Oral Microbiome and CPT1A Function in Fatty Acid Metabolism in Oral Cancer
Abstract
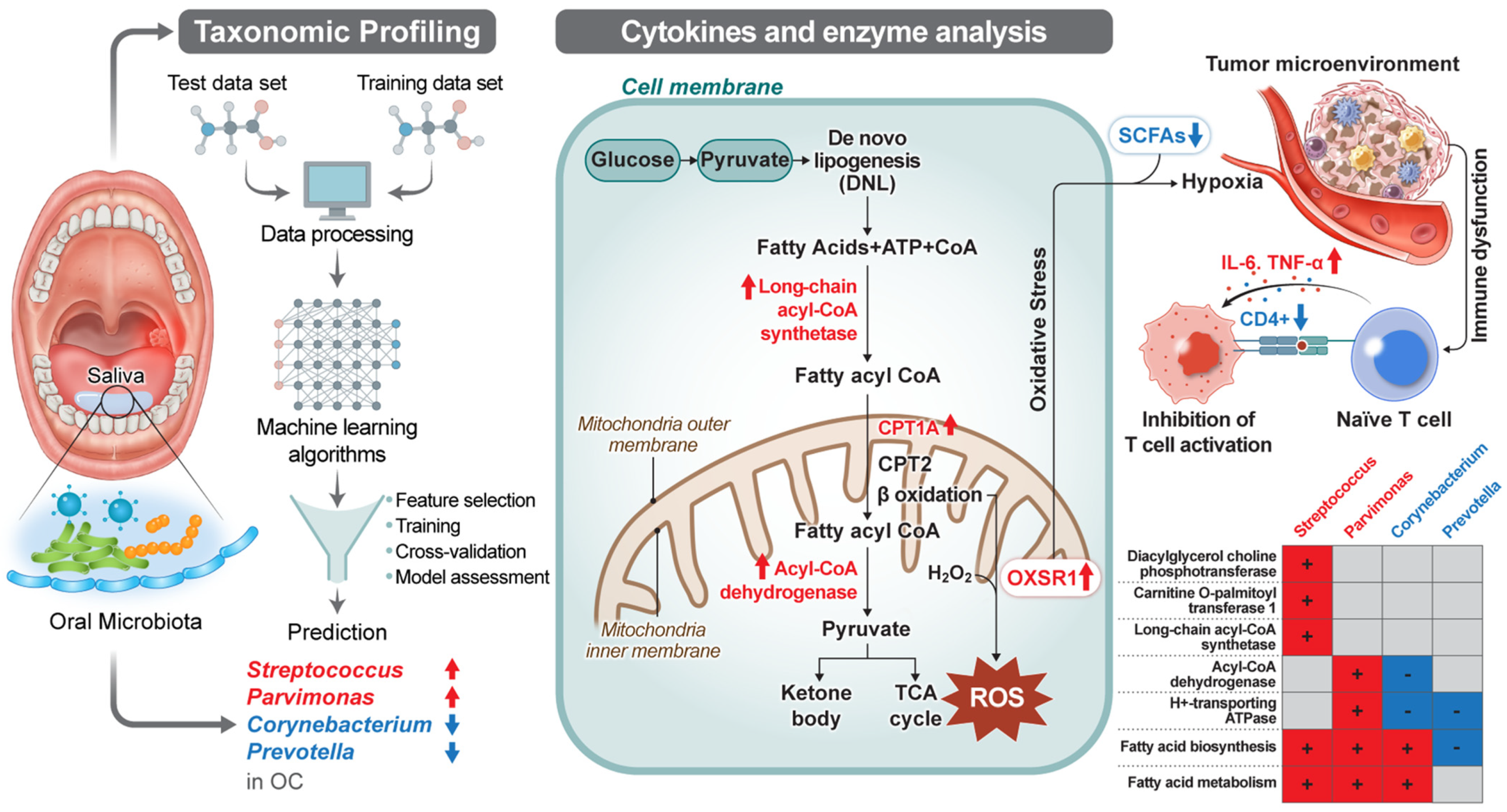
:1. Introduction
2. Results
2.1. Demographic Characteristics of Oral Cancer Patients and Controls
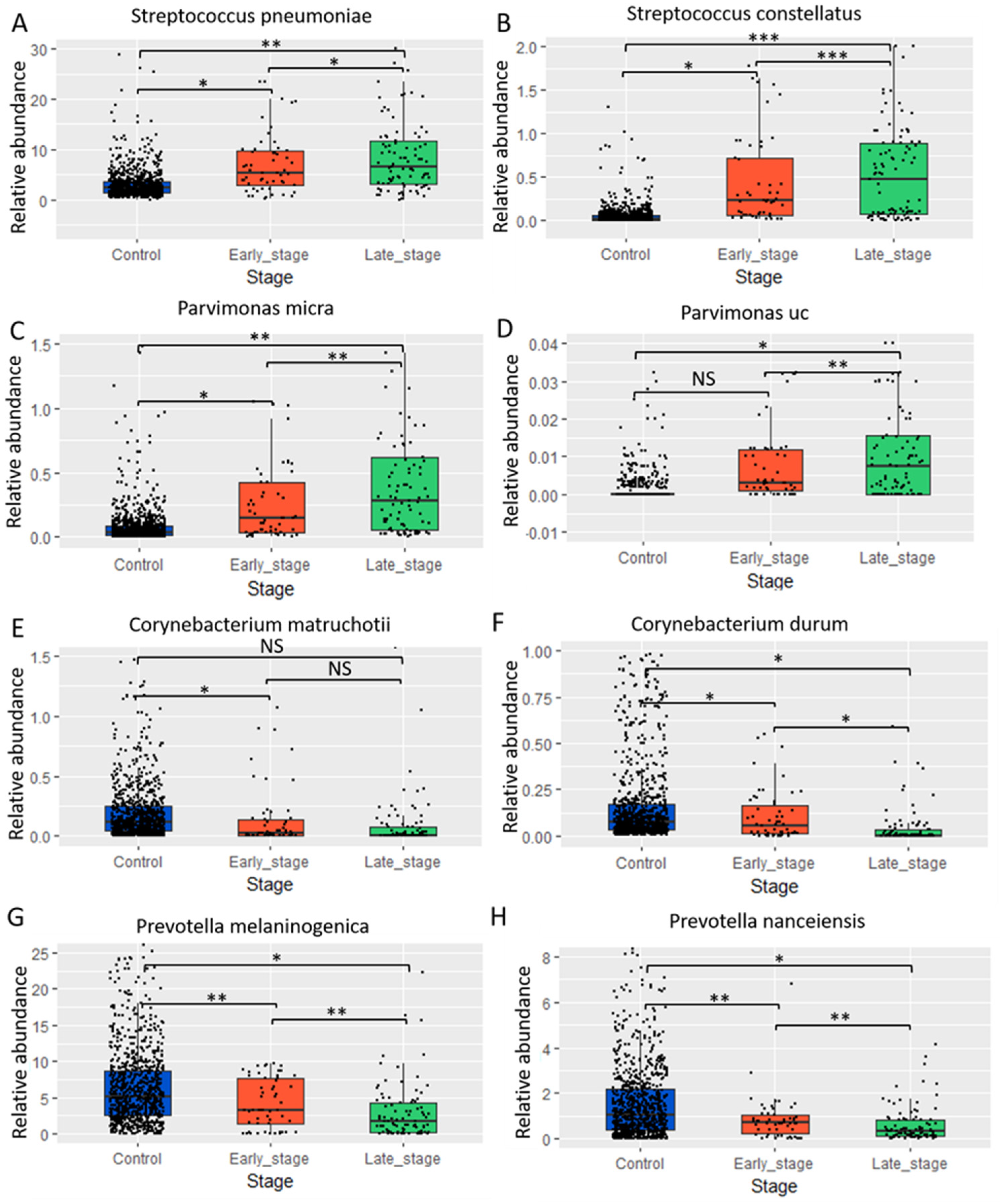
2.2. Contrasting Oral Microbiome Compositions in Oral Cancer Patient and Control Groups: Taxonomic Analyses and Identification of Cancer-Associated Genera
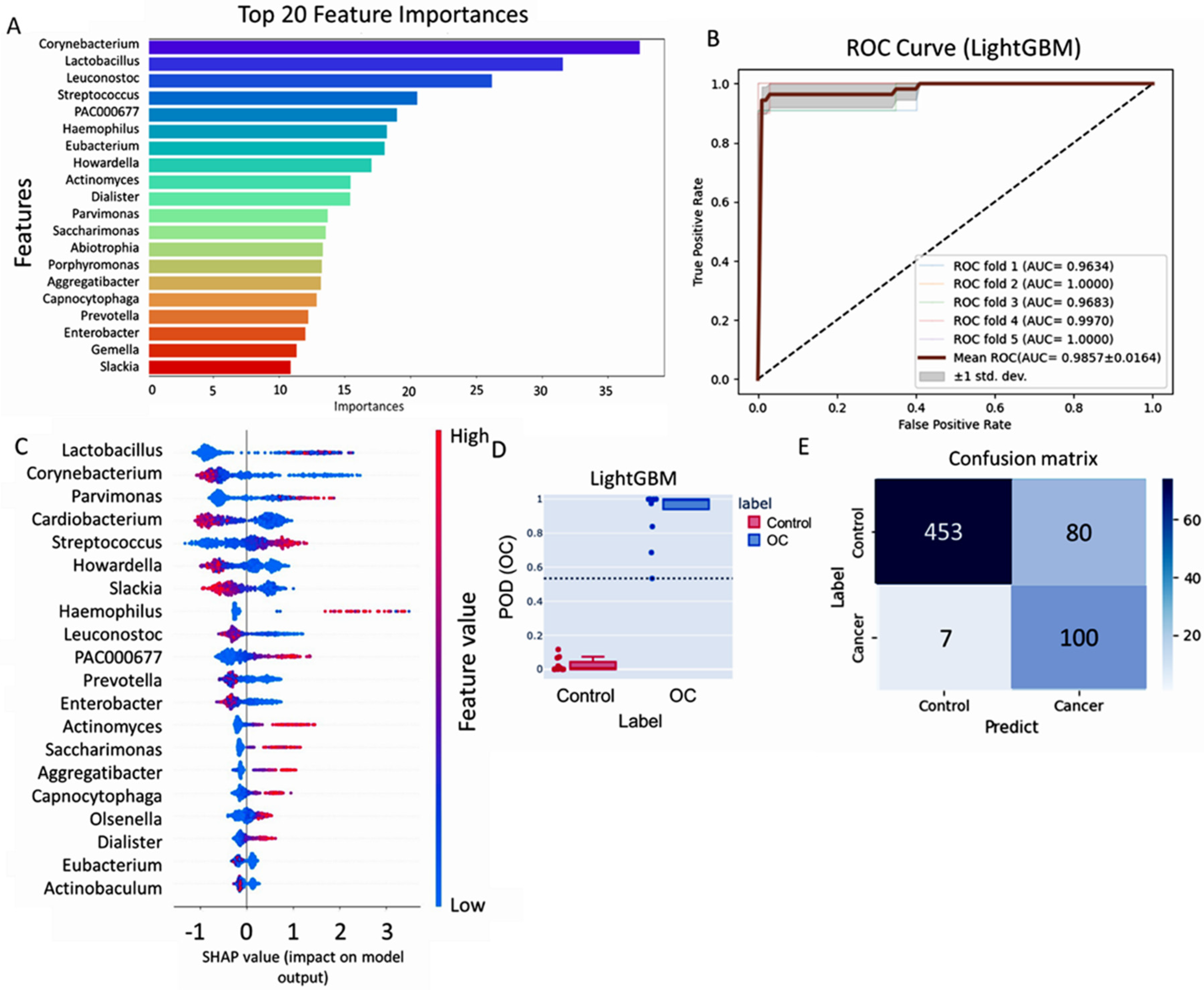
2.3. Machine-Learning Analysis Identified Microbial Biomarkers for Oral Cancer Risk: LightGBM Model Accuracy and Microbiome Associations
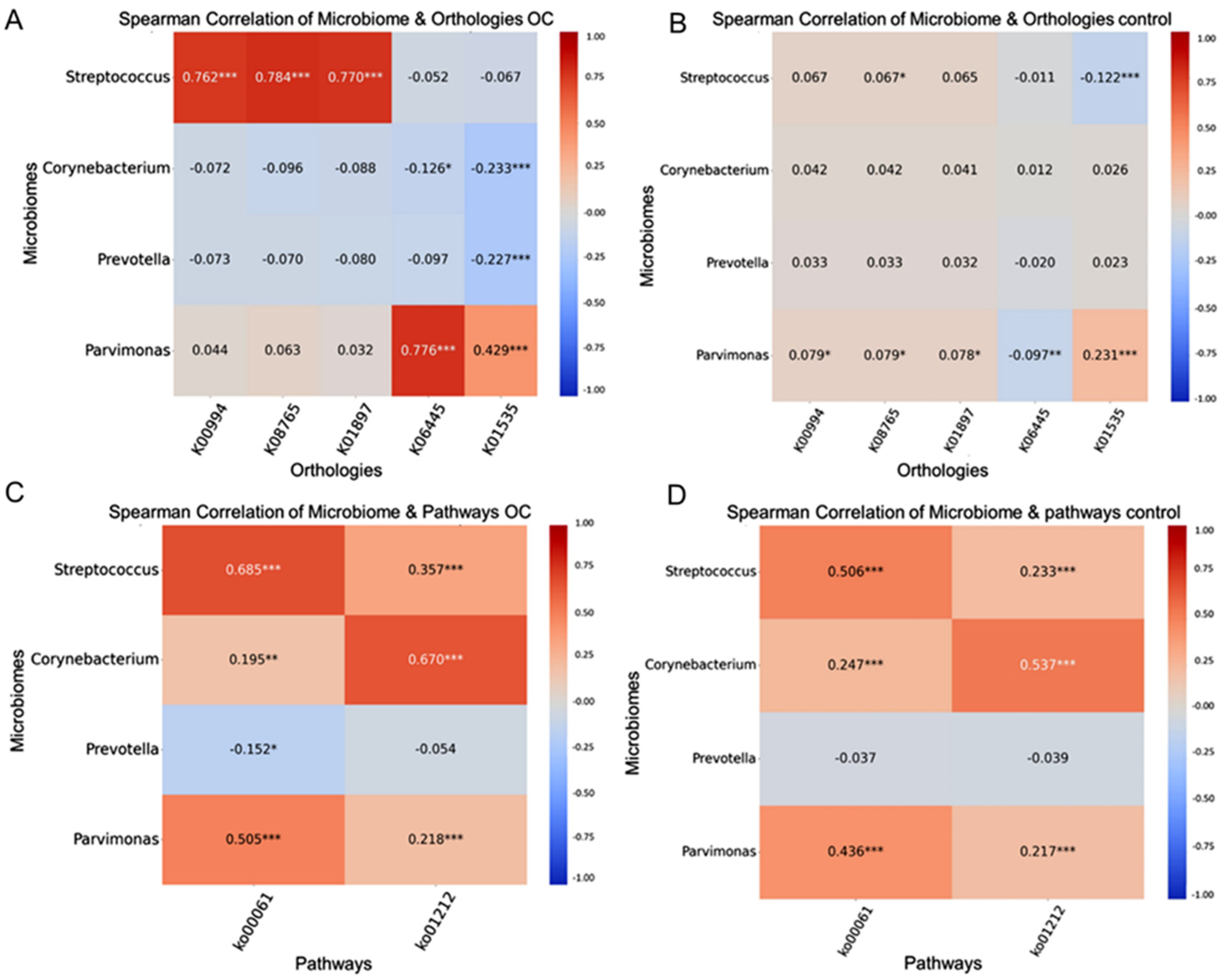
2.4. Integration of the Microbiome and Functional-Pathway Analysis Revealed Potential Biomarkers and Associations in Oral Cancer
2.5. Microbiome Associations with Disease-Free and Overall Survival in Oral Cancer
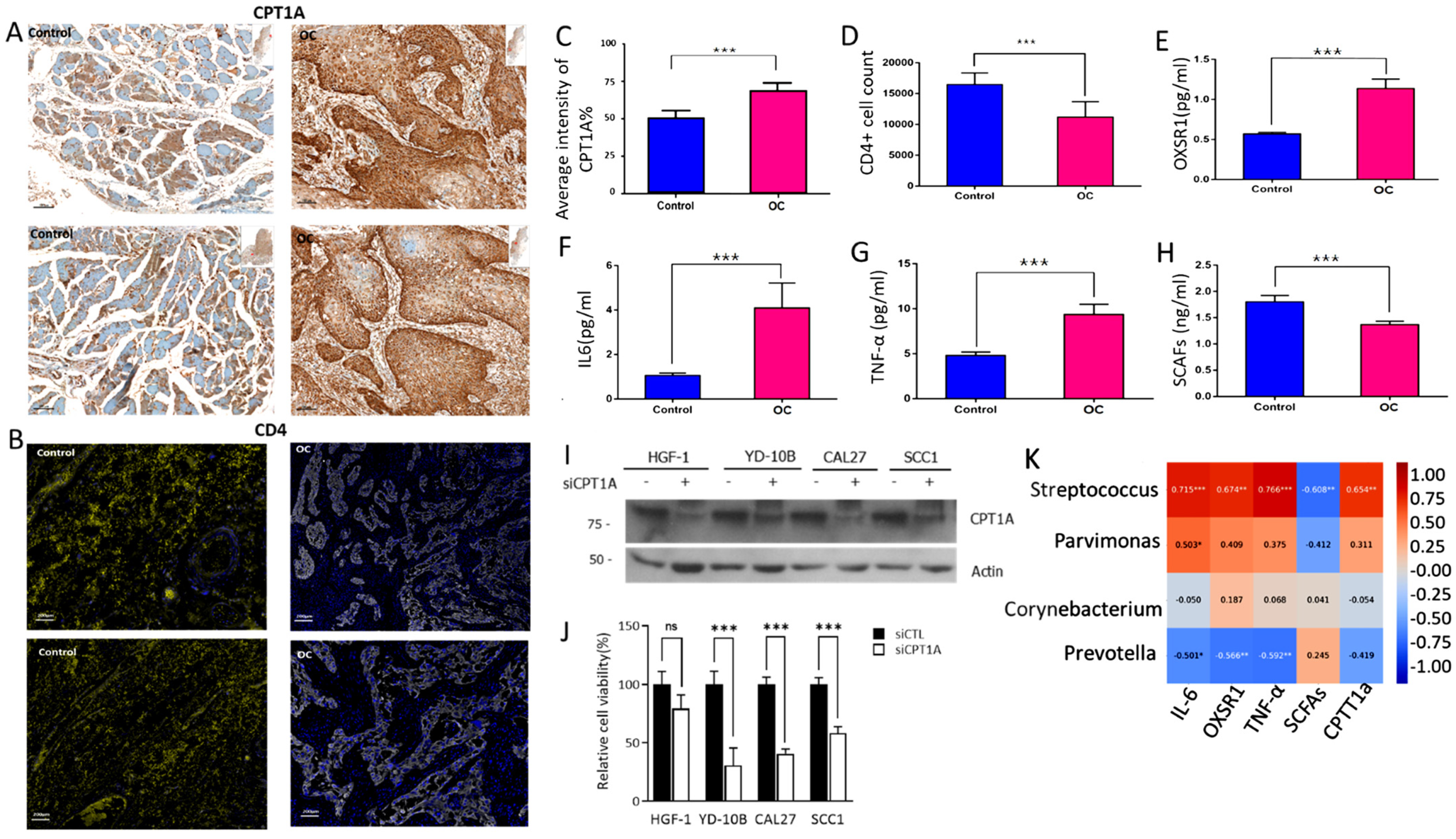
2.6. Oral Microbiome Modulation of Cytokine and Immune Pathways: Implications for Cancer Onset
3. Discussion
4. Materials and Methods
4.1. Participant Characteristics
4.2. Saliva and Blood Sample Collection
4.3. Oral Microbiome Characterization Based on 16S rRNA Gene Amplification and Sequencing
4.4. Functional Homology Inferences: Predicting Orthologs
4.5. Cell Lines
4.6. Small-Interfering RNA (siRNA) Experiments
4.7. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Assay
4.8. Western Blotting
4.9. Immunohistochemistry (IHC) Analysis of CPT1A and CD4+T-Helper Cell Counts
4.10. Antibodies and Reagents
4.11. Measuring Oral Microbial Signals, Including Cytokine Levels (OXSR1, CPT1A, SCFAs, IL6, and TNF-α)
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Louredo, B.; Vargas, P.; Pérez-De-Oliveira, M.; Lopes, M.; Kowalski, L.; Curado, M. Epidemiology and survival outcomes of lip, oral cavity, and oropharyngeal squamous cell carcinoma in a southeast Brazilian population. Med. Oral Patol. Oral Y Cirugia Bucal 2022, 27, e274–e284. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Yang, Y.; Li, M.; Li, C.; Zhou, Z.; Tang, G.; Wu, L.; Yao, Y.; Shen, X.; Hou, Z.; et al. LncRNA IFITM4P promotes immune escape by up-regulating PD-L1 via dual mechanism in oral carcinogenesis. Mol. Ther. 2022, 30, 1564–1577. [Google Scholar] [CrossRef] [PubMed]
