The Critical Role of Phenylpropanoid Biosynthesis Pathway in Lily Resistance Against Gray Mold
Abstract
1. Introduction
2. Results
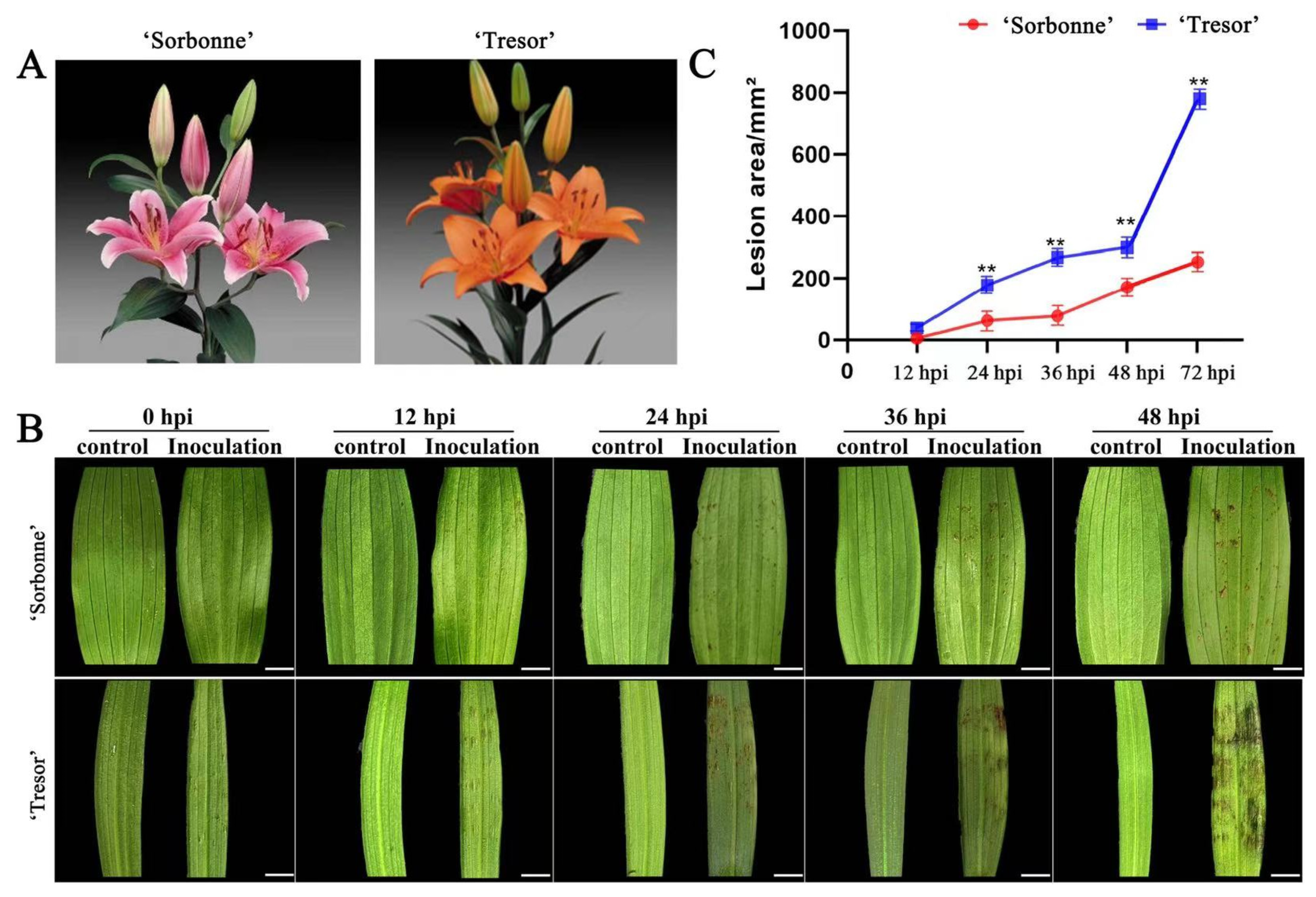
2.1. Leaf Phenotypes of Two Lily Cultivars