent fractionated colonies of Aspergillus parasiticus. Arch. Microbiol. 2004, 181, 204–214. [Google Scholar] [CrossRef]
- Menke, J.; Weber, J.; Broz, K.; Kistler, H.C. Cellular development associated with induced mycotoxin synthesis in the filamentous fungus Fusarium graminearum. PLoS ONE 2013, 8, e63077. [Google Scholar] [CrossRef]
- Kistler, H.C.; Broz, K. Cellular compartmentalization of secondary metabolism. Front. Microbiol. 2015, 6, 68. [Google Scholar] [CrossRef]
- Boenish, M.J.; Broz, K.L.; Purvine, S.O.; Chrisler, W.B.; Nicora, C.D.; Connolly, L.R.; Freitag, M.; Baker, S.E.; Kistler, H.C. Structural reorganization of the fungal endoplasmic reticulum upon induction of mycotoxin biosynthesis. Sci. Rep. 2017, 7, 44296. [Google Scholar] [CrossRef]
- Ames, B.D.; Walsh, C.T. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry 2010, 49, 3351–3365. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.D.; Haynes, S.W.; Gao, X.; Evans, B.S.; Kelleher, N.L.; Tang, Y.; Walsh, C.T. Complexity generation in fungal peptidyl alkaloid biosynthesis: Oxidation of fumiquinazoline A to the heptacyclic hemiaminal fumiquinazoline C by the flavoenzyme Af12070 from Aspergillus fumigatus. Biochemistry 2011, 50, 8756–8769. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.Y.; Keller, N.P. Spatial and temporal control of fungal natural product synthesis. Nat. Prod. Rep. 2014, 31, 1277–1286. [Google Scholar] [CrossRef]
- Aharonowitz, Y.; Bergmeyer, J.; Cantoral, J.M.; Cohen, G.; Demain, A.L.; Fink, U.; Kinghorn, J.; Kleinkauf, H.; MacCabe, A.; Palissa, H.; et al. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase, the multienzyme integrating the four primary reactions in beta-lactam biosynthesis, as a model peptide synthetase. Biotechnology 1993, 11, 807–810. [Google Scholar]
- Müller, W.H.; van der Krift, T.P.; Krouwer, A.J.; Wösten, H.A.; van der Voort, L.H.; Smaal, E.B.; Verkleij, A.J. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 1991, 10, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Van der Lende, T.R.; van de Kamp, M.; van den Berg, M.; Sjollema, K.; Bovenberg, R.A.; Veenhuis, M.; Koning, W.N.; Driessen, A.J.M. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase, that mediates the first committed step in penicillin biosynthesis, is a cytosolic enzyme. Fungal Genet. Biol. 2002, 37, 49–55. [Google Scholar] [CrossRef]
- Martín, J.F. Transport systems, intracellular traffic of intermediates and secretion of β-lactam antibiotics in fungi. Fungal Biol. Biotechnol. 2020, 7, 6. [Google Scholar] [CrossRef]
- Campos, C.; Lázaro-Rodríguez, T.G.; Fragoso-Soriano, R.; Fernández, F.J. Vesicular transport and secretion of penicillin G in Penicillium rubens P2-32-T. Arch. Microbiol. 2020, 202, 1257–1262. [Google Scholar] [CrossRef]
- Alvarez, E.; Cantoral, J.M.; Barredo, J.L.; Díez, B.; Martín, J.F. Purification to homogeneity and characterization of the acyl-CoA:6-APA acyltransferase of Penicillium chrysogenum. Antimicrob. Agents Chemother. 1987, 31, 1675–1682. [Google Scholar] [CrossRef]
- Aplin, R.T.; Baldwin, J.E.; Roach, P.L.; Robinson, C.V.; Schofield, C.J. Investigations into the post-translational modification and mechanism of isopenicillin N: Acyl-CoA acyltransferase using electrospray mass spectrometry. Biochem. J. 1993, 1294, 357–363. [Google Scholar] [CrossRef]
- Tobin, M.B.; Cole, S.C.; Miller, J.R.; Baldwin, J.E.; Sutherland, J.D. Amino-acid substitutions in the cleavage site of acyl-coenzyme A: Isopenicillin N acyltransferase from Penicillium chrysogenum: Effect on proenzyme cleavage and activity. Gene 1995, 162, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Koetsier, M.J.; Jekel, P.A.; van den Berg, M.A.; Bovenberg, R.A.; Janssen, D.B. Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem. J. 2009, 417, 467–476. [Google Scholar] [CrossRef]
- Koetsier, M.J.; Gombert, A.K.; Fekken, S.; Bovenberg, R.A.; van den Berg, M.A.; Kiel, J.A.W.; Jekel, P.A.; Janssen, D.B.; Pronk, J.T.; van der Klei, I.; et al. The Penicillium chrysogenum aclA gene encodes a broad-substrate-specificity acyl-coenzyme A ligase involved in activation of adipic acid, a side-chain precursor for cephem antibiotics. Fungal Genet. Biol. 2010, 47, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.H.; Essers, J.; Humbel, B.M.; Verkleij, A.J. Enrichment of Penicillium chrysogenum microbodies by isopycnic centrifugation in nycodenz as visualized with immuno-electron microscopy. Biochim. Biophys. Acta 1995, 1245, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Spröte, P.; Hynes, M.J.; Hortschansky, P.; Shelesty, E.; Scharf, D.H.; Wolke, S.M.; Brakhage, A.A. Identification of the novel penicillin biosynthesis gene aatB of Aspergillus nidulans and its putative evolutionary relationship to this fungal secondary metabolism gene cluster. Mol. Microbiol. 2008, 70, 445–461. [Google Scholar] [CrossRef] [PubMed]