- Bettendorf, O.; Piffko, J.; Bankfalvi, A. Prognostic and predictive factors in oral squamous cell cancer: Important tools for planning individual therapy? Oral Oncol. 2004, 40, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Z.; Xie, L.; Shang, Z.-J. Burden of Oral Cancer on the 10 Most Populous Countries from 1990 to 2019: Estimates from the Global Burden of Disease Study 2019. Int. J. Environ. Res. Public Health 2022, 19, 875. [Google Scholar] [CrossRef]
- Stasiewicz, M.; Karpiński, T.M. The oral microbiota and its role in carcinogenesis. Semin. Cancer Biol. 2022, 86, 633–642. [Google Scholar] [CrossRef]
- Sedghi, L.; DiMassa, V.; Harrington, A.; Lynch, S.V.; Kapila, Y.L. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontology 2000 2021, 87, 107–131. [Google Scholar] [CrossRef]
- Granato, D.C.; Neves, L.X.; Trino, L.D.; Carnielli, C.M.; Lopes, A.F.; Yokoo, S.; Pauletti, B.A.; Domingues, R.R.; Sá, J.O.; Persinoti, G.; et al. Meta-omics analysis indicates the saliva microbiome and its proteins associated with the prognosis of oral cancer patients. Biochim. Biophys. Acta BBA-Proteins Proteom. 2021, 1869, 140659. [Google Scholar] [CrossRef]
- Karpiński, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef]
- Gallimidi, A.B.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef]
- Asoudeh-Fard, A.; Barzegari, A.; Dehnad, A.; Bastani, S.; Golchin, A.; Omidi, Y. Lactobacillus plantarum induces apoptosis in oral cancer KB cells through upregulation of PTEN and downregulation of MAPK signalling pathways. BioImpacts 2017, 7, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.R.; Lam, Y.K.; Uhlig, H.H. Short-chain fatty acids: Linking diet, the microbiome and immunity. Nat. Rev. Immunol. 