The Goat Cytotoxic T Lymphocyte-Associated Antigen-4 Gene: mRNA Expression and Association Analysis of Insertion/Deletion Variants with the Risk of Brucellosis
Abstract
:1. Introduction
2. Results
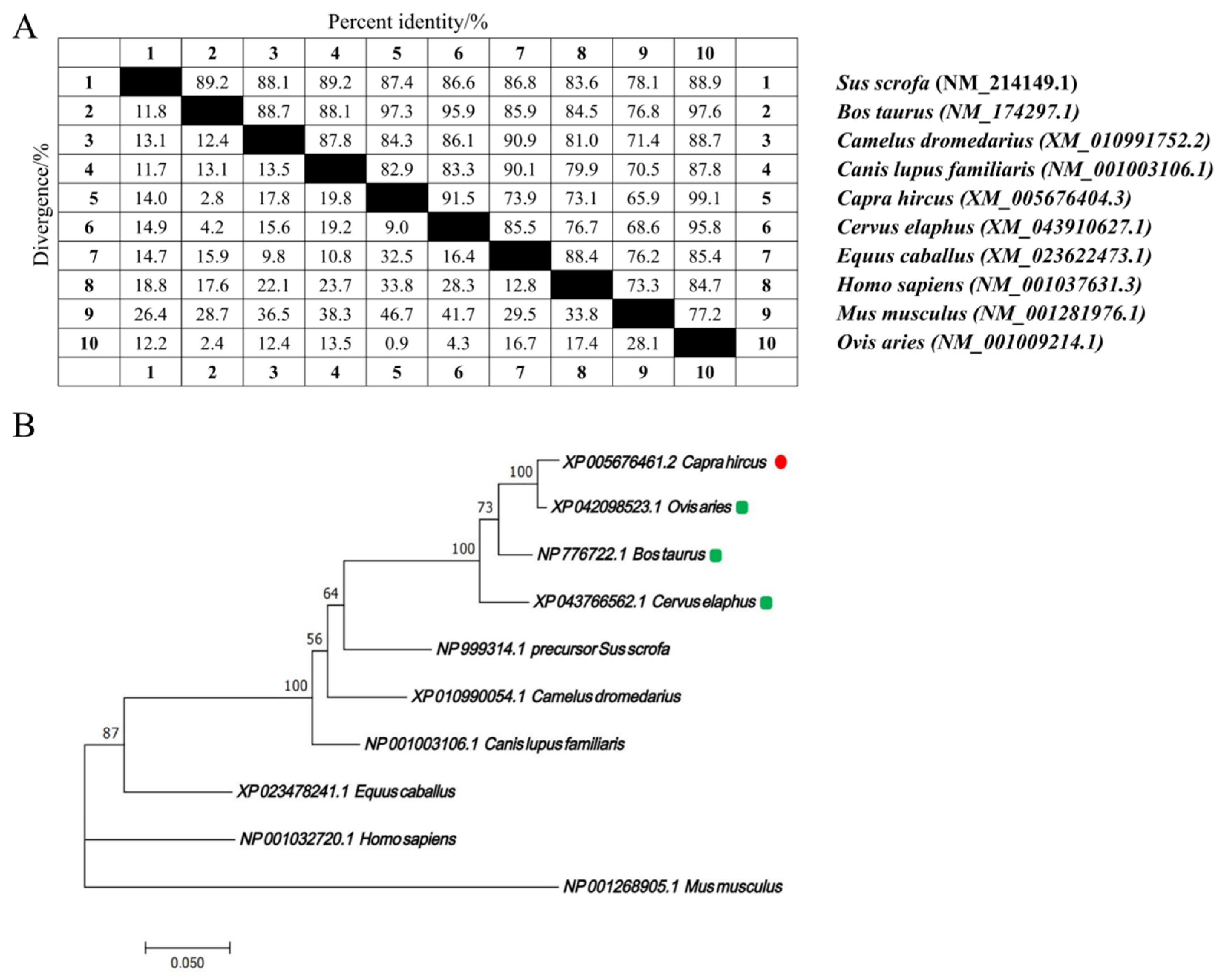
2.1. Gene Conservation Analysis and mRNA Expression
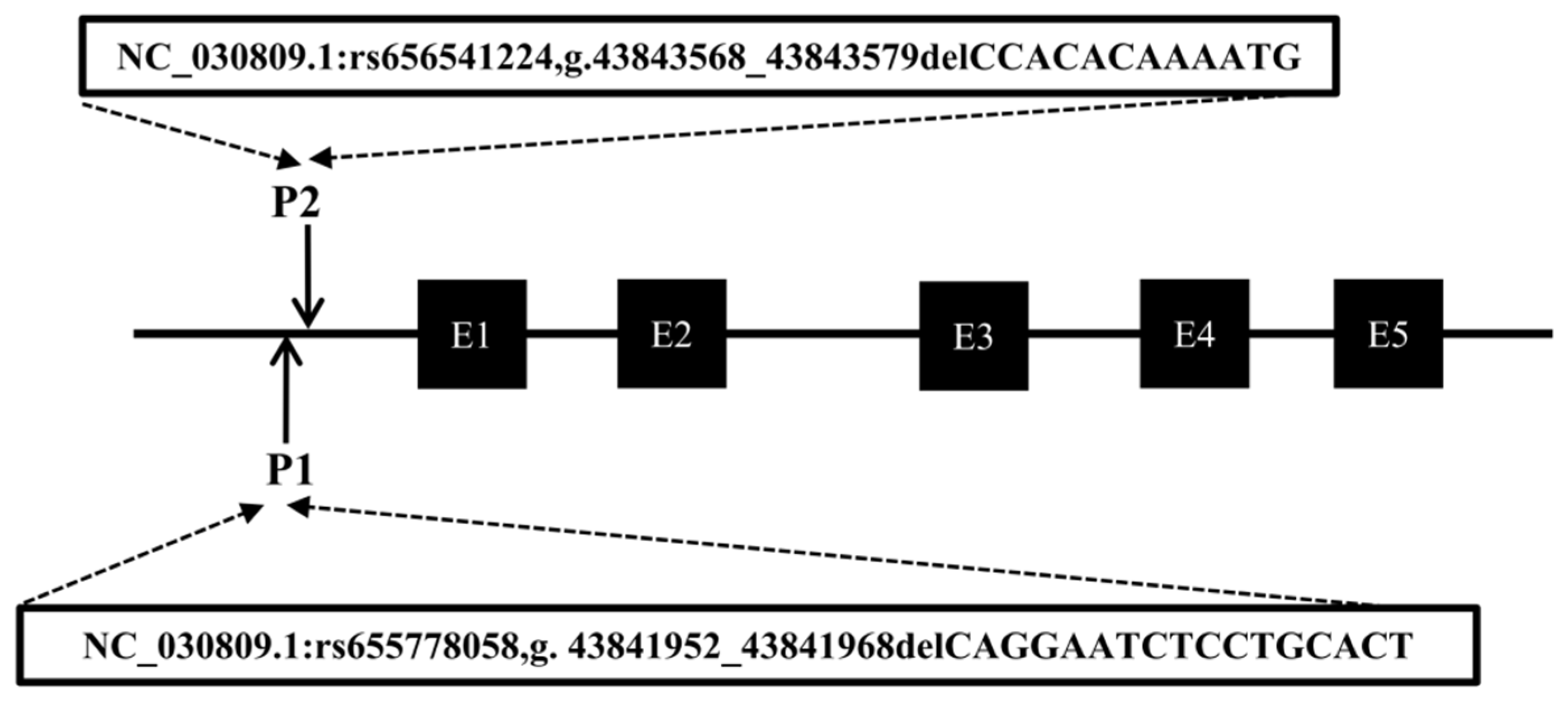
2.2. Identification of Insertion/Deletion Variants in the CTLA4 Gene
2.3. Analysis of Genetic Parameters of CTLA4 in Goats
2.4. Association Analysis of CTLA4 Gene Polymorphisms with the Risk of Brucellosis in Goats
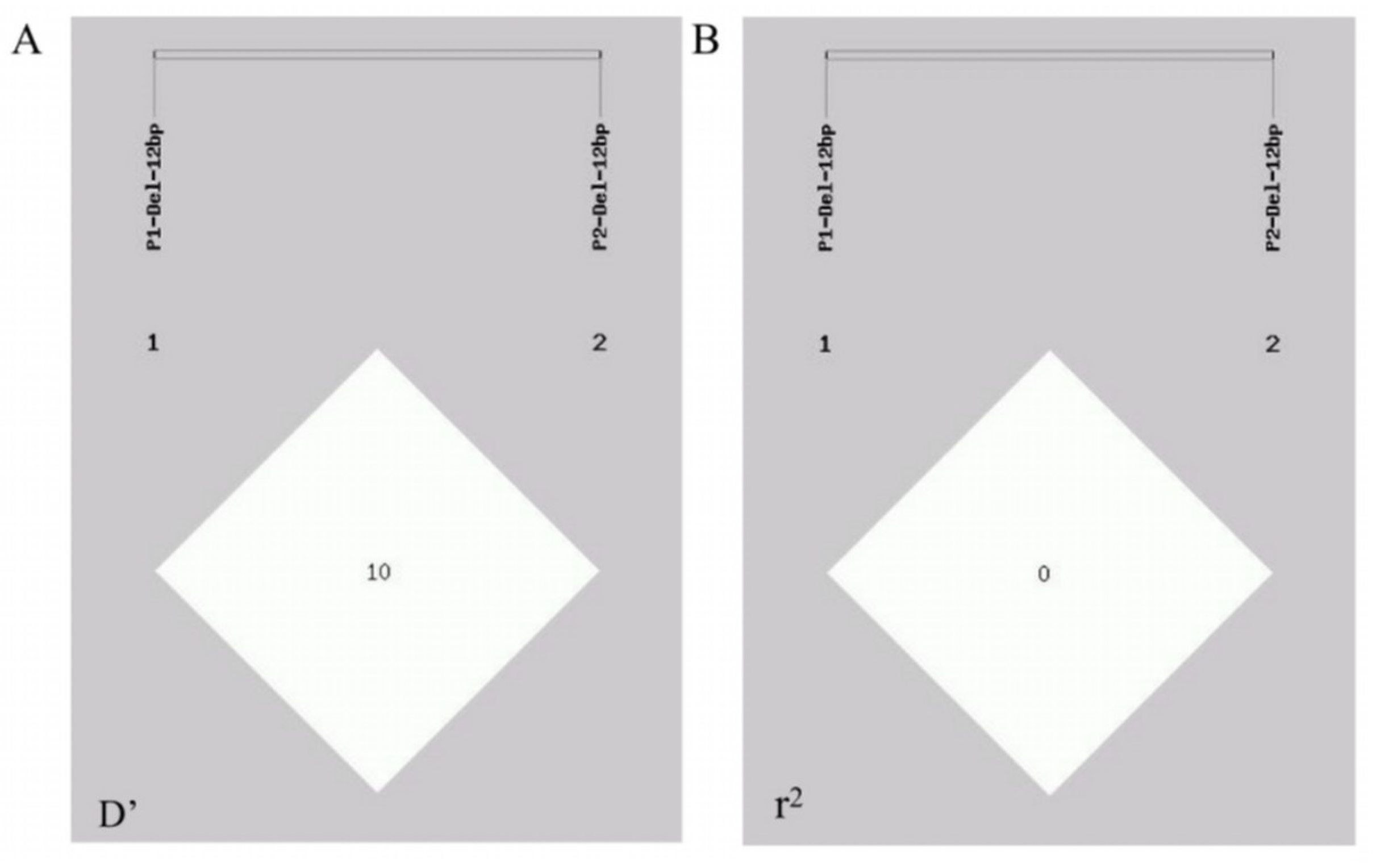
2.5. Linkage Disequilibrium (LD) and Haplotype Association Analysis
2.6. LPS Stimulates Changes in CTLA4 and Cytokine mRNA Expression Levels in Peripheral Blood Monocytes and/or Macrophages
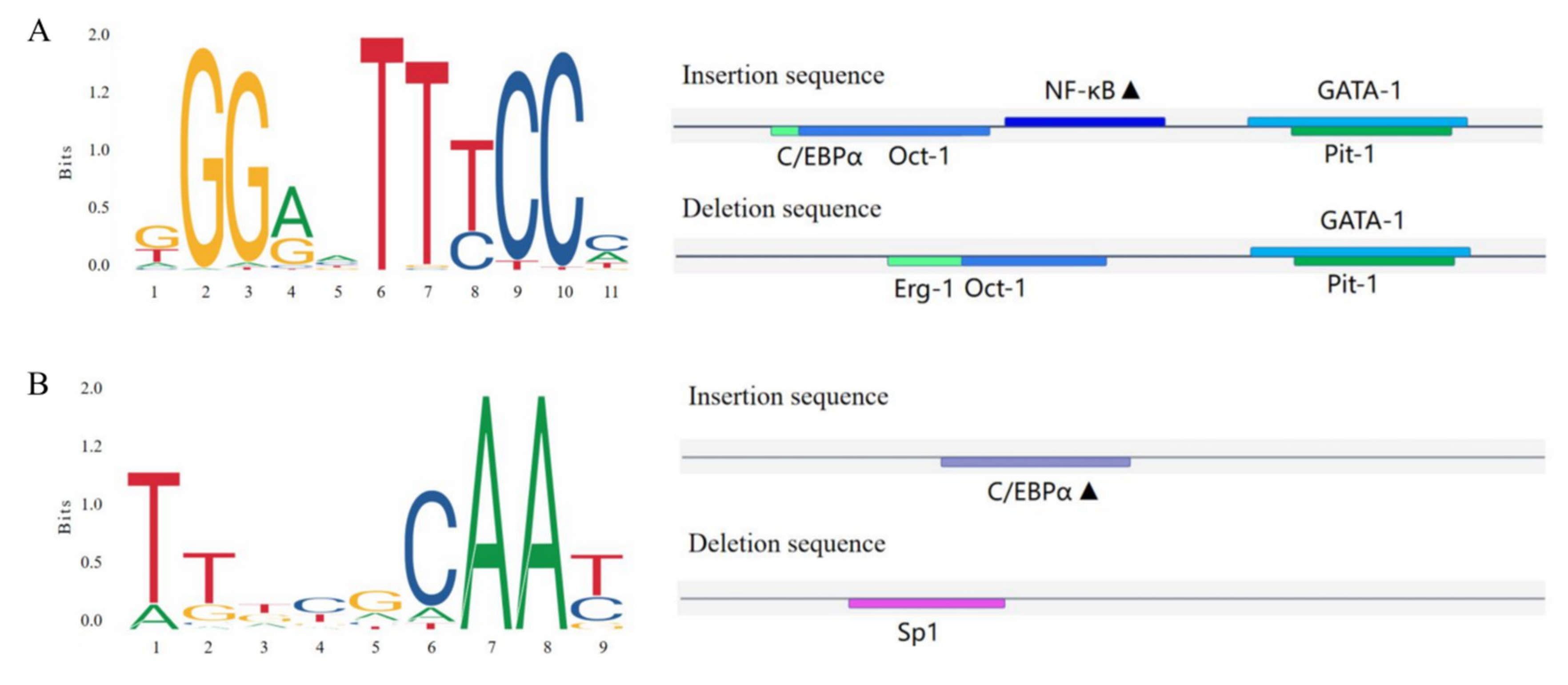
2.7. Predicted Binding of Transcription Factors
3. Discussion
4. Materials and Methods
4.1. Samples and Data Collection
4.2. DNA Extraction, Primer Design, and Genotyping
4.3. RNA Extraction, cDNA Synthesis, and qRT-PCR
4.4. Bioinformatic Analysis
4.5. LPS In Vitro Stimulation of Peripheral Blood Monocytes and/or Macrophages
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, T.; Zhu, Y.; Shi, J.; Shang, K.; Yin, Z.; Shi, H.; He, Y.; Ding, J.; Zhang, F. The development of a human Brucella mucosal vaccine: What should be considered? Life Sci. 2024, 355, 122986. [Google Scholar] [CrossRef]
- Li, Y.; Tan, D.; Xue, S.; Shen, C.; Ning, H.; Cai, C.; Liu, Z. Prevalence, distribution and risk factors for brucellosis infection in goat farms in Ningxiang, China. BMC Vet. Res. 2021, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Siengsanan-Lamont, J.; Kong, L.; Heng, T.; Khoeun, S.; Tum, S.; Selleck, P.W.; Gleeson, L.J.; Blacksell, S.D. Risk mapping using serologic surveillance for selected One Health and transboundary diseases in Cambodian goats. PLoS Neglected Trop. Dis. 2023, 17, e0011244. [Google Scholar] [CrossRef]
- Rossetti, C.A.; Arenas-Gamboa, A.M.; Maurizio, E. Caprine brucellosis, A historically neglected disease with significant impact on public health. PLoS Neglected Trop. Dis. 2017, 11, e0005692. [Google Scholar] [CrossRef]
- De Massis, F.; Di Girolamo, A.; Petrini, A.; Pizzigallo, E.; Giovannini, A. Correlation between animal and human brucellosis in Italy during the period 1997–2002. Clin. Microbiol. Infect. 2005, 11, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Sibhat, B.; Tessema, T.S.; Nile, E.; Asmare, K. Brucellosis in Ethiopia: A comprehensive review of literature from the year 2000–2020 and the way forward. Transbound. Emerg. Dis. 2022, 69, e1231–e1252. [Google Scholar] [CrossRef]