- García-Estrada, C.; Barreiro, C.; Jami, M.-S.; Martín-González, J.; Martín, J.F. The inducers 1,3-diaminopropane and spermidine cause the reprogramming of metabolism in Penicillium chrysogenum, leading to multiple vesicles and penicillin overproduction. J. Proteom. 2013, 85, 129–159. [Google Scholar] [CrossRef] [PubMed]
- Ullán, R.V.; Casqueiro, J.; Bañuelos, O.; Fernández, F.J.; Gutiérrez, S.; Martín, J.F. A novel epimerization system in fungal secondary metabolism involved in the conversion of isopenicillin N into penicillin N in Acremonium chrysogenum. J. Biol. Chem. 2002, 277, 46216–46225. [Google Scholar] [CrossRef]
- Hopper, M.; Gentzsch, C.; Schorgendorfer, K. Structure and localization of cyclosporine synthetase, the key enzyme of cyclosporine biosynthesis in Tolypocladium inflatum. Arch. Microbiol. 2001, 176, 285–293. [Google Scholar] [CrossRef]
- Fernández-Aguado, M.; Teijeira, F.; Martín, J.F.; Ullán, R.V. A vacuolar membrane protein affects drastically the biosynthesis of the ACV tripeptide and the beta-lactam pathway of Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2013, 97, 795–808. [Google Scholar] [CrossRef]
- Roze, L.V.; Chanda, A.; Linz, J.E. Compartmentalization and molecular traffic in secondary metabolism: A new understanding of established cellular processes. Fungal Genet. Biol. 2011, 48, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Maggio-Hall, L.A.; Wilson, R.A.; Keller, N.P. Fundamental contribution of beta-oxidation to polyketide mycotoxin production in planta. Mol. Plant Microbe Interact. 2005, 18, 783–793. [Google Scholar] [CrossRef]
- Fernández-Aguado, M.; Ullán, R.V.; Teijeira, F.; Rodríguez-Castro, R.; Martín, J.F. The transport of phenylacetic acid across the peroxisomal membrane is mediated by the PaaT protein in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2013, 97, 3073–3084. [Google Scholar] [CrossRef]
- Chiou, C.H.; Lee, L.W.; Owens, S.A.; Whallon, J.H.; Klomparens, K.L.; Townsend, C.A.; Linz, J.E. Distribution and sub-cellular localization of the aflatoxin enzyme versicolorin B synthase in time-fractionated colonies of Aspergillus parasiticus. Arch. Microbiol. 2004, 182, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Roze, L.V.; Linz, J.E. A possible role for exocytosis in aflatoxin export in Aspergillus parasiticus. Eukaryot. Cell 2010, 9, 1724–1727. [Google Scholar] [CrossRef] [PubMed]
- Linz, J.E.; Chanda, A.V.; Hong, S.Y.; Whitten, D.A.; Wilkerson, C.; Roze, L.V. Proteomic and biochemical evidence support a role for transport vesicles and endosomes in stress response and secondary metabolism in Aspergillus parasiticus. J. Proteome Res. 2012, 11, 767–775. [Google Scholar] [CrossRef]
- Linz, J.E.; Wee, J.M.; Roze, L.V. Aflatoxin Biosynthesis; Regulation and subcellular localization. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.F., García-Estrada, C., Zeilinger, S., Eds.; Springer: New York, NY, USA, 2014; Volume 1, pp. 89–110. [Google Scholar]
- Chang, P.K. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Genet. Genom. 2003, 268, 711–719. [Google Scholar] [CrossRef]
- Molloy, S. Fungal physiology: Reaching the right location. Nat. Rev. Microbiol. 2014, 12, 396–397. [Google Scholar] [CrossRef]
- Salogiannis, J.; Egan, M.J.; Reck-Peterson, S.L. Peroxisomes move by hitchhiking on early endosomes using the novel linker protein PxdA. J. Cell Biol. 2016, 212, 289–296. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Luo, Z.; Li, R.; Zhang, Y.; Kalaji, H.M.; Quiang, S.; Chen, S. Recent Advances in Alternaria Phytotoxins: A Review of Their Occurrence, Structure, Bioactivity, and Biosynthesis. J. Fungi 2022, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Lazarow, P.B. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 2006, 1763, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Eisendle, M.; Oberegger, H.; Zadra, I.; Haas, H. The siderophore system is essential for viability of Aspergillus nidulans: Functional analysis of two genes encoding l-ornithine N5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 2003, 49, 359–375. [Google Scholar] [CrossRef] [PubMed]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef]