2024, 24, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.B.; Alimova, Y.; Myers, T.M.; Ebersole, J.L. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch. Oral Biol. 2011, 56, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, E.; Governa, V.; Garzon, J.F.G.; Mele, V.; Amicarella, F.; Muraro, M.G.; Trella, E.; Galati-Fournier, V.; Oertli, D.; Däster, S.R.; et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018, 67, 1984–1994. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Elsabagh, M.; Zhang, Y.; Ma, Y.; Jin, Y.; Wang, M.; Wang, H.; Jiang, H. Effects of the Gut Microbiota and Barrier Function on Melatonin Efficacy in Alleviating Liver Injury. Antioxidants 2022, 11, 1727. [Google Scholar] [CrossRef]
- Leonov, G.E.; Varaeva, Y.R.; Livantsova, E.N.; Starodubova, A.V. The Complicated Relationship of Short-Chain Fatty Acids and Oral Microbiome: A Narrative Review. Biomedicines 2023, 11, 2749. [Google Scholar] [CrossRef]
- Halczy-Kowalik, L.; Drozd, A.; Stachowska, E.; Drozd, R.; Żabski, T.; Domagała, W. Fatty acids distribution and content in oral squamous cell carcinoma tissue and its adjacent microenvironment. PLoS ONE 2019, 14, e0218246. [Google Scholar] [CrossRef]
- Cheng, X.-M.; Hu, Y.-Y.; Yang, T.; Wu, N.; Wang, X.-N. Reactive Oxygen Species and Oxidative Stress in Vascular-Related Diseases. Oxidative Med. Cell. Longev. 2022, 2022, 7906091. [Google Scholar] [CrossRef]
- Miyazaki, T.; Nakagawa, H. Beneficial effects of antioxidative lactic acid bacteria. AIMS Microbiol. 2017, 3, 1–7. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Yeh, Y.-M.; Yu, H.-Y.; Chin, C.-Y.; Hsu, C.-W.; Liu, H.; Huang, P.-J.; Hu, S.-N.; Liao, C.-T.; Chang, K.-P.; et al. Oral Microbiota Community Dynamics Associated with Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zheng, H.J.; Zhang, C.P. The Oral Microbiota May Have Influence on Oral Cancer. Front. Cell. Infect. Microbiol. 2019, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Delgado, R.Z.R.; Frias-Lopez, J. The Oral Microbiome and Cancer. Front. Immunol. 2020, 11, 591088. [Google Scholar] [CrossRef]