- Miller, C.N.; Smith, E.P.; Cundiff, J.A.; Knodler, L.A.; Bailey Blackburn, J.; Lupashin, V.; Celli, J. A Brucella Type IV Effector Targets the COG Tethering Complex to Remodel Host Secretory Traffic and Promote Intracellular Replication. Cell Host Microbe 2017, 22, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yan, H.; Wang, K.; Cui, Y.; Chen, R.; Liu, J.; Zhu, H.; Qu, L.; Pan, C. Goat SPEF2, Expression profile, indel variants identification and association analysis with litter size. Theriogenology 2019, 139, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Haque, N.; Bari, M.S.; Hossain, M.A.; Muhammad, N.; Ahmed, S.; Rahman, A.; Hoque, S.M.; Islam, A. An overview of Brucellosis. Mymensingh Med. J. 2011, 20, 742–747. [Google Scholar]
- Ran, X.; Chen, X.; Wang, M.; Cheng, J.; Ni, H.; Zhang, X.X.; Wen, X. Brucellosis seroprevalence in ovine and caprine flocks in China during 2000–2018: A systematic review and meta-analysis. BMC Vet. Res. 2018, 14, 393. [Google Scholar] [CrossRef]
- Ran, X.; Cheng, J.; Wang, M.; Chen, X.; Wang, H.; Ge, Y.; Ni, H.; Zhang, X.X.; Wen, X. Brucellosis seroprevalence in dairy cattle in China during 2008–2018: A systematic review and meta-analysis. Acta Trop. 2019, 189, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.; Dohoo, I.; Lindahl, J.; Verdugo, C.; Akuku, I.; Grace, D. Prevalence of tuberculosis, brucellosis and trypanosomiasis in cattle in Tanzania: A systematic review and meta-analysis. Anim. Health Res. Rev. 2016, 17, 16–27. [Google Scholar] [CrossRef]
- Njeru, J.; Wareth, G.; Melzer, F.; Henning, K.; Pletz, M.W.; Heller, R.; Neubauer, H. Systematic review of brucellosis in Kenya: Disease frequency in humans and animals and risk factors for human infection. BMC Public Health 2016, 16, 853. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.O. Brucellosis in food-producing animals in Mosul, Iraq: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0235862. [Google Scholar] [CrossRef] [PubMed]
- Bamaiyi, P.H.; Hassan, L.; Khairani-Bejo, S.; Zainal Abidin, M. The economic impact attributable to brucellosis among goat farms in Peninsula Malaysia and cost benefit analysis. Res. Opin. Anim. Vet. Sci. 2015, 5, 57–64. [Google Scholar]
- Sulima, M.; Venkataraman, K. Economic losses due to Brucella melitensis infection in sheep and goats. Tamilnadu J. Vet. Anim. Sci. 2010, 6, 191–192. [Google Scholar]
- Godfroid, J.; Al Dahouk, S.; Pappas, G.; Roth, F.; Matope, G.; Muma, J.; Marcotty, T.; Pfeiffer, D.; Skjerve, E. A “One Health” surveillance and control of brucellosis in developing countries, moving away from improvisation. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 241–248. [Google Scholar] [CrossRef]
- Hou, Q.; Sun, X.; Zhang, J.; Liu, Y.; Wang, Y.; Jin, Z. Modeling the transmission dynamics of sheep brucellosis in Inner Mongolia Autonomous Region, China. Math. Biosci. 2013, 242, 51–58. [Google Scholar] [CrossRef]