after Infection by B. elliptica
2.2. Transcriptome Profile of Lily in Response to B. elliptica Infection
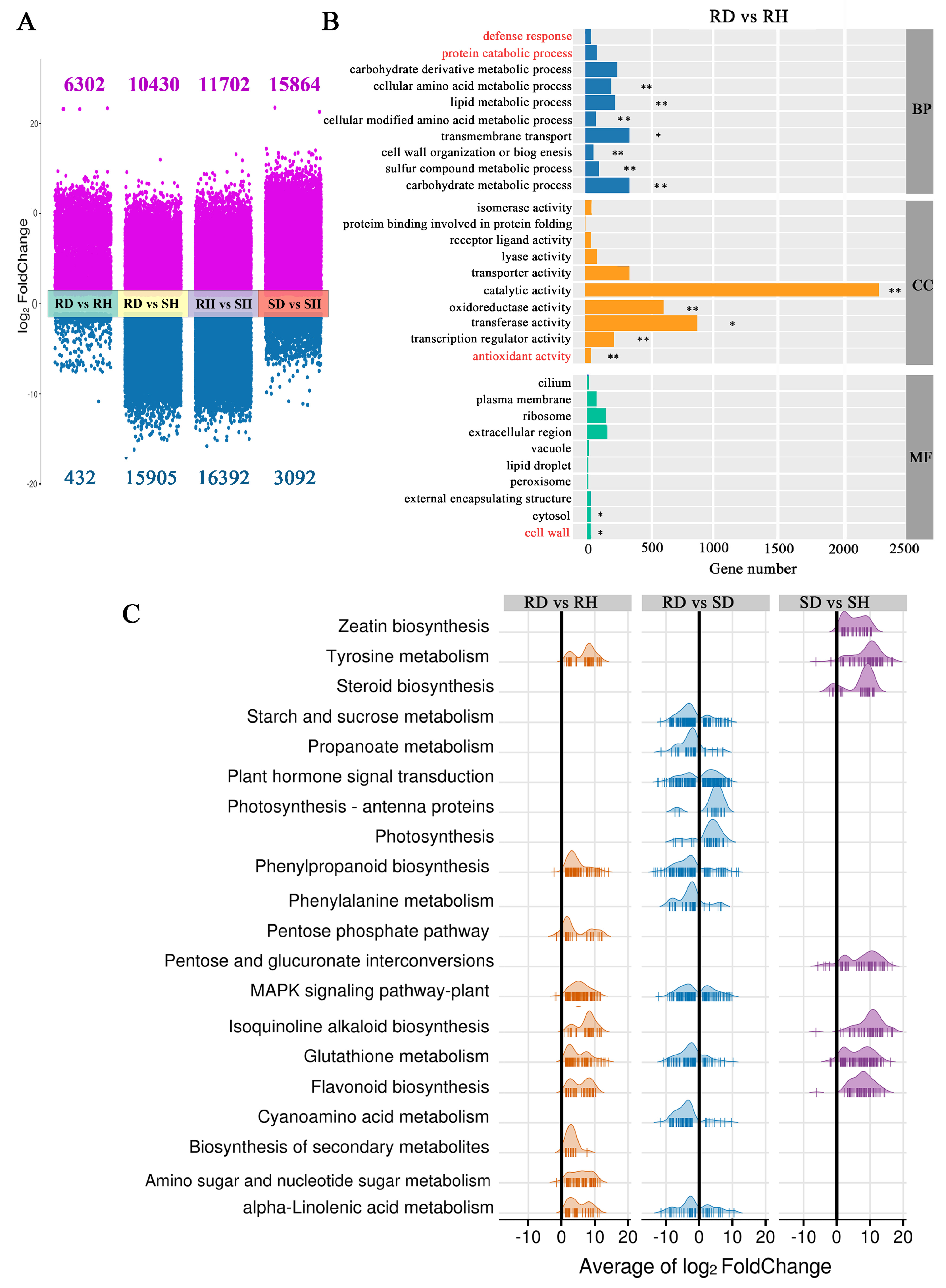
2.2.1. Overview and Analysis of Transcriptome Sequencing Data