- Chocolatewala, N.; Chaturvedi, P.; Desale, R. The role of bacteria in oral cancer. Indian J. Med Paediatr. Oncol. 2010, 31, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.; Do, T.; Devine, D. Oral biofilms: Molecular analysis, challenges, and future prospects in dental diagnostics. Clin. Cosmet. Investig. Dent. 2013, 5, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.; Al-Hebshi, N.N.; Speicher, D.J.; Perera, I.; Johnson, N.W. Emerging role of bacteria in oral carcinogenesis: A review with special reference to perio-pathogenic bacteria. J. Oral Microbiol. 2016, 8, 32762. [Google Scholar] [CrossRef]
- KEGG. Fatty Acid Biosynthesis-Corynebacterium glutamicum ATCC 13032 (Bielefeld). Available online: https://www.genome.jp/pathway/cgb00061 (accessed on 11 April 2023).
- KEGG. KEGG Fatty Acid Biosynthesis Prevotella intermedia. Available online: https://www.genome.jp/pathway/pit00061+M00083 (accessed on 11 April 2023).
- Son, M.-Y.; Cho, H.-S. Anticancer Effects of Gut Microbiota-Derived Short-Chain Fatty Acids in Cancers. J. Microbiol. Biotechnol. 2023, 33, 849–856. [Google Scholar] [CrossRef]
- Provenzano, J.C.; Rôças, I.N.; Tavares, L.F.D.; Neves, B.C.; Siqueira, J.F., Jr. Short-chain fatty acids in infected root canals of teeth with apical periodontitis before and after treatment. J. Endod. 2015, 41, 831–835. [Google Scholar] [CrossRef]
- Akhtar, M.; Chen, Y.; Ma, Z.; Zhang, X.; Shi, D.; Khan, J.A.; Liu, H. Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation. Anim. Nutr. 2022, 8, 350–360. [Google Scholar] [CrossRef]
- Sun, J.; Tang, Q.; Yu, S.; Xie, M.; Xie, Y.; Chen, G.; Chen, L. Role of the oral microbiota in cancer evolution and progression. Cancer Med. 2020, 9, 6306–6321. [Google Scholar] [CrossRef]