- Olucha, J.; Meneely, K.M.; Chilton, A.S.; Lamb, A.L. Two structures of an N-hydroxylating flavoprotein monooxygenase: Ornithine hydroxylase from Pseudomonas aeruginosa. J. Biol. Chem. 2011, 286, 31789–31798. [Google Scholar] [CrossRef]
- Yasmin, S.; Alcazar-Fuoli, L.; Grundlinger, M.; Puempel, T.; Cairns, T.; Blatzer, M.; López, J.F.; Grimalt, J.O.; Bignell, E.; Haas, H. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. USA 2011, 109, 497–504. [Google Scholar] [CrossRef]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Kim, H.S.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar] [CrossRef]
- Pestka, J.J.; Smolinski, A.T. Deoxynivalenol: Toxicology and potential effects on humans. J. Toxicol. Environ. Health B 2005, 8, 39–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, S.; Zhu, G.; Zhao, Z.; Wang, H.; Zhou, M.; Zhao, X.; Wu, A. Identification of Candidate Genes Associated with Trichothecene Biosynthesis in Fusarium graminearum Species Complex Combined with Transcriptomic and Proteomic Analysis. Microorganisms 2022, 10, 1479. [Google Scholar] [CrossRef]
- Kimura, M.; Anzai, H.; Yamaguchi, I. Microbial toxins in plant-pathogen interactions: Biosynthesis, resistance mechanisms, and significance. J. Gen. Appl. Microbiol. 2001, 47, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Menke, J.; Dong, Y.; Kistler, H.C. Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation . Mol. Plant Microbe Interact. 2012, 25, 1408–1418. [Google Scholar] [CrossRef]
- Fernández-Aguado, M.; Martín, J.F.; Rodríguez-Castro, R.; García-Estrada, C.; Albillos, S.M.; Teijeira, F.; Ullan, R.V. New insights into the isopenicillin N transport in Penicillium chrysogenum. Metabolic. Eng. 2014, 22, 89–103. [Google Scholar] [CrossRef]
- Teijeira, F.; Ullán, R.V.; Guerra, S.M.; García-Estrada, C.; Vaca, I.; Martín, J.F. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem. J. 2009, 418, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Regueira, T.B.; Kildgaard, K.R.; Hansen, B.G.; Mortensen, U.H.; Hertweck, C.; Nielsen, J. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl. Environ. Microbiol. 2011, 77, 3035–3043. [Google Scholar] [CrossRef]
- Del-Cid, A.; Gil-Durán, C.; Vaca, I.; Rojas-Aedo, J.F.; García-Rico, R.O.; Levicán, G.; Chávez, R. Identification and Functional Analysis of the Mycophenolic Acid Gene Cluster of Penicillium roqueforti. PLoS ONE 2016, 11, e0147047. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, S.; Qiu, L.; Qi, F.; Li, Z.; Yang, Y.; Huang, S.; Bai, F.; Liu, C.; Wan, X.; et al. Functional characterization of MpaG0, the O-methyltransferase involved in the biosynthesis of mycophenolic acid. ChemBioChem 2015, 16, 565–569. [Google Scholar] [CrossRef]
- Lim, F.Y.; Ames, B.; Walsh, C.T.; Keller, N.P. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell Microbiol. 2014, 16, 1267–1283. [Google Scholar] [CrossRef] [PubMed]
- Resende, D.I.S.P.; Boonpothing, P.; Sousa, E.; Kijjoa, A.; Pinto, M. Chemistry of the fumiquinazolines and structurally related alkaloids. Nat. Prod. Rep. 2019, 26, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Franzen, A.J.; Cunha, M.M.L.; Miranda, K.; Hentschel, J.; Plattner, H.; da Silva, M.B.; Salgado, C.G.; de Souza, W.; Rozental, S. Ultrastructural characterization of melanosomes of the human pathogenic fungus Fonsecaea pedrosoi. J. Struct. Biol. 2008, 162, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F. Interaction of calcium responsive proteins and transcriptional factors with the PHO regulon in yeasts and fungi. Front. Cell Dev. Biol. 2023, 11, 1225774. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Torres, G.; Lin, X. Laccases involved in 1,8-dihydroxynaphthalene melanin biosynthesis in Aspergillus fumigatus are regulated by developmental factors and copper homeostasis. Eukaryot. Cell 2013, 12, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
- Eisenman, H.C.; Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef]
- Schumacher, J. DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 2016, 99, 729–748. [Google Scholar] [CrossRef]
- Ao, J.; Bandyopadhyay, S.; Free, S.J. Characterization of the Neurospora crassa DHN melanin biosynthetic pathway in developing ascospores and peridium cells. Fungal Biol. 2019, 123, 1–9. [Google Scholar] [CrossRef]
- Fujii, I.; Yasukoa, Y.; Tsai, H.-F.; Chang, Y.C.; Known-Chung, K.J.; Ebizuka, Y. Hydrolytic polyketide shortening by Ayg1p, a novel enzyme involved in fungal melanin biosynthesis. J. Biol. Chem. 2004, 279, 44613–44620. [Google Scholar] [CrossRef]