- Blasco, J.M.; Molina-Flores, B. Control and eradication of Brucella melitensis infection in sheep and goats. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 95–104. [Google Scholar] [CrossRef]
- Li, T.; Tong, Z.; Huang, M.; Tang, L.; Zhang, H.; Chen, C. Brucella melitensis M5-90Δbp26 as a potential live vaccine that allows for the distinction between natural infection and immunization. Can. J. Microbiol. 2017, 63, 719–729. [Google Scholar] [CrossRef]
- Pascual, D.W.; Yang, X.; Wang, H.; Goodwin, Z.; Hoffman, C.; Clapp, B. Alternative strategies for vaccination to brucellosis. Microbes Infect. 2018, 20, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Wijayanti, D.; Zhang, S.; Yang, Y.; Bai, Y.; Akhatayeva, Z.; Pan, C.; Zhu, H.; Qu, L.; Lan, X. Goat SMAD family member 1 (SMAD1): mRNA expression, genetic variants, and their associations with litter size. Theriogenology 2022, 193, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Agusti, A.; Gulsvik, A.; Bakke, P.; Coxson, H.; Lomas, D.A.; Silverman, E.K.; Pillai, S.G.; ICGN investigators. CTLA4 gene polymorphisms are associated with chronic bronchitis. Eur. Respir. J. 2009, 34, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.S.K. The link between circulating follicular helper T cells and autoimmunity. Nat. Rev. Immunol. 2022, 22, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Ghorbaninezhad, F.; Masoumi, J.; Bakhshivand, M.; Baghbanzadeh, A.; Mokhtarzadeh, A.; Kazemi, T.; Aghebati-Maleki, L.; Shotorbani, S.S.; Jafarlou, M.; Brunetti, O.; et al. CTLA-4 silencing in dendritic cells loaded with colorectal cancer cell lysate improves autologous T cell responses in vitro. Front. Immunol. 2022, 13, 931316. [Google Scholar] [CrossRef]
- Louthrenoo, W.; Kasitanon, N.; Wongthanee, A.; Kuwata, S.; Takeuchi, F. CTLA-4 polymorphisms in Thai patients with rheumatoid arthritis, systemic lupus erythematosus, and systemic sclerosis. Int. J. Rheum. Dis. 2021, 24, 1378–1385. [Google Scholar] [CrossRef]
- Skendros, P.; Pappas, G.; Boura, P. Cell-mediated immunity in human brucellosis. Microbes Infect. 2011, 13, 134–142. [Google Scholar] [CrossRef]
- Sun, H.L.; Du, X.F.; Tang, Y.X.; Li, G.Q.; Yang, S.Y.; Wang, L.H.; Li, X.W.; Ma, C.J.; Jiang, R.M. Impact of immune checkpoint molecules on FoxP3+ Treg cells and related cytokines in patients with acute and chronic brucellosis. BMC Infect. Dis. 2021, 21, 1025. [Google Scholar] [CrossRef]
- Eskandari-Nasab, E.; Moghadampour, M.; Najibi, H.; Hadadi-Fishani, M. Investigation of CTLA-4 and CD86 gene polymorphisms in Iranian patients with brucellosis infection. Microbiol. Immunol. 2014, 58, 135–141. [Google Scholar] [CrossRef]