- Yang, W.; Yu, T.; Huang, X.; Bilotta, A.J.; Xu, L.; Lu, Y.; Sun, J.; Pan, F.; Zhou, J.; Zhang, W.; et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020, 11, 4457. [Google Scholar] [CrossRef] [PubMed]
- Fidelle, M.; Rauber, C.; Alves Costa Silva, C.; Tian, A.L.; Lahmar, I.; de La Varende, A.M.; Zhao, L.; Thelemaque, C.; Lebhar, I.; Messaoudene, M.; et al. A microbiota-modulated checkpoint directs immunosuppressive intestinal T cells into cancers. Science 2023, 380, eabo2296. [Google Scholar] [CrossRef]
- Zaugg, K.; Yao, Y.; Reilly, P.T.; Kannan, K.; Kiarash, R.; Mason, J.; Huang, P.; Sawyer, S.K.; Fuerth, B.; Faubert, B.; et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011, 25, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Maishi, N.; Akahori, E.; Hasebe, A.; Takeda, R.; Matsuda, A.Y.; Hida, Y.; Nam, J.; Onodera, Y.; Kitagawa, Y.; et al. The oral bacterium Streptococcus mutans promotes tumor metastasis by inducing vascular inflammation. Cancer Sci. 2022, 113, 3980–3994. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Stoykova, G.E.; Salzmann-Sullivan, M.; Dzieciatkowska, M.; Liebman, L.N.; Deep, G.; Schlaepfer, I.R. CPT1A Supports Castration-Resistant Prostate Cancer in Androgen-Deprived Conditions. Cells 2019, 8, 1115. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Qin, J.; Wu, J.; Dai, X.; Xu, J. OSR1 phosphorylates the Smad2/3 linker region and induces TGF-β1 autocrine to promote EMT and metastasis in breast cancer. Oncogene 2021, 40, 68–84. [Google Scholar] [CrossRef]
- Guan, L.; Chen, Y.; Wang, Y.; Zhang, H.; Fan, S.; Gao, Y.; Jiao, T.; Fu, K.; Sun, J.; Yu, A.; et al. Effects of carnitine palmitoyltransferases on cancer cellular senescence. J. Cell. Physiol. 2018, 234, 1707–1719. [Google Scholar] [CrossRef]
- Pacilli, A.; Calienni, M.; Margarucci, S.; D’apolito, M.; Petillo, O.; Rocchi, L.; Pasquinelli, G.; Nicolai, R.; Koverech, A.; Calvani, M.; et al. Carnitine-Acyltransferase System Inhibition, Cancer Cell Death, and Prevention of Myc-Induced Lymphomagenesis. J. Natl. Cancer Inst. 2013, 105, 489–498. [Google Scholar] [CrossRef]