- Sugareva, V.; Hartl, A.; Brock, M.; Hubner, K.; Rohde, M.; Heinekamp, T.; Brakhage, A.A. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch. Microbiol. 2006, 186, 345–355. [Google Scholar] [CrossRef]
- Cundliffe, E.; Demain, A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 2010, 37, 643–672. [Google Scholar] [CrossRef] [PubMed]
- Skellam, E. Subcellular localization of fungal specialized metabolites. Fungal Biol. Biotechnol. 2022, 9, 11–35. [Google Scholar] [CrossRef] [PubMed]
- Young, C.; McMillan, L.; Telfer, E.; Scott, B. Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli. Mol. Microbiol. 2001, 39, 754–764. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zong, Y.; Shang, Y.; Zhang, Z.; Xu, X.; Wang, X.; Long, M.; Tian, S. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019, 21, 1124–1139. [Google Scholar] [CrossRef] [PubMed]
- Tannous, J.; El Khoury, R.; Snini, S.P.; Lippi, Y.; Khoury, A.-E.; Atoui, A.; Lteif, R.; Oswald, I.P.; Puel, O. Sequencing, physical organization and kinetic expression of the patulin biosynthetic gene cluster from Penicillium expansum. Int. J. Food Microbiol. 2014, 189, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Snini, S.P.; Tadrist, S.; Laffitte, J.; Jamin, E.L.; Oswald, I.P.; Puel, O. The gene patG involved in the biosynthesis pathway of patulin, a food-bourne mycotoxin, encodes a 6-methylsalicylic acid decarboxylase. Int. J. Food Microbiol. 2014, 171, 77–83. [Google Scholar] [CrossRef]
- Urquhart, A.S.; Hu, J.; Chooi, Y.H.; Idnurm, A. The fungal gene cluster for biosynthesis of the antibacterial agent viriditoxin. Fungal Biol. Biotechnol. 2019, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Talesh, N.; Horio, T.; Araujo-Bazán, L.; Xiaowei Dou, X.; Espeso, E.A.; Peñalva, M.A.; Osmani, S.A.; Oakley, B.R. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 2008, 19, 1439–1449. [Google Scholar] [CrossRef]
- Horio, T.; Oakley, B.R. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol. Biol. Cell 2005, 16, 918–926. [Google Scholar] [CrossRef]
- Egan, M.J.; McClintock, M.A.; Reck-Peterson, S.L. Microtubule based transport in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 637–645. [Google Scholar] [CrossRef]
- Higuchi, Y.; Ashwin, P.; Roger, Y.; Steinberg, G. Early endosome motility spatially organizes polysome distribution. J. Cell Biol. 2014, 204, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, S.C.; Schuster, M.; Bielska, E.; Dagdas, G.; Kilaru, S.; Meadows, B.R.; Schrader, M.; Steinberg, G. Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J. Cell Biol. 2015, 211, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Baumann, S.; Pohlmann, T.; Jungbluth, M.; Brachmann, A.; Feldbrügge, M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 2012, 125, 2740–2752. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; Schuster, M.; Roger, Y.; Berepiki, A.; Soanes, D.M.; Talbot, N.J.; Steinberg, G. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J. Cell Biol. 2014, 204, 989–1007. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Liras, P.; Sánchez, S. Modulation of Gene Expression in Actinobacteria by Translational Modification of Transcriptional Factors and Secondary Metabolite Biosynthetic Enzymes. Front. Microbiol. 2021, 12, 630694. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.D.; Triplett, L.; Chen, X.L. Emerging roles of posttranslational modifications in plant-pathogenic fungi and bacteria. Annu. Rev. Phytopathol. 2021, 59, 99–124. [Google Scholar] [CrossRef]
- Resh, M.D. Trafficking and signalling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006, 2, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.F.; Junmei Wan, J.; Bailey, A.O.; Sun, B.; Kuchar, J.A.; Green, W.N.; Phinney, B.S.; Yates, J.R., III; Davis, N.G. Global Analysis of Protein Palmitoylation in Yeast. Cell 2006, 125, 1003–1013. [Google Scholar] [CrossRef]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef]
- Martín, J.F.; van den Berg, M.A.; Ver Loren van Themaat, E.; Liras, P. Sensing and transduction of nutritional and chemical signals in filamentous fungi: Impact on cell development and secondary metabolites biosynthesis. Biotechnol. Adv. 2019, 37, 107392. [Google Scholar] [CrossRef]
- O’Brien, P.J.; Zatz, M. Acylation of bovine rhodopsin by [3H]palmitic acid. J. Biol. Chem. 1984, 259, 5054–5057. [Google Scholar] [CrossRef]