- Cui, Y.; Yan, H.; Wang, K.; Xu, H.; Zhang, X.; Zhu, H.; Liu, J.; Qu, L.; Lan, X.; Pan, C. Insertion/Deletion Within the KDM6A Gene Is Significantly Associated With Litter Size in Goat. Front. Genet. 2018, 9, 91. [Google Scholar] [CrossRef]
- Baldwin, C.L.; Goenka, R. Host immune responses to the intracellular bacteria Brucella, does the bacteria instruct the host to facilitate chronic infection? Crit. Rev. Immunol. 2011, 26, 407–442. [Google Scholar] [CrossRef] [PubMed]
- Chen, L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 2004, 4, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Khattri, R.; Auger, J.A.; Griffin, M.D.; Sharpe, A.H.; Bluestone, J.A. Lymphoproliferative disorder in CTLA-4 knockout mice is characterized by CD28-regulated activation of Th2 responses. J. Immunol. 1999, 162, 5784–5791. [Google Scholar] [CrossRef] [PubMed]
- Peggs, K.S.; Quezada, S.A.; Chambers, C.A.; Korman, A.J.; Allison, J.P. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J. Exp. Med. 2009, 206, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhu, Y.; Yin, Z.; Shi, J.; Shang, K.; Tian, T.; Shi, H.; Ding, J.; Zhang, F. Design a novel of Brucellosis preventive vaccine based on IgV_CTLA-4 and multiple epitopes via immunoinformatics approach. Microb. Pathog. 2024, 195, 106909. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Diaz, J.M.; Zambrano-Román, M.; Padilla-Gutiérrez, J.R.; Valle, Y.; Muñoz-Valle, J.F.; Valdés-Alvarado, E. Association of CTLA-4 (AT)n Variants in Basal Cell Carcinoma and Squamous Cell Carcinoma Patients from Western Mexico. Curr. Issues Mol. Biol. 2024, 46, 8368–8375. [Google Scholar] [CrossRef]
- Akhtar, M.S. The Variant Allele Frequency of CTLA-4 rs11571317 (−658 C/T) Polymorphism With Colorectal Cancer Susceptibility in the Saudi Population and Other Ethnic Groups. Cureus 2023, 15, e50091. [Google Scholar] [CrossRef]
- Ghoneim, M.E.; Sheashaa, H.; Wafa, E.; Awadalla, A.; Ahmed, A.E.; Sobh, M.; Shokeir, A.A. Impact of CD 28, CD86, CTLA-4 and PD-1 genes polymorphisms on acute renal allograft rejection and graft survival among Egyptian recipients. Sci. Rep. 2024, 14, 2047. [Google Scholar] [CrossRef]
- Hasenauer, F.C.; Rossi, U.A.; Caffaro, M.E.; Raschia, M.A.; Maurizio, E.; Poli, M.A.; Rossetti, C.A. Association of TNF rs668920841 and INRA111 polymorphisms with caprine brucellosis: A case-control study of candidate genes involved in innate immunity. Genomics 2020, 112, 3925–3932. [Google Scholar] [CrossRef]
- Omar, A.I.; Alam, M.B.B.; Notter, D.R.; Zhao, S.; Faruque, M.O.; Thi, T.N.T.; Yin, L.; Li, J.; Azmal, S.A.; Du, X. Association of single nucleotide polymorphism in NLRC3, NLRC5, HIP1, and LRP8 genes with fecal egg counts in goats naturally infected with Haemonchus contortus. Trop. Anim. Health Prod. 2020, 52, 1583–1598. [Google Scholar] [CrossRef]