- Liang, K. Mitochondrial CPT1A: Insights into structure, function, and basis for drug development. Front. Pharmacol. 2023, 14, 1160440. [Google Scholar] [CrossRef]
- Fingerhut, R.; Röschinger, W.; Muntau, A.C.; Dame, T.; Kreischer, J.; Arnecke, R.; Superti-Furga, A.; Troxler, H.; Liebl, B.; Olgemöller, B.; et al. Hepatic Carnitine Palmitoyltransferase I Deficiency: Acylcarnitine Profiles in Blood Spots Are Highly Specific. Clin. Chem. 2001, 47, 1763–1768. [Google Scholar] [CrossRef]
- Innes, A.M.; Seargeant, E.L.; Balachandra, K.; Roe, C.R.; Wanders, A.R.J.; Ruiter, J.P.N.; Casiro, O.; Grewar, A.D.; Greenberg, C.R. Hepatic Carnitine Palmitoyltransferase I Deficiency Presenting as Maternal Illness in Pregnancy. Pediatr. Res. 2000, 47, 43. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.A.; Sinclair, G.; McIntosh, S.; Bamforth, F.; Thompson, R.; Sobol, I.; Osborne, G.; Corriveau, A.; Santos, M.; Hanley, B.; et al. Carnitine palmitoyltransferase 1A (CPT1A) P479L prevalence in live newborns in Yukon, Northwest Territories, and Nunavut. Mol. Genet. Metab. 2010, 101, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.M.; Yeung, M.H.Y.; Wong, A.N.N.; Tsang, H.F.; Yu, A.C.S.; Yim, A.K.Y.; Wong, S.C.C. Targeted Sequencing Approach and Its Clinical Applications for the Molecular Diagnosis of Human Diseases. Cells 2023, 12, 493. [Google Scholar] [CrossRef] [PubMed]
- CJ Bioscience. Available online: https://www.cjbioscience.com/ (accessed on 20 June 2023).
- Nouri, Z.; Choi, S.W.; Choi, I.J.; Ryu, K.W.; Woo, S.M.; Park, S.-J.; Lee, W.J.; Choi, W.; Jung, Y.-S.; Myung, S.-K.; et al. Exploring Connections between Oral Microbiota, Short-Chain Fatty Acids, and Specific Cancer Types: A Study of Oral Cancer, Head and Neck Cancer, Pancreatic Cancer, and Gastric Cancer. Cancers 2023, 15, 2898. [Google Scholar] [CrossRef] [PubMed]

| Discovery Data Set (n = 637) | Validation Data Set (n = 385) | ||||||
|---|---|---|---|---|---|---|---|