- Liang, X.; Nazarian, A.; Erdjument-Bromage, H.; Bornmann, W.; Tempst, P.; Resh, M.D. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J. Biol. Chem. 2001, 276, 30987–30994. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, L.V.; Serebryakova, M.V.; Baratova, L.A.; Veit, M. S acylation of the hemagglutinin of influenza viruses: Mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 2008, 82, 9288–9292. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.J.; Mitchell, D.A.; Dong, X.; Deschenes, R.J. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 6775–6787. [Google Scholar] [CrossRef] [PubMed]
- Lobo, S.; Greentree, W.K.; Linder, M.E.; Deschenes, R.J. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 41268–41273. [Google Scholar] [CrossRef]
- Grassie, M.A.; McCallum, J.F.; Guzzi, F.; Magee, A.I.; Milligan, G.; Parenti, M. The palmitoylation status of the G-protein G(o)1 alpha regulates its activity of interaction with the plasma membrane. Biochem. J. 1994, 302, 913–920. [Google Scholar] [CrossRef]
- Nichols, C.B.; Ost, K.S.; Grogan, D.P.; Pianalto, K.; Hasan, S.; Alspaugh, J.A. Impact of protein palmitoylation on the virulence potential of Cryptococcus neoformans. Eukaryot. Cell 2015, 14, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Tirado, F.H.; Peng, T.; Yang, M.; Hang, H.; Doering, T.L. A single protein S-acyl transferase acts through diverse substrates to determine Cryptococcal morphology, stress tolerance, and pathogenic outcome. PLoS Pathog. 2015, 11, e1004908. [Google Scholar] [CrossRef]
- González Montoro, A.; Chumpen Ramírez, S.; Valdez-Taubas, J. The canonical DHHC motif is not absolutely required for the activity of the yeast S-acyltransferases Swf1 and Pfa4. J. Biol. Chem. 2015, 290, 22448–22449. [Google Scholar] [CrossRef]
- Hagiwara, D.; Kondo, A.; Fujioka, T.; Abe, K. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr. Genet. 2008, 54, 325–338. [Google Scholar] [CrossRef]
- Greaves, J.; Prescott, G.R.; Gorleku, O.A.; Chamberlain, L.H. The fat controller: Roles of palmitoylation in intracellular protein trafficking and targeting to membrane microdomains. Mol. Membr. Biol. 2009, 26, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Greaves, J.; Chamberlain, K.H. Palmitoylation-dependent protein sorting. J. Cell Biol. 2007, 176, 249–254. [Google Scholar] [CrossRef]
- Dietrich, L.E.; Ungermann, C. On the mechanism of protein palmitoylation. EMBO Rep. 2004, 5, 1053–1057. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. The PenV vacuolar membrane protein that controls penicillin biosynthesis is a putative member of a subfamily of stress-gated transient receptor calcium channels. Curr. Res. Biotechnol. 2021, 3, 317–322. [Google Scholar] [CrossRef]
- Valdez-Taubas, J.; Pelham, H. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 2005, 24, 2524–2532. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; John Peter, A.T.; Meiringer, C.; Subramanian, K.; Ungermann, C. Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic 2009, 10, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, Q.; Sun, C.; Song, J.; Gao, L.; Zhang, S.; Muñoz, A.; Read, N.D.; Lu, L. Palmitoylation of the Cysteine Residue in the DHHC Motif of a Palmitoyl Transferase Mediates Ca2+ Homeostasis in Aspergillus. PLoS Genet. 2016, 12, e1005977. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.A.; Mitchell, G.; Ling, Y.; Budde, C.; Deschenes, R.J. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 2010, 285, 38104–38114. [Google Scholar] [CrossRef]
- Chapa-y-Lazo, B.; Allwood, E.G.; Smaczynska-De Rooij, I.I.; Snape, M.L.; Ayscough, K.R. Yeast endocytic adaptor AP-2 binds the stress sensor Mid2 and functions in polarized cell responses. Traffic 2014, 15, 546–557. [Google Scholar] [CrossRef]
- Fortwendel, J.R.; Juvvadi, P.R.; Rogg, L.E.; Asfaw, Y.G.; Burns, K.A.; Randell, S.H.; Steinbach, W.J. Plasma membrane localization is required for RasA mediated polarized morphogenesis and virulence of Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 966–977. [Google Scholar] [CrossRef]
- Al Abdallah, Q.; Martin-Vicente, A.; Souza, A.C.O.; Ge, W.; Fortwendel, J.R. C-terminus proteolysis and palmitoylation cooperate for optimal plasma membrane localization of RasA in Aspergillus. Front. Microbiol. 2018, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yun, Y.; Lou, Y.; Abubakar, Y.S.; Guo, P.; Wang, S.; Li, C.; Feng, Y.; Adnan, M.; Zhou, J.; et al. FgAP-2 complex is essential for pathogenicity and polarised growth and regulates the apical localisation of membrane lipid flippases in Fusarium graminearum. Cell Microbiol. 2019, 21, e13041. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Wang, C.Y.; Zhang, X.; Wang, Q.; Lin, L. VdNUC-2, the key regulator of phosphate responsive signalling pathway, is required for Verticillium dahliae infection. PLoS ONE 2015, 10, e0145190. [Google Scholar] [CrossRef]
