Unraveling the Role of RNA-Binding Proteins, with a Focus on RPS5, in the Malignant Progression of Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
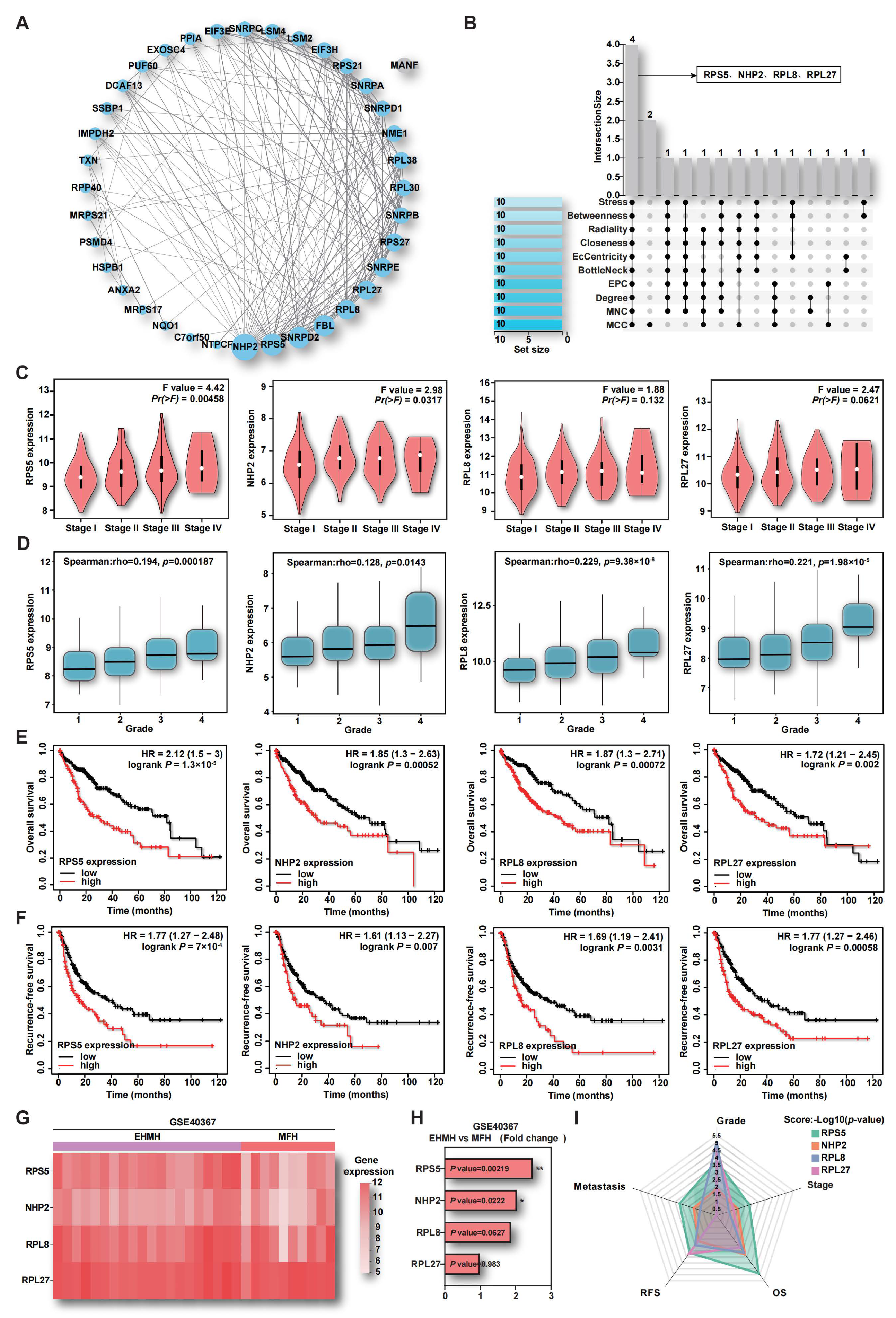
2.1. Aberrant Expression of RBPs in HCC
2.2. The Up-Regulation of RPS5 in Tumor Samples Indicates HCC Malignancy
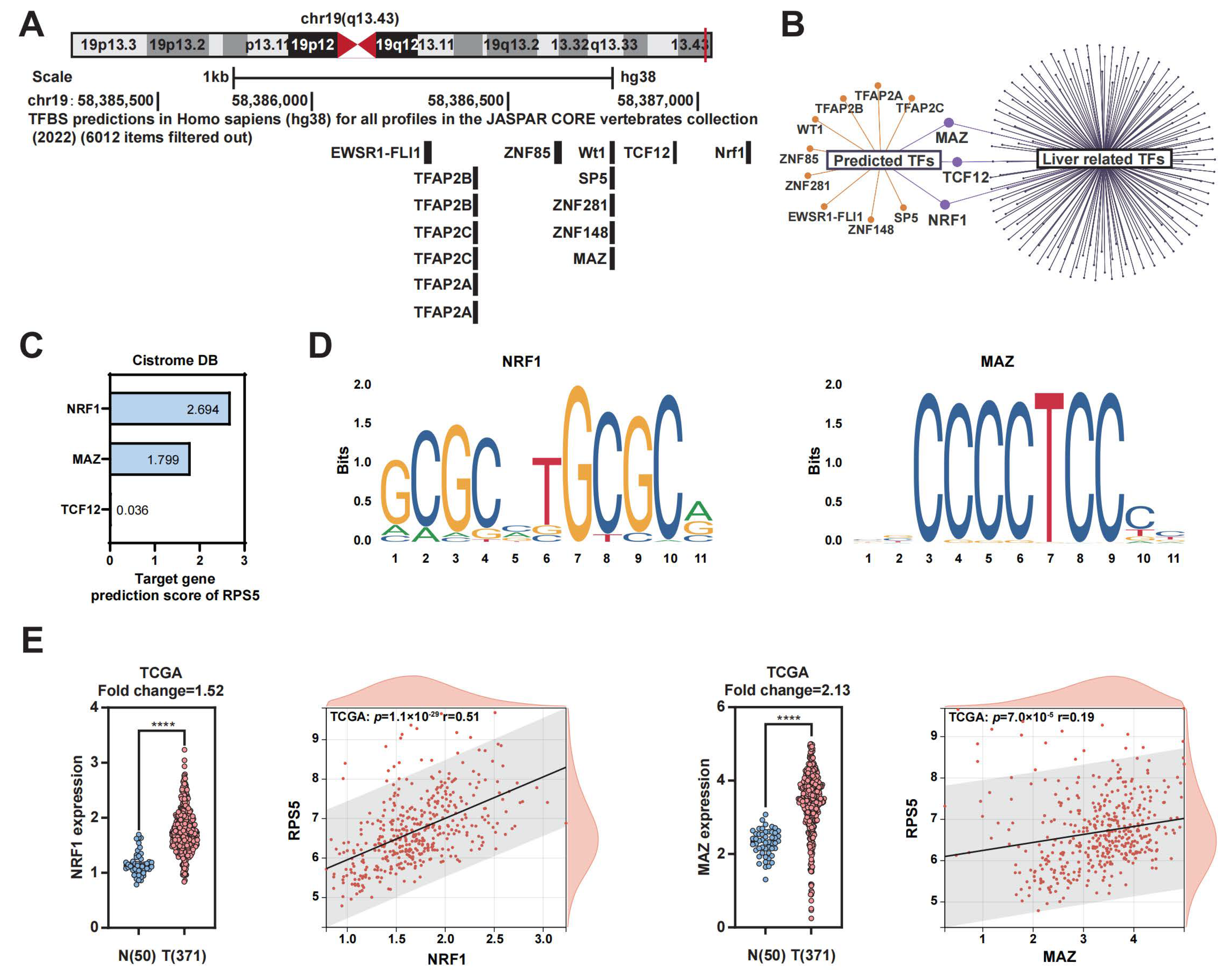
2.3. NRF1 and MAZ Are Potential Transcription Factors of RPS5 in HCC
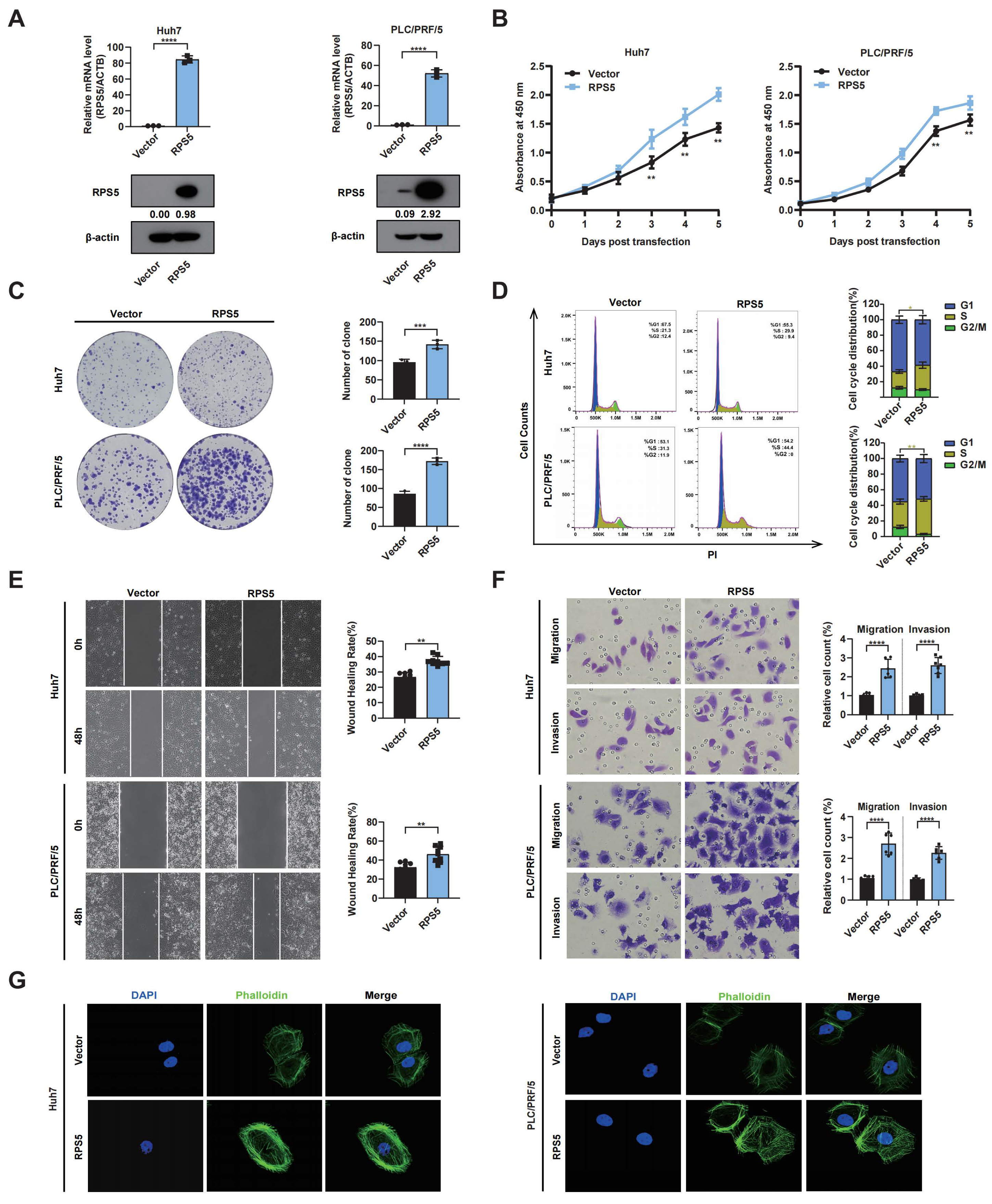
2.4. RPS5 Promotes the Progression of Liver Cancer by Regulating Cell Cycle
2.5. Experimental Confirmation of the Frequent Up-Regulation of RPS5 in HCC
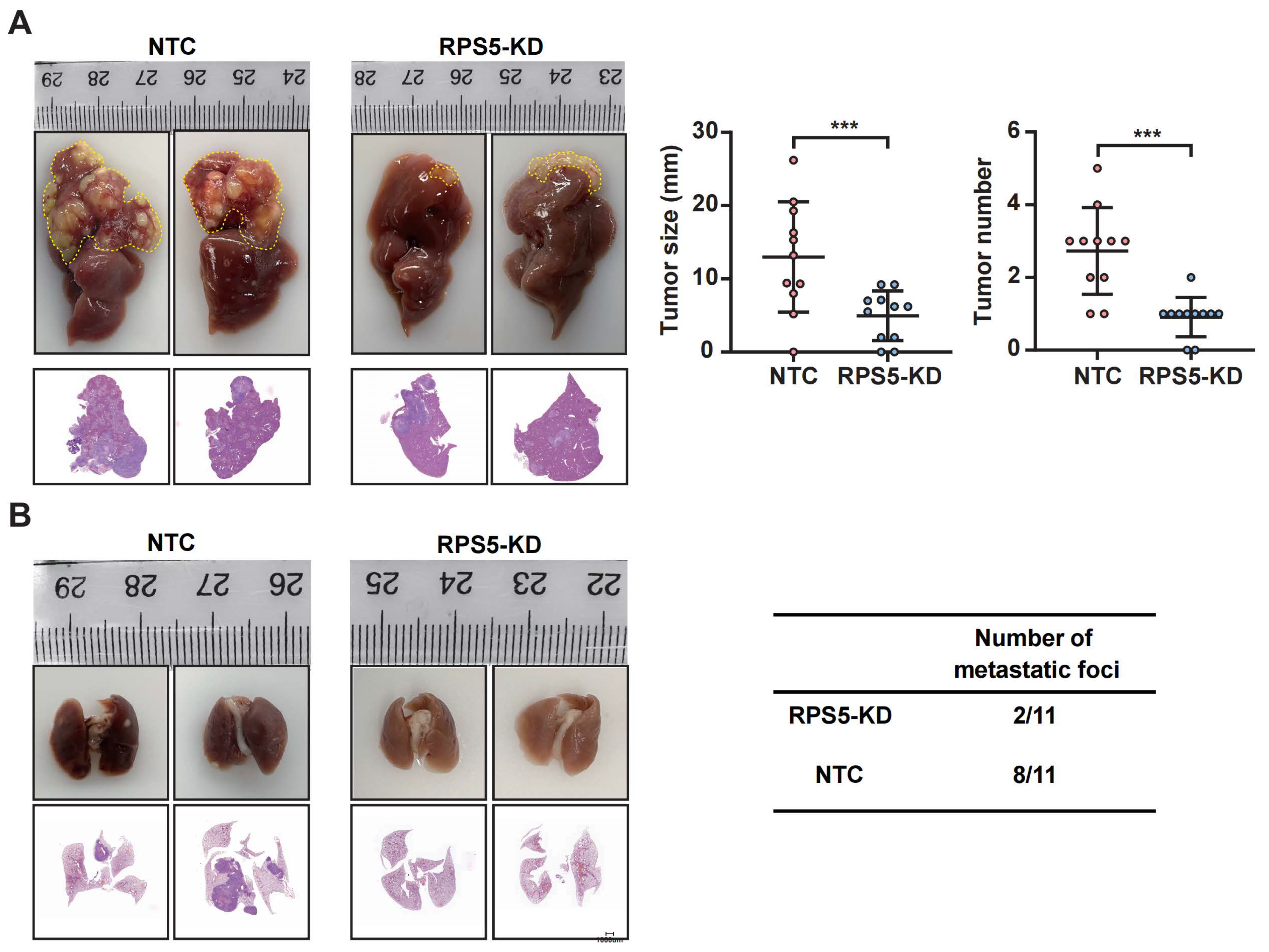
2.6. RPS5 Promotes Hepatic Tumorigenesis In Vitro and In Vivo
3. Discussion
4. Materials and Methods
4.1. Data Downloading and Comparative Gene Expression Analysis
4.2. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.3. Cell Culture
4.4. Clinical Samples
4.5. Animal Experiment
4.6. Construction of the RPS5 shRNA Lentivirus
4.7. Statistical Analysis and Chart Plotting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular Carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e1. [Google Scholar] [CrossRef] [PubMed]