2.2.2. Identification and Functional Enrichment Analysis of DEGs in Response to B. elliptica Infection
2.3. Characteristics of the Metabolome of Lily in Response to B. elliptica Infection
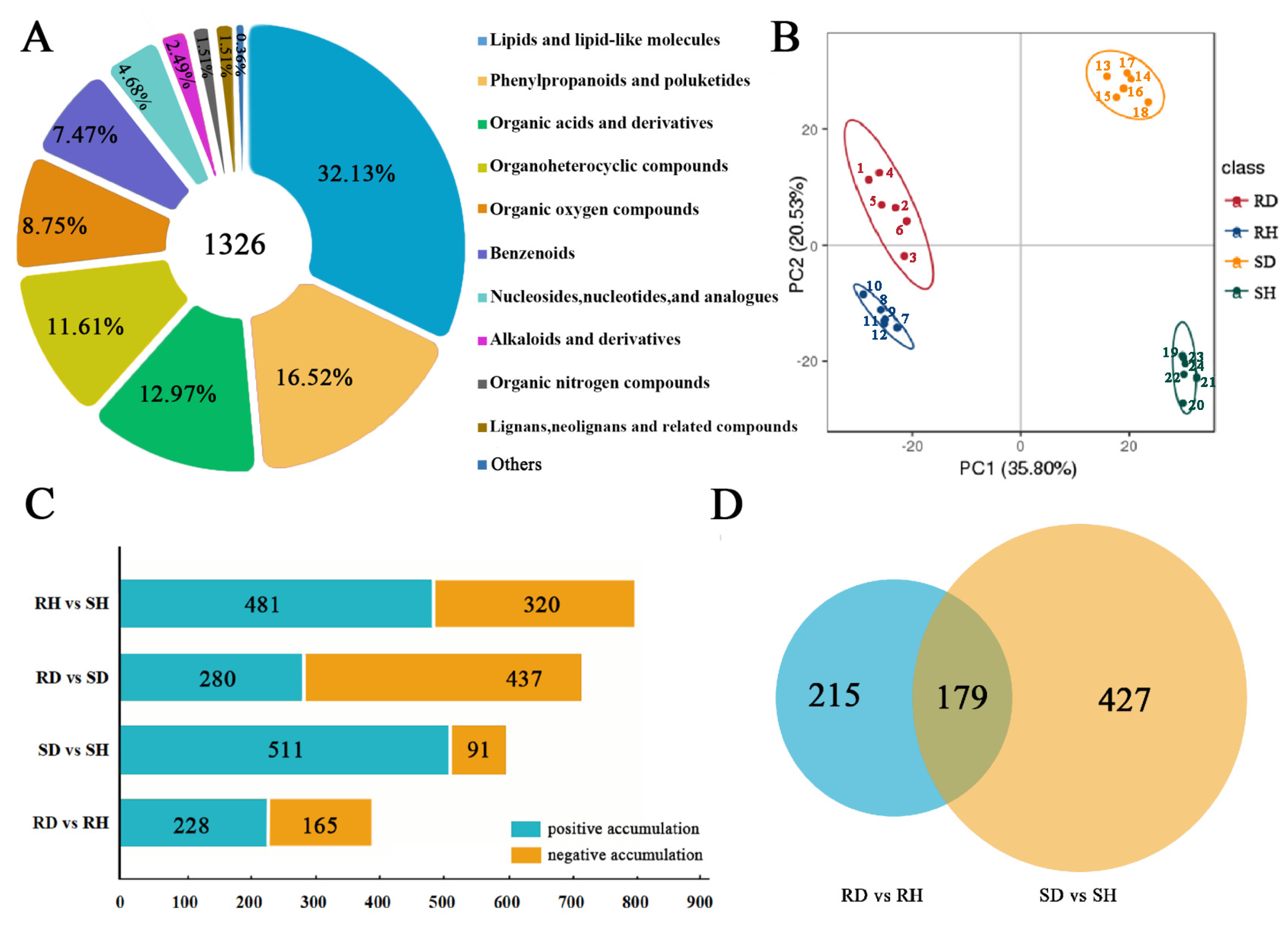