- Wan, J.; Roth, A.F.; Bailey, A.O.; Davis, N.G. Palmitoylated proteins: Purification and identification. Nat. Protoc. 2007, 2, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Gao, Y.; Wang, J.; Wang, H.; Lou, J.; Bi, Y.; Yan, Y.; Li, D.; Song, F. Palmitoyl Transferase FonPAT2-Catalyzed Palmitoylation of the FonAP-2 Complex Is Essential for Growth, Development, Stress Response, and Virulence in Fusarium oxysporum sp. niveum. Microbiol. Spectr. 2023, 11, e0386122. [Google Scholar] [CrossRef] [PubMed]
- Turrà, D.; Di Pietro, A. Chemotropic sensing in fungus-plant interactions. Curr. Opin. Plant. Biol. 2015, 26, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.J.; Collins, B.M.; Evans, P.R. Adaptors for clathrin coats: Structure and function. Annu. Rev. Cell. Dev. Biol. 2004, 20, 153–191. [Google Scholar] [CrossRef]
- Knafler, H.C.; Smaczynska-de Rooij, I.I.; Walker, L.A.; Lee, K.K.; Gow, N.A.R.; Ayscough, K.R. AP-2-dependent endocytic recycling of the chitin synthase Chs3 regulates polarized growth in Candida albicans. mBio 2019, 10, e02421-18. [Google Scholar] [CrossRef]
- de León, N.; Hoya, M.; Curto, M.A.; Moro, S.; Yanguas, F.; Doncel, C.; Valdivieso, M.H. The AP-2 complex is required for proper temporal and spatial dynamics of endocytic patches in fission yeast. Mol. Microbiol. 2016, 100, 409–424. [Google Scholar] [CrossRef]
- Martzoukou, O.; Amillis, S.; Zervakou, A.; Christoforidis, S.; Diallinas, G. The AP-2 complex has a specialized clathrin-independent role in apical endocytosis and polar growth in fungi. elife 2017, 6, e20083. [Google Scholar] [CrossRef]
- Charron, G.; Zhang, M.M.; Yount, J.S.; Wilson, J.; Raghavan, A.S.; Shamir, E.; Hang, H.C. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 2009, 131, 4967–4975. [Google Scholar] [CrossRef] [PubMed]
- Godio, R.P.; Fouces, R.; Martín, J.F. A squalene epoxidase is involved in biosynthesis of both the antitumor compound clavaric acid and sterols in the basidiomycete H. sublateritium. Chem. Biol. 2007, 14, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Abe, I.; Prestwich, G.D. Squalene epoxidase and oxidosqualene:lanosterol cyclase; key enzymes in cholesterol biosynthesis. In Comprehensive Natural Products Chemistry; Barton, D.H.R., Nakanishi, K., Eds.; Elsevier: Oxford, UK, 1999; Volume 2, pp. 267–298. [Google Scholar]
- Zhang, S.; Zheng, H.; Long, N.; Carbo, N.; Chen, P.; Aguilar, P.S.; Lu, L. FigA, a putative homolog of low-affinity calcium system member Fig1 in Saccharomyces cerevisiae, is involved in growth and asexual and sexual development in Aspergillus nidulans. Eukaryot. Cell 2014, 13, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F. Vacuolal and Peroxisomal Calcium Ion Transporters in Yeasts and Fungi: Key Role in the Translocation of Intermediates in the Biosynthesis of Fungal Metabolites. Genes 2022, 13, 1450. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ortiz, P.; Espeso, E.A. Spatiotemporal dynamics of the calcineurin target CrzA. Cell Signal. 2017, 29, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, K.W. Acidic calcium stores of Saccharomyces cerevisiae. Cell Calcium 2011, 50, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Zelter, A.; Bencina, M.; Bowman, B.J.; Yarden, O.; Read, N.D. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet. Biol. 2004, 41, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, M.; Nastase, K.K.; Cunningham, K.W. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002, 21, 2343–2353. [Google Scholar] [CrossRef]
- Yoshimoto, H.; Saltsman, K.; Gasch, A.P.; Li, H.X.; Ogawa, N.; Botstein, D.; O’Brown, P.; Cyert, M.S. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 31079–31088. [Google Scholar] [CrossRef]
- Kaur, R.; Castaño, I.; Cormack, B.P. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: Roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 2004, 48, 1600–1613. [Google Scholar] [CrossRef]
- Hickey, P.C.; Read, N.D. Imaging living cells of Aspergillus in vitro. Med. Mycol. 2009, 47 (Suppl. 1), S110–S119. [Google Scholar] [CrossRef] [PubMed]
- Pantazopoulou, A.; Peñalva, M.A. Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol. Biol. Cell 2009, 20, 4335–4347. [Google Scholar] [CrossRef] [PubMed]