- DiLeo, M.F.; Nair, A.; Kardos, M.; Husby, A.; Saastamoinen, M. Demography and environment modulate the effects of genetic diversity on extinction risk in a butterfly metapopulation. Proc. Natl. Acad. Sci. USA 2024, 121, e2309455121. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.F.; Waller, D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002, 17, 230–241. [Google Scholar] [CrossRef]
- Quéméré, E.; Rossi, S.; Petit, E.; Marchand, P.; Merlet, J.; Game, Y.; Galan, M.; Gilot-Fromont, E. Genetic epidemiology of the Alpine ibex reservoir of persistent and virulent brucellosis outbreak. Sci. Rep. 2020, 10, 4400. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, I.; Sharma, A.; Singh, R.; Deb, S.M.; Singh, D.K.; Mitra, A. Association of microsatellite (GT)n polymorphism at 3′UTR of NRAMP1 with the macrophage function following challenge with Brucella LPS in buffalo (Bubalus bubalis). Vet. Microbiol. 2008, 129, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Vacca, G.M.; Pazzola, M.; Pisano, C.; Carcangiu, V.; Diaz, M.L.; Nieddu, M.; Robledo, R.; Mezzanotte, R.; Dettori, M.L. Chromosomal localisation and genetic variation of the SLC11A1 gene in goats (Capra hircus). Vet. J. 2011, 190, 60–65. [Google Scholar] [CrossRef]
- Capparelli, R.; Alfano, F.; Amoroso, M.G.; Borriello, G.; Fenizia, D.; Bianco, A.; Roperto, S.; Roperto, F.; Iannelli, D. Protective effect of the Nramp1 BB genotype against Brucella abortus in the water buffalo (Bubalus bubalis). Infect. Immun. 2007, 75, 988–996. [Google Scholar] [CrossRef]
- Lou, L.; Bao, W.; Liu, X.; Song, H.; Wang, Y.; Zhang, K.; Gao, W.; Li, H.; Tu, Z.; Wang, S. An Autoimmune Disease-Associated Risk Variant in the TNFAIP3 Gene Plays a Protective Role in Brucellosis That Is Mediated by the NF-κB Signaling Pathway. J. Clin. Microbiol. 2018, 56, e01363. [Google Scholar] [CrossRef]
- Martins, G.A.; Tadokoro, C.E.; Silva, R.B.; Silva, J.S.; Rizzo, L.V. CTLA-4 blockage increases resistance to infection with the intracellular protozoan Trypanosoma cruzi. J. Immunol. 2004, 172, 4893–4901. [Google Scholar] [CrossRef]
- Wang, P.H.; Wu, M.F.; Hsu, C.Y.; Lin, S.Y.; Chang, Y.N.; Lee, H.S.; Wei, Y.F.; Shu, C.C. The Dynamic Change of Immune Checkpoints and CD14+ Monocytes in Latent Tuberculosis Infection. Biomedicines 2021, 9, 1479. [Google Scholar] [CrossRef]
- Huang, D.W.; Wang, J.X.; Liu, Q.Y.; Chu, M.X.; Di, R.; He, J.N.; Cao, G.L.; Fang, L.; Feng, T.; Li, N. Analysis on DNA sequence of TSHB gene and its association with reproductive seasonality in goats. Mol. Biol. Rep. 2013, 40, 1893–1904. [Google Scholar] [CrossRef]