2.3.1. Quality Control of Metabolomic Data
2.3.2. Identification and Functional Enrichment Analysis of DAMs in Response to B. elliptica Infection
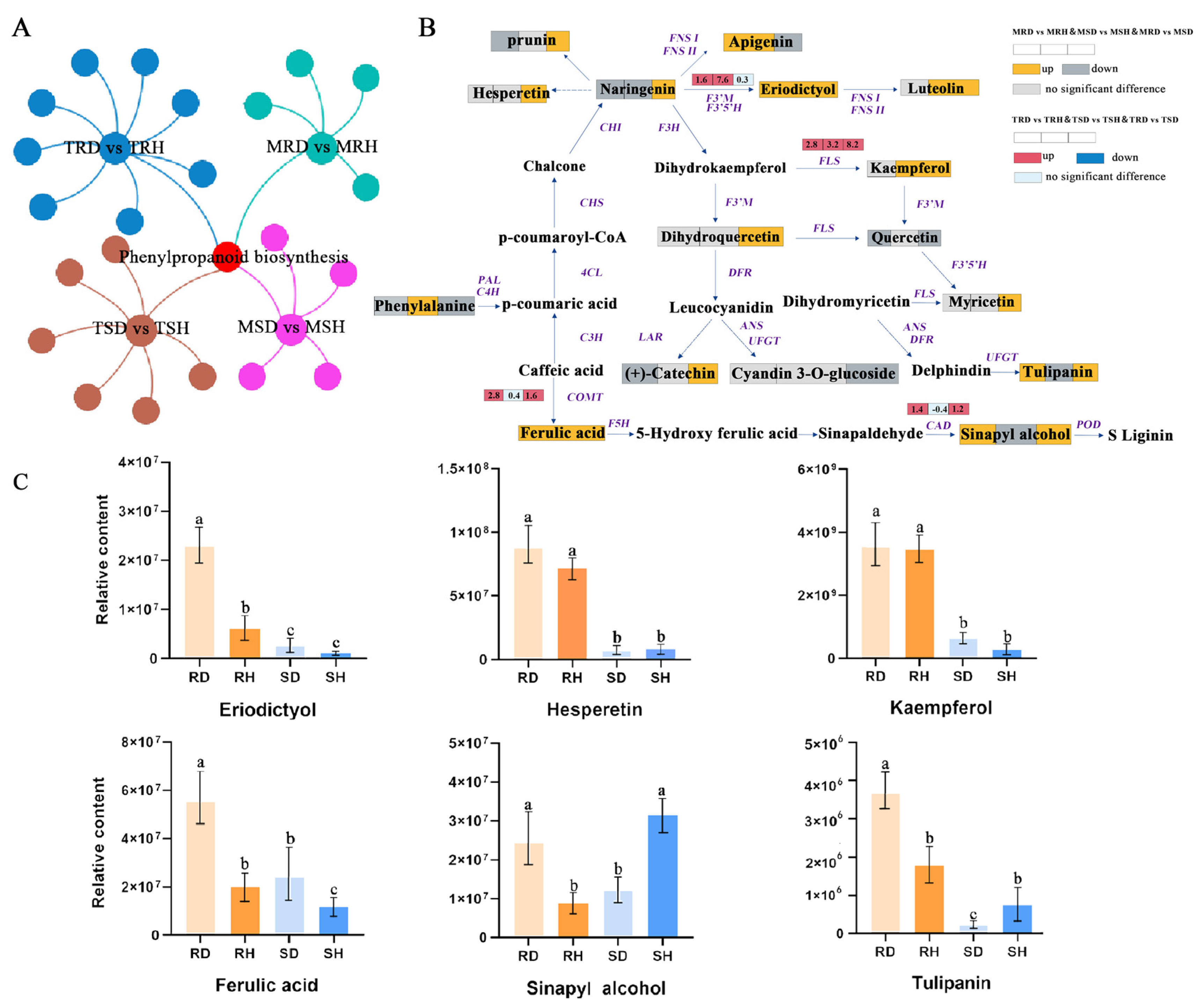