- Ulengin-Talkish, I.; Parson, M.A.H.; Jenkins, M.L.; Roy, J.; Shih, A.Z.L.; St-Denis, N.; Gulyas, G.; Balla, T.; Gingras, A.-C.; Várnai, P.; et al. Palmitoylation targets the calcineurin phosphatase to the phosphatidylinositol 4-kinase complex at the plasma membrane. Nat. Commun. 2021, 12, 6064. [Google Scholar] [CrossRef] [PubMed]
- Bond, R.; Ly, N.; Cyert, M.S. The unique C terminus of the calcineurin isoform CNAβ1 confers non-canonical regulation of enzyme activity by Ca2+ and calmodulin. J. Biol. Chem. 2017, 292, 16709–16721. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Cyert, M.S. Identifying New Substrates and Functions for an Old Enzyme: Calcineurin. Cold Spring Harb. Perspect. Biol. 2020, 12, a035436. [Google Scholar]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef]
- Greaves, J.; Chamberlain, L.H. DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends Biochem. Sci. 2011, 36, 245–253. [Google Scholar] [CrossRef]
- Varghese, R.; Dalvi, Y.B.; Lamrood, P.Y.; Shinde, B.P.; Nair, C.K.K. Historical and current perspectives on therapeutic potential of higher basidiomycetes: An overview. 3 Biotech 2019, 9, 362. [Google Scholar] [CrossRef]
- Bölker, M.; Basse, C.W.; Schirawski, J. Ustilago maydis secondary metabolism—From genomics to biochemistry. Fungal Genet. Biol. 2008, 45, S88–S93. [Google Scholar] [CrossRef]
| Producer Fungus | Final Product | Enzyme (s) in the Pathway | Supporting Evidence | References |
|---|---|---|---|---|
| Penicillium chrysogenum Aspergillus nidulans | Penicillin | Isopenicillin N acyltransferase Phenylacetyl-CoA ligase | Immunoelectron microscopy PTS1 sequences in both enzymes Peroxisome-less mutants Located in purified peroxisomes | [8,12,13] |
| Acremonium chrysogenum | Cephalosporin C | Isopenicillin N-CoA ligase Isopenicillinyl N-CoA epimerase | PTS1 targeting sequences | [8,14] |
| Aspergillus parasiticus Aspergillus flavus | Aflatoxin | AflA, B, C (PKS complex) HypC: anthrone oxidase AflJ (early times) | Norsolorinic acid accumulation PTS1 sequences | [15,16] |
| Aspergillus nidulans | Sterigmatocystin | AflA, B, C | Accumulation of norsolorinic acid | [15] |
| Alternaria alternata | AK toxins | AK1: carboxyl activating enzyme AK2: α,β hydrolase AK3: Enoyl-CoA hydratase | PTS1 targeting sequence Peroxisome deficient mutants Fluorescent labelled enzymes | [9,17,18,19] |
| Penicillium brevicompactum | Mycophenolic acid | PbACL891: Acyl-CoA ligase MpaH’: Acyl-CoA hydrolase | Fluorescent labelled enzymes PTS1 targeting sequences | [20] |
| Aspergillus fumigatus Botrytis cinerea | Melanin | BcPKS12: Sclerotia polyketide synthase BcPKS13: Conidia polyketide synthase BcYGH1 PK trimming down hydrolase | PTS1 sequences Fluorescence labelled enzymes | [21,22,23] |
| Penicillium patxilli | Patxilin | PaxG: Geranyl-Geranyl-PP synthase B | PTS1 targeting sequence Patxilin negative mutants Fluorescent labelled enzymes | [24] |
| Aspergillus fumigatus | Fusarinine | SidI: mevalonyl-CoA ligase SidH: mevalonyl-CoA dehydratase SidF: Anhidromevalonyl-CoA transferase | C-terminal PTS1 (SidH, SidF) N-terminal PTS2 (SidI) | [25] |
| Producer Fungus | Final Product | Enzyme (s) in the Pathway | Supporting Evidence | References |
|---|---|---|---|---|
| A. Vesicles or Endosomes | ||||
| Aspergillus parasiticus Aspergillus flavus | Aflatoxins | AflM (Ver-1): NADPH-dep. reductase AflD-(Nor-1): NADPH-dep. ketoreductase. AflP (OmtA): Methyl transferase AflJ: Transcriptional Co-activator (late times) | Fluorescence labelled enzymes | [16,26,27,28] |
| Fusarium graminearum | Trichothecenes | Hmr1: Hydroximethylglutaryl-CoA reductase Tri5: Trichodiene synthase Tri1: Calonectrin oxygenase Tri12: transporter | Tri12 mutants Fluorescence labelled enzymes | [29,30,31] |
| Aspergillus fumigatus | Fumiquilazoline C | FmqA: Trimodular NRPS FmqE: Transporter | Fluorescent labelled enzyme Microscopic vacuole stain | [32,33,34] |
| B. Cell Wall | ||||
| Aspergillus fumigatus | Fumiquinazoline C | FMQD: Fimiquinazoline oxidoreductase | Fluorescent labelled enzyme | [4,34] |
| Aspergillus fumigatus | Melanin | Abr1, Abr2 Laccases 3 | Fluorescent Labelled enzyme | [23] |
| Botrytis cinerea | Melanin | BcSCD1: Scytolone dehydratase BcBRN1/2: Trihydroxynaphthalene reductase 2 LaccaseI 3 | Fluorescent labelled enzyme | [21] |
| Yeast/Fungi | Important Features | References |
|---|---|---|
| Yeasts | ||