- Jin, J.; Hu, H.; Li, H.S.; Yu, J.; Xiao, Y.; Brittain, G.C.; Zou, Q.; Cheng, X.; Mallette, F.A.; Watowich, S.S.; et al. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity 2014, 40, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Malle, E.K.; Zammit, N.W.; Walters, S.N.; Koay, Y.C.; Wu, J.; Tan, B.M.; Villanueva, J.E.; Brink, R.; Loudovaris, T.; Cantley, J.; et al. Nuclear factor κB-inducing kinase activation as a mechanism of pancreatic β cell failure in obesity. J. Exp. Med. 2015, 212, 1239–1254. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Bhargavan, B.; Woollard, S.M.; Kanmogne, G.D. Toll-like receptor-3 mediates HIV-1 transactivation via NFκB and JNK pathways and histone acetylation, but prolonged activation suppresses Tat and HIV-1 replication. Cell Signal. 2016, 28, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Zong, Q.; Wang, H.; Wu, S.; Cai, D.; Bao, W. C/EBPα Epigenetically Modulates TFF1 Expression via mC-6 Methylation in the Jejunum Inflammation Induced by a Porcine Coronavirus. Front. Immunol. 2022, 13, 881289. [Google Scholar] [CrossRef]
- Peña-Martínez, E.G.; Rodríguez-Martínez, J.A. Decoding Non-coding Variants: Recent Approaches to Studying Their Role in Gene Regulation and Human Diseases. Front. Biosci. (Sch. Ed.) 2024, 16, 4. [Google Scholar] [CrossRef]
- Schokker, D.; Peters, T.H.; Hoekman, A.J.; Rebel, J.M.; Smits, M.A. Differences in the early response of hatchlings of different chicken breeding lines to Salmonella enterica serovar Enteritidis infection. Poult. Sci. 2012, 91, 346–353. [Google Scholar] [CrossRef]
- Kim, Y.C.; Won, S.Y.; Jeong, B.H. Absence of single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the prion protein gene (PRNP) in a large sampling of various chicken breeds. BMC Genom. 2019, 20, 922. [Google Scholar] [CrossRef]
- Rahman, S.U.; Zhu, L.; Cao, L.; Zhang, Y.; Chu, X.; Feng, S.; Li, Y.; Wu, J.; Wang, X. Prevalence of Caprine brucellosis in Anhui province, China. Vet. World 2019, 12, 558–564. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4703. [Google Scholar] [CrossRef]
- Shrivastava, K.; Kumar, P.; Sahoo, N.R.; Kumar, A.; Khan, M.F.; Kumar, A.; Prasad, A.; Patel, B.H.; Nasir, A.; Bhushan, B.; et al. Genotyping of major histocompatibility complex Class II DRB gene in Rohilkhandi goats by polymerase chain reaction-restriction fragment length polymorphism and DNA sequencing. Vet. World 2015, 8, 1183–1188. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7, Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, G.; Zhang, J.; Zhang, X.; Cui, M.; Guo, Y.; Liu, G.; Li, G.; Feng, J.; Lian, Z. Transgenic cloned sheep overexpressing ovine toll-like receptor 4. Theriogenology 2013, 80, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3333. [Google Scholar] [CrossRef]
- Sepanjnia, A.; Eskandari-Nasab, E.; Moghadampour, M.; Tahmasebi, A.; Dahmardeh, F. TGFβ1 genetic variants are associated with an increased risk of acute brucellosis. Infect. Dis. 2015, 47, 458–464. [Google Scholar] [CrossRef]