2.4. Integrated Analysis of Transcriptomic and Metabolomic Data Revealed the Importance of Phenylpropanoid Biosynthesis for Lily Resistance to B. elliptica
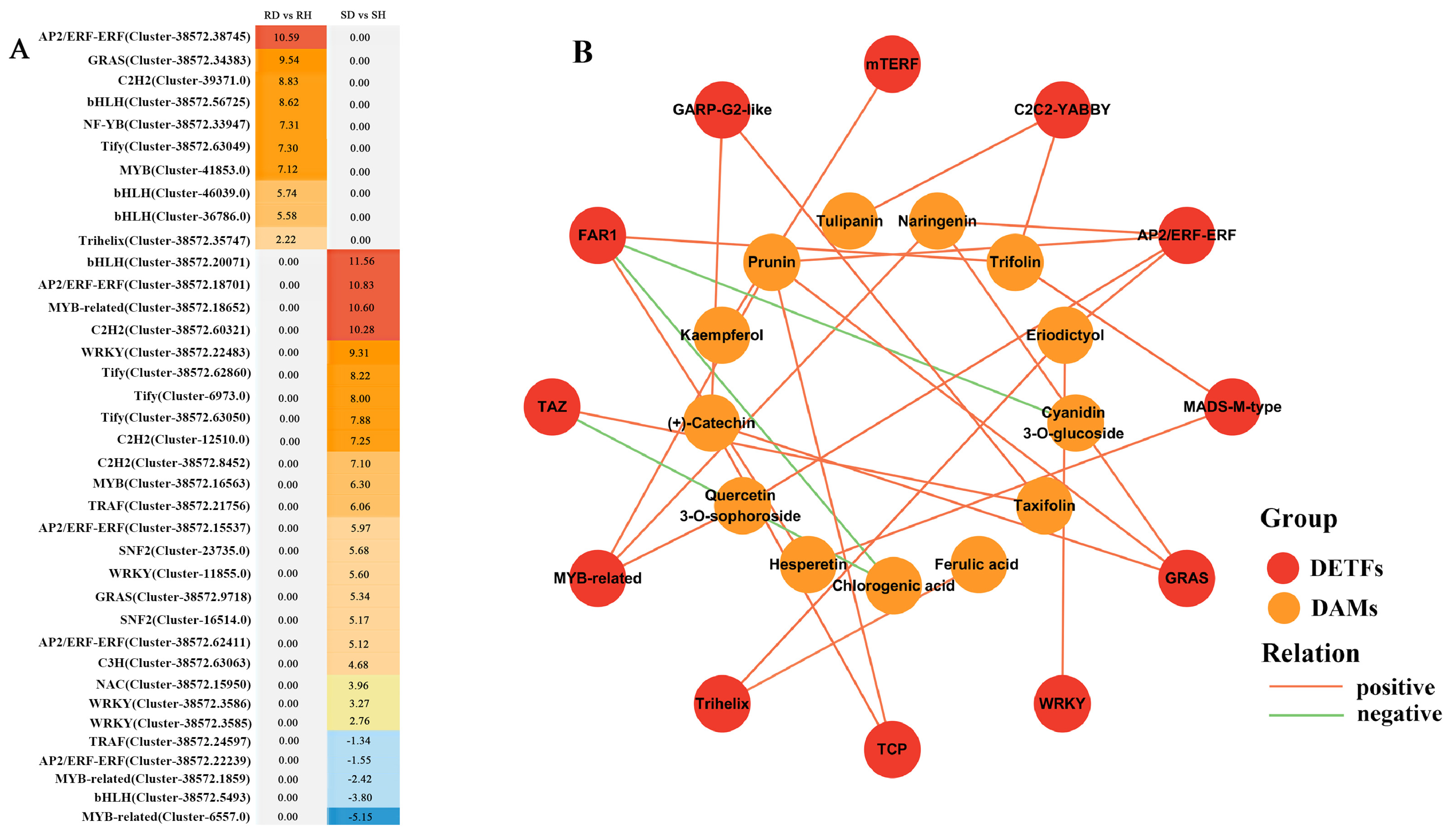