| Saccharomyces cerevisiae | 1. S. cerevisiae contains seven palmitoyl transferases. 2. Some PATs are self-palmitoylated before transferring the acyl group to other substrate proteins. 3. The best-known PATs contain a DHHC motif essential for their activity. The cysteine is the site for S-acylation. 4. A cysteine near a transmembrane domain is the site for S-acylation in PATs that lack the DHHC motif. 5. At least 47 palmitoylated proteins are known. 6. The palmitoylated proteins include heterotrimeric G proteins alpha subunits, SNARE proteins, amino acid permeases, membrane phosphatases, Inositol-4-phosphate kinase. 7. The SNARE proteins and the amino acid permeases are acylated by dedicated PATs. | [108,115,116,117,118,119,120,121,122,123,125,128] |
| Fungi: Ascomycetes | ||
| Aspergillus nidulans | 1.The model PAT Akr1 contains 737 amino acids a DHHC motif and 5 transmembrane domains and is well conserved in Ascomycetes. 2. Akr1 is located in the Golgi system. 3. Akr1 is self-palmitoylated. 4. The akr1 gene is regulated by calcium/calmodulin through a calcineurin-dependent regulatory element located upstream of the gene. | [120,127,144] |
| Aspergillus fumigatus | 1. A. fumigatus contains 234 palmitoylated proteins, 99 of them fully confirmed. 2. The Akr1 PAT acylates four melanin biosynthetic enzymes. 3. Palmitoylation of three of the melanin biosynthetic enzymes is triggered by conidiation inducing signals. 4. Palmitoylation of two early enzymes of the melanin pathway is essential for their localization in endosomes. 5. PAT4 palmitoylates the RAS protein. 6. The PAT4 defective mutants show reduced growth polarity and decreased pathogenicity. | [22,23,122,130,131] |
| Fusarium oxysporum var niveum | 1. F. oxysporum var. niveum contains six palmitoyl transferases (PAT1 to PAT6) and 211 palmitoylated proteins. 2. The six PATs contain the DHHC motif. Only PAT1, 2 and 4 show in vitro PAT activity, and are self-palmitoylated. 3. Palmitoylation affects growth, differentiation, cell wall stress and sensitivity to metal ions stress. 4. PAT mutants show reduced virulence in watermelon plants. 5. The palmitoylated proteins include components of the cargo adaptor protein complex (AP-2) which plays a key role in the localization of the membrane endocytic process in hyphae tips. | 134,135] |
| Fusarium graminearum | 1. Component proteins of the cargo adaptor FgAP-2 are palmitoylated. 2. The FgAP-2 complex regulates growth, polarity and apical localization of lipid flippases during endocytosis. 3. Palmitoylation of the FgAP-2 components plays important roles in the early stages of pathogenicity in wheat infection. | [132] |
| Fungi: Basidiomycetes | ||
| Cryptococcus neoformans | 1. C. neoformans has seven palmitoyl transferases. 2. The PATs are not essential for growth, although mutants in PAT3 and PAT 4 show distinct degrees of temperature sensitivity. 3. PAT4 is involved in acylation of the RAS GTPase. 4. Palmitoylation of the RAS protein is required for its proper localization in the cell membrane. 5. Mutants in the RAS protein show pronounced morphological defects in both the yeast and mycelial form. 6. Palmitoylation by PAT4 affects the pathogenicity in humans. | [118,119,131,132] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín, J.F.; Liras, P. Targeting of Specialized Metabolites Biosynthetic Enzymes to Membranes and Vesicles by Posttranslational Palmitoylation: A Mechanism of Non-Conventional Traffic and Secretion of Fungal Metabolites. Int. J. Mol. Sci. 2024, 25, 1224. https://doi.org/10.3390/ijms25021224
Martín JF, Liras P. Targeting of Specialized Metabolites Biosynthetic Enzymes to Membranes and Vesicles by Posttranslational Palmitoylation: A Mechanism of Non-Conventional Traffic and Secretion of Fungal Metabolites. International Journal of Molecular Sciences. 2024; 25(2):1224. https://doi.org/10.3390/ijms25021224
Chicago/Turabian StyleMartín, Juan F., and Paloma Liras. 2024. "Targeting of Specialized Metabolites Biosynthetic Enzymes to Membranes and Vesicles by Posttranslational Palmitoylation: A Mechanism of Non-Conventional Traffic and Secretion of Fungal Metabolites" International Journal of Molecular Sciences 25, no. 2: 1224. https://doi.org/10.3390/ijms25021224
APA StyleMartín, J. F., & Liras, P. (2024). Targeting of Specialized Metabolites Biosynthetic Enzymes to Membranes and Vesicles by Posttranslational Palmitoylation: A Mechanism of Non-Conventional Traffic and Secretion of Fungal Metabolites. International Journal of Molecular Sciences, 25(2), 1224. https://doi.org/10.3390/ijms25021224