| Locus | Serological Results | Sizes | Genotypic Frequencies | Allelic Frequencies | Population Parameters | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | II | ID | DD | I | D | Ho | He | Ne | PIC | p Value | ||
| P1 | Cases | 398 | 0.668 (266) | 0.271 (108) | 0.060 (24) | 0.804 | 0.196 | 0.685 | 0.315 | 1.460 | 0.266 | p < 0.05 |
| Control | 406 | 0.586 (238) | 0.328 (133) | 0.086 (35) | 0.750 | 0.250 | 0.625 | 0.375 | 1.600 | 0.305 | p < 0.05 | |
| Sum | 804 | 0.627 (504) | 0.300 (241) | 0.073 (59) | 0.777 | 0.223 | 0.653 | 0.347 | 1.531 | 0.287 | p < 0.05 | |
| P2 | Cases | 416 | 0.728 (303) | 0.236 (98) | 0.036 (15) | 0.846 | 0.154 | 0.740 | 0.260 | 1.352 | 0.227 | p > 0.05 |
| Control | 422 | 0.782 (274) | 0.694 (132) | 0.038 (16) | 0.806 | 0.194 | 0.687 | 0.313 | 1.456 | 0.264 | p > 0.05 | |
| Sum | 838 | 0.689 (577) | 0.274 (230) | 0.037 (31) | 0.826 | 0.174 | 0.712 | 0.288 | 1.404 | 0.246 | p > 0.05 | |
| Locus | Genotype/Allele | Frequencies | χ2 | p Value | |
|---|---|---|---|---|---|
| Cases | Control | ||||
| P1 | II | 0.668 (266) | 0.586 (238) | 6.121 | 0.047 |
| ID | 0.271 (108) | 0.328 (133) | |||
| DD | 0.060 (24) | 0.086 (35) | |||
| I | 0.804 (640) | 0.750 (609) | 6.764 | 0.009 | |
| D | 0.196 (156) | 0.250 (203) | |||
| P2 | II | 0.728 (303) | 0.782 (274) | 6.473 | 0.039 |
| ID | 0.236 (98) | 0.694 (132) | |||
| DD | 0.036 (15) | 0.038 (16) | |||
| I | 0.846 (704) | 0.806 (680) | 4.769 | 0.029 | |
| D | 0.154 (128) | 0.194 (164) | |||
| Locus | Genetic Models | Genotype/Allele | Frequencies | p Value | OR (95% CI) | |
|---|---|---|---|---|---|---|
| Cases | Control | |||||
| P1 | Codominant | II | 0.668 (266) | 0.586 (238) | 1.00 | |
| ID | 0.271 (108) | 0.328 (133) | 0.042 | 1.337 (0.968~1.846) | ||
| DD | 0.060 (24) | 0.086 (35) | 0.081 | 1.501 (0.853~2.643) | ||
| Dominant | II | 0.668 (266) | 0.586 (238) | 1.00 | ||
| ID + DD | 0.332 (132) | 0.414 (168) | 0.016 | 1.422 (1.067~1.896) | ||
| Recessive | II + ID | 0.940 (374) | 0.914 (371) | 1.00 | ||
| DD | 0.060 (24) | 0.086 (35) | 0.161 | 1.470 (0.858~2.520) | ||
| Alleles | I | 0.804 (640) | 0.750 (609) | 1.00 | ||
| D | 0.196 (156) | 0.250 (203) | 0.009 | 1.303 (1.028~1.652) | ||
| P2 | Codominant | II | 0.728 (303) | 0.782 (274) | 1.00 | |
| ID | 0.236 (98) | 0.694 (132) | 0.011 | 1.512 (1.096~2.086) | ||
| DD | 0.036 (15) | 0.038 (16) | 0.654 | 1.186 (0.556~2.532) | ||
| Dominant | II | 0.728 (303) | 0.782 (274) | 1.00 | ||
| ID + DD | 0.272 (113) | 0.218 (148) | 0.014 | 1.448 (1.079~1.944) | ||
| Recessive | II + ID | 0.964 (401) | 0.962 (406) | 1.00 | ||
| DD | 0.036 (15) | 0.038 (16) | 0.887 | 1.054 (0.514~2.160) | ||
| Alleles | I | 0.846 (704) | 0.806 (680) | 1.00 | ||
| D | 0.154 (128) | 0.194 (164) | 0.029 | 1.326 (1.029~1.710) | ||