- Müller-McNicoll, M.; Neugebauer, K.M. How Cells Get the Message: Dynamic Assembly and Function of mRNA-Protein Complexes. Nat. Rev. Genet. 2013, 14, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A Brave New World of RNA-Binding Proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Ma, C.; Du, C.; Huang, Y.; Xu, H.; Li, C.; Cheng, X.; Hao, R.; Xu, Y. RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function. Cells 2022, 11, 2357. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A Census of Human RNA-Binding Proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef]
- Zhang, K.; Barry, A.E.; Lamm, R.; Patel, K.; Schafer, M.; Dang, H. The Role of RNA Binding Proteins in Hepatocellular Carcinoma. Adv. Drug Deliv. Rev. 2022, 182, 114114. [Google Scholar] [CrossRef]
- Wang, S.; Sun, Z.; Lei, Z.; Zhang, H.-T. RNA-Binding Proteins and Cancer Metastasis. Semin. Cancer Biol. 2022, 86, 748–768. [Google Scholar] [CrossRef] [PubMed]
- Lachiondo-Ortega, S.; Delgado, T.C.; Baños-Jaime, B.; Velázquez-Cruz, A.; Díaz-Moreno, I.; Martínez-Chantar, M.L. Hu Antigen R (HuR) Protein Structure, Function and Regulation in Hepatobiliary Tumors. Cancers 2022, 14, 2666. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Chao, W.; Zhong, S.; Ren, A.-J. Eukaryotic Ribosomal Protein S5 of the 40S Subunit: Structure and Function. Int. J. Mol. Sci. 2023, 24, 3386. [Google Scholar] [CrossRef] [PubMed]
- Bhat, P.; Shwetha, S.; Sharma, D.K.; Joseph, A.P.; Srinivasan, N.; Das, S. The Beta Hairpin Structure within Ribosomal Protein S5 Mediates Interplay between Domains II and IV and Regulates HCV IRES Function. Nucleic Acids Res. 2015, 43, 2888–2901. [Google Scholar] [CrossRef] [PubMed]
- Lumsden, T.; Bentley, A.A.; Beutler, W.; Ghosh, A.; Galkin, O.; Komar, A.A. Yeast Strains with N-Terminally Truncated Ribosomal Protein S5: Implications for the Evolution, Structure and Function of the Rps5/Rps7 Proteins. Nucleic Acids Res. 2010, 38, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Galkin, O.; Bentley, A.A.; Gupta, S.; Compton, B.-A.; Mazumder, B.; Kinzy, T.G.; Merrick, W.C.; Hatzoglou, M.; Pestova, T.V.; Hellen, C.U.T.; et al. Roles of the Negatively Charged N-Terminal Extension of Saccharomyces Cerevisiae Ribosomal Protein S5 Revealed by Characterization of a Yeast Strain Containing Human Ribosomal Protein S5. RNA 2007, 13, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A Census of Human Transcription Factors: Function, Expression and Evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Ruvkun, G.; Lehrbach, N. Regulation and Functions of the ER-Associated Nrf1 Transcription Factor. Cold Spring Harb. Perspect. Biol. 2023, 15, a041266. [Google Scholar] [CrossRef]
- Sekine, H.; Motohashi, H. Roles of CNC Transcription Factors NRF1 and NRF2 in Cancer. Cancers 2021, 13, 541. [Google Scholar] [CrossRef]
- Song, J.; Ugai, H.; Nakata-Tsutsui, H.; Kishikawa, S.; Suzuki, E.; Murata, T.; Yokoyama, K.K. Transcriptional Regulation by Zinc-Finger Proteins Sp1 and MAZ Involves Interactions with the Same Cis-Elements. Int. J. Mol. Med. 2003, 11, 547–553. [Google Scholar] [CrossRef]
- Li, Y.; Chao, Y.; Fang, Y.; Wang, J.; Wang, M.; Zhang, H.; Ying, M.; Zhu, X.; Wang, H. MTA1 Promotes the Invasion and Migration of Non-Small Cell Lung Cancer Cells by Downregulating miR-125b. J. Exp. Clin. Cancer Res. 2013, 32, 33. [Google Scholar] [CrossRef]