| Variable | OC (N = 104) | Control (N = 533) | p Value | OC (N = 53) | Control (N = 332) | p Value | |
| Gender | Male | 67 (64.4) | 254 (47.7) | 0.003 | 29 (54.7) | 105 (31.6) | 0.002 |
| Female | 37 (35.6) | 279 (52.3) | 24 (45.3) | 227 (68.4) | |||
| Age | I (Age < 50) | 16 (15.4) | 91 (17.1) | 0.000 | 5 (9.40) | 40 (12.0) | 0.000 |
| II (Age 45–60) | 28 (26.9) | 241 (45.2) | 5 (9.40) | 142 (42.8) | |||
| III (Age 60–70) | 19 (18.3) | 168 (31.5) | 17 (32.1) | 140 (42.2) | |||
| IV (Age 70<) | 41 (39.4) | 33 (6.20) | 26 (49.1) | 10 (3.00) | |||
| BMI | BMI < 18.5 | 17 (16.4) | 16 (3.00) | 0.000 | 2 (3.70) | 7 (2.10) | |
| BMI 18.5–23 | 30 (28.8) | 190 (35.6) | 11 (20.8) | 127 (38.3) | |||
| BMI 23–25 | 19 (18.3) | 137 (25.7) | 16 (30.2) | 90 (27.1) | 0.118 | ||
| BMI ≥ 25 | 38 (36.5) | 186 (34.9) | 23 (43.4) | 106 (31.9) | |||
| Unknown | 0 (0.00) | 4 (0.80) | 1 (1.90) | 2 (0.60) | |||
| Smoking | Non smoker | 55 (52.9) | 298 (55.9) | 0.010 | 29 (54.7) | 227 (68.4) | 0.206 |
| Currentsmoker | 23 (22.1) | 58 (10.9) | 5 (9.40) | 17 (5.10) | |||
| Ex-smoker | 25 (24.0) | 160 (30.0) | 18 (34.0) | 80 (24.1) | |||
| Unknown | 1 (1.00) | 17 (3.20) | 1 (1.90) | 8 (2.40) | |||
| Drinking | Non drinker | 42 (40.3) | 119 (22.3) | 0.000 | 35 (66.0) | 97 (29.3) | <0.0001 |
| Currentdrinker | 35 (33.7) | 318 (59.7) | 17 (32.1) | 186 (56.0) | |||
| Ex-drinker | 26 (25.0) | 69 (12.9) | 0 (0.00) | 35 (10.5) | |||
| Unknown | 1 (1.00) | 27 (5.10) | 1 (1.90) | 14 (4.20) | |||
| Tstage | T1 | 16 (15.4) | 0.000 | 14 (26.4) | 0.0017 | ||
| T2 | 21 (20.2) | 16 (30.2) | |||||
| T3 | 26 (25.0) | 1 (1.90) | |||||
| T4 | 34 (32.7) | 16 (30.2) | |||||
| Unknown | 7 (6.70) | 6 (11.3) | |||||
| Nstage | N0 | 62 (59.7) | 0.000 | 36 (67.9) | <0.0001 | ||
| N1 | 13 (12.5) | 5 (9.40) | |||||
| N2 | 12 (11.5) | 7 (13.2) | |||||
| N3 | 10 (9.60) | 1 (1.90) | |||||
| Unknown | 7 (6.70) | 4 (7.60) | |||||
| Stage | 1 | 17 (16.4) | 0.000 | 13 (24.5) | 0.002 | ||
| 2 | 11 (10.6) | 13 (24.5) | |||||