| Haplotypic Names | Haplotypic Types | Haplotypic Frequencies | p Value | OR (95% CI) | |
|---|---|---|---|---|---|
| Cases | Control | ||||
| Hap1 | IP1IP2 | 529 (0.692) | 476 (0.609) | 1.00 | |
| Hap2 | IP1DP2 | 84 (0.110) | 107 (0.137) | 0.029 | 1.416 (1.037–1.933) |
| Hap3 | DP1IP2 | 117 (0.154) | 149 (0.191) | 0.012 | 1.415 (1.078–1.858) |
| Hap4 | DP1DP2 | 34 (0.044) | 50 (0.064) | 0.034 | 1.634 (1.039–2.571) |
| Primer | Primer Sequence (5′-3′) | Length (bp) | Tm (°C) | Function |
|---|---|---|---|---|
| P1 | F: ATTCCATCACCCACAG R: ATTACCCAGTTATCCTTC | 285 | 59 | Indel detection |
| P2 | F: CACAGCCATACCCATTC R: GCACCATAAGGAGCAA | 291 | 59 | Indel detection |
| GAPDH | F: AAAGTGGACATCGTCGCCAT R: CCGTTCTCTGCCTTGACTGT | 116 | 60 | Internal control |
| CTLA4 | F: GGCAGCAGTTAGTTCAGGGT R: GCTCTGTTGGGGGCATTTTC | 165 | 60 | qPCR |
| IL-6 | F: TTCAGTCCACTCGCTGTCTC R: TGCTTGGGGTGGTGTCATTC | 106 | 60 | qPCR |
| IL-10 | F: TGATGCCACAGGCTGAGAAC R: TTCGCAGGGCAGAAAACGAT | 119 | 60 | qPCR |
| IL-12 | F: AACCAGACCCACCCAAGAAC R: GTGGCATGTGACTTTGGCTG | 197 | 60 | qPCR |
| IFN-γ | F: AGATCCAGCGCAAAGCCATA R: TCTCCGGCCTCGAAAGAGAT | 110 | 60 | qPCR |
| TNF-α | F: AACCCATCTACCAGGGAGGG R: AGTAGACCTGCCCAGACTCA | 108 | 60 | qPCR |
| TGF-β | F: AACAATTCCTGGCGCTACCT R: ACTGAGGCGAAAGCCCTCTA | 135 | 60 | qPCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Liu, X.; Ren, Z.; Du, X.; Li, N.; Song, X.; Wu, W.; Qu, L.; Zhu, H.; Hua, J. The Goat Cytotoxic T Lymphocyte-Associated Antigen-4 Gene: mRNA Expression and Association Analysis of Insertion/Deletion Variants with the Risk of Brucellosis. Int. J. Mol. Sci. 2024, 25, 10948. https://doi.org/10.3390/ijms252010948
Wang C, Liu X, Ren Z, Du X, Li N, Song X, Wu W, Qu L, Zhu H, Hua J. The Goat Cytotoxic T Lymphocyte-Associated Antigen-4 Gene: mRNA Expression and Association Analysis of Insertion/Deletion Variants with the Risk of Brucellosis. International Journal of Molecular Sciences. 2024; 25(20):10948. https://doi.org/10.3390/ijms252010948
Chicago/Turabian StyleWang, Congliang, Xiaoyu Liu, Zhaofei Ren, Xiaomin Du, Na Li, Xiaoyue Song, Weiwei Wu, Lei Qu, Haijing Zhu, and Jinlian Hua. 2024. "The Goat Cytotoxic T Lymphocyte-Associated Antigen-4 Gene: mRNA Expression and Association Analysis of Insertion/Deletion Variants with the Risk of Brucellosis" International Journal of Molecular Sciences 25, no. 20: 10948. https://doi.org/10.3390/ijms252010948
APA StyleWang, C., Liu, X., Ren, Z., Du, X., Li, N., Song, X., Wu, W., Qu, L., Zhu, H., & Hua, J. (2024). The Goat Cytotoxic T Lymphocyte-Associated Antigen-4 Gene: mRNA Expression and Association Analysis of Insertion/Deletion Variants with the Risk of Brucellosis. International Journal of Molecular Sciences, 25(20), 10948. https://doi.org/10.3390/ijms252010948