- Cornelius, V.A.; Naderi-Meshkin, H.; Kelaini, S.; Margariti, A. RNA-Binding Proteins: Emerging Therapeutics for Vascular Dysfunction. Cells 2022, 11, 2494. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Wu, Q.; Zhao, H.; Xie, R.; He, X.; Gu, H. Unraveling the Role of RNA-Binding Proteins, with a Focus on RPS5, in the Malignant Progression of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2024, 25, 773. https://doi.org/10.3390/ijms25020773
Zhou C, Wu Q, Zhao H, Xie R, He X, Gu H. Unraveling the Role of RNA-Binding Proteins, with a Focus on RPS5, in the Malignant Progression of Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2024; 25(2):773. https://doi.org/10.3390/ijms25020773
Chicago/Turabian StyleZhou, Chongyang, Qiumin Wu, Haibei Zhao, Ruixi Xie, Xin He, and Huiying Gu. 2024. "Unraveling the Role of RNA-Binding Proteins, with a Focus on RPS5, in the Malignant Progression of Hepatocellular Carcinoma" International Journal of Molecular Sciences 25, no. 2: 773. https://doi.org/10.3390/ijms25020773
APA StyleZhou, C., Wu, Q., Zhao, H., Xie, R., He, X., & Gu, H. (2024). Unraveling the Role of RNA-Binding Proteins, with a Focus on RPS5, in the Malignant Progression of Hepatocellular Carcinoma. International Journal of Molecular Sciences, 25(2), 773. https://doi.org/10.3390/ijms25020773