| 3 | 25 (24.0) | 3 (5.70) | |||||
| 4 | 46 (44.2) | 19 (35.9) | |||||
| Unknown | 5 (4.80) | 5 (9.40) | |||||
| Grade | Poor | 14 (13.4) | 0.000 | 0 (0.00) | <0.001 | ||
| Moderate | 43 (35.3) | 4 (7.60) | |||||
| Well | 35 (30.7) | 37 (69.8) | |||||
| Unknown | 12 (20.6) | 12 (22.6) | |||||
| DFS | Univariate Cox Proportional | Multivariate Cox Proportional | ||||
|---|---|---|---|---|---|---|
| Microbiome | HR | 95%CI | p-Value | HR | 95%CI | p-Value |
| Streptococcus | 2.30 | (1.10–5.10) | 0.035 | 2.85 | (1.29–6.32) | 0.009 |
| Parvimonas | 2.52 | (1.26–5.42) | 0.023 | 2.17 | (0.98–4.79) | 0.05 |
| Prevotella | 0.29 | (0.26–0.91) | 0.007 | 0.28 | (0.11–0.69) | 0.006 |
| Corynebacterium | 0.28 | (0.43–1.30) | 0.016 | 0.26 | (1.35–6.03) | 0.01 |
| Overall survival | ||||||
| Streptococcus | 5.10 | (1.90–13.0) | 0.001 | 5.76 | (2.17–15.3) | 0.0004 |
| Parvimonas | 3.20 | (1.40–7.30) | 0.005 | 2.90 | (1.26–6.66) | 0.0119 |
| Prevotella | 0.29 | (0.12–0.67) | 0.004 | 0.30 | (0.12–0.72) | 0.0074 |
| Corynebacterium | 0.35 | (0.14–0.87) | 0.024 | 0.36 | (0.14–0.91) | 0.0318 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Praveen, Z.; Choi, S.-W.; Lee, J.H.; Park, J.Y.; Oh, H.J.; Kwon, I.J.; Park, J.H.; Kim, M.K. Oral Microbiome and CPT1A Function in Fatty Acid Metabolism in Oral Cancer. Int. J. Mol. Sci. 2024, 25, 10890. https://doi.org/10.3390/ijms252010890
Praveen Z, Choi S-W, Lee JH, Park JY, Oh HJ, Kwon IJ, Park JH, Kim MK. Oral Microbiome and CPT1A Function in Fatty Acid Metabolism in Oral Cancer. International Journal of Molecular Sciences. 2024; 25(20):10890. https://doi.org/10.3390/ijms252010890
Chicago/Turabian StylePraveen, Zeba, Sung-Weon Choi, Jong Ho Lee, Joo Yong Park, Hyun Jun Oh, Ik Jae Kwon, Jin Hee Park, and Mi Kyung Kim. 2024. "Oral Microbiome and CPT1A Function in Fatty Acid Metabolism in Oral Cancer" International Journal of Molecular Sciences 25, no. 20: 10890. https://doi.org/10.3390/ijms252010890