2.5. Transcriptional Network Regulating Phenylpropanoid Biosynthesis during Lily Defense Response to B. elliptica
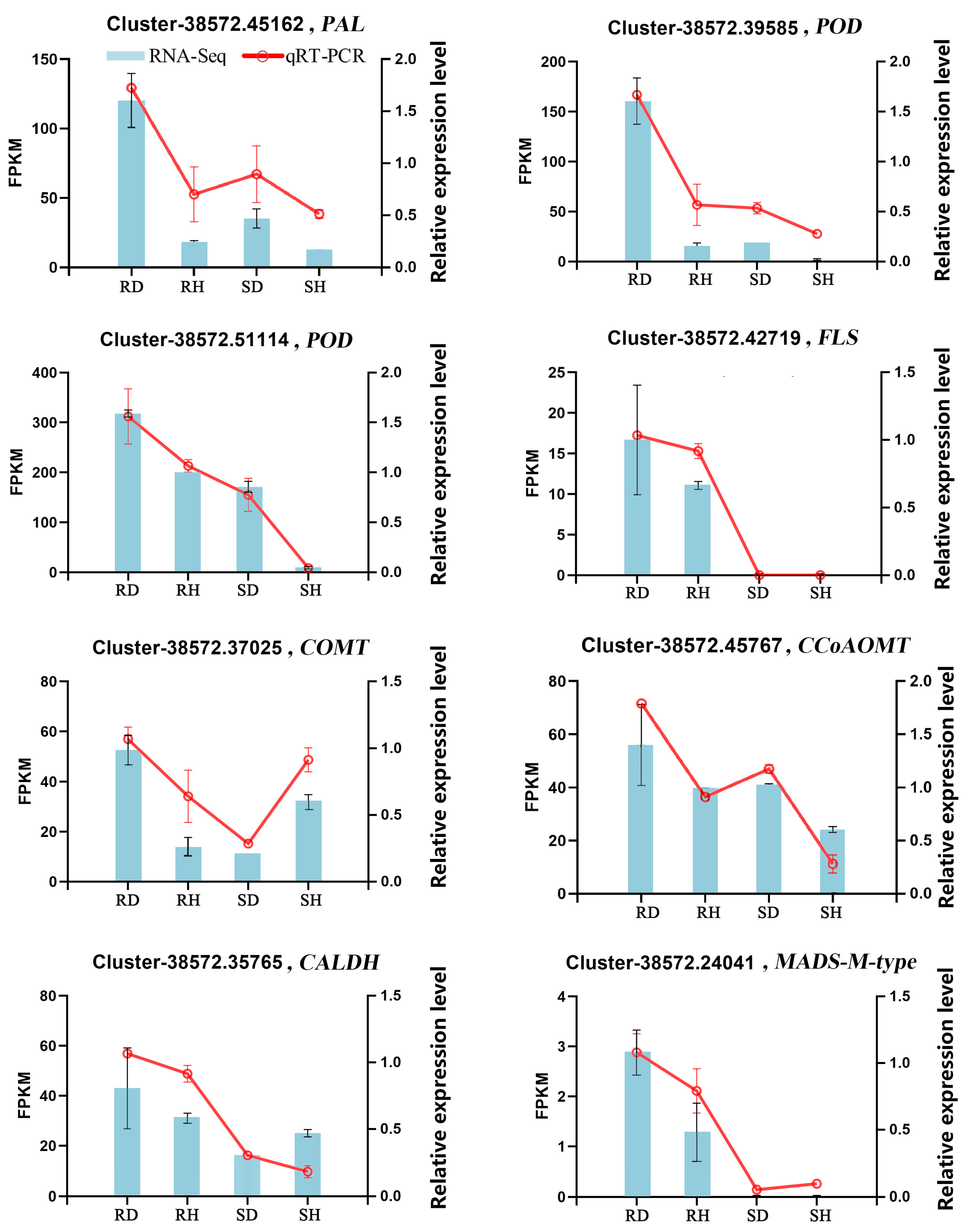
2.6. qRT-PCR Validation of DEGs Associated with Phenylpropanoid Biosynthesis in Lily
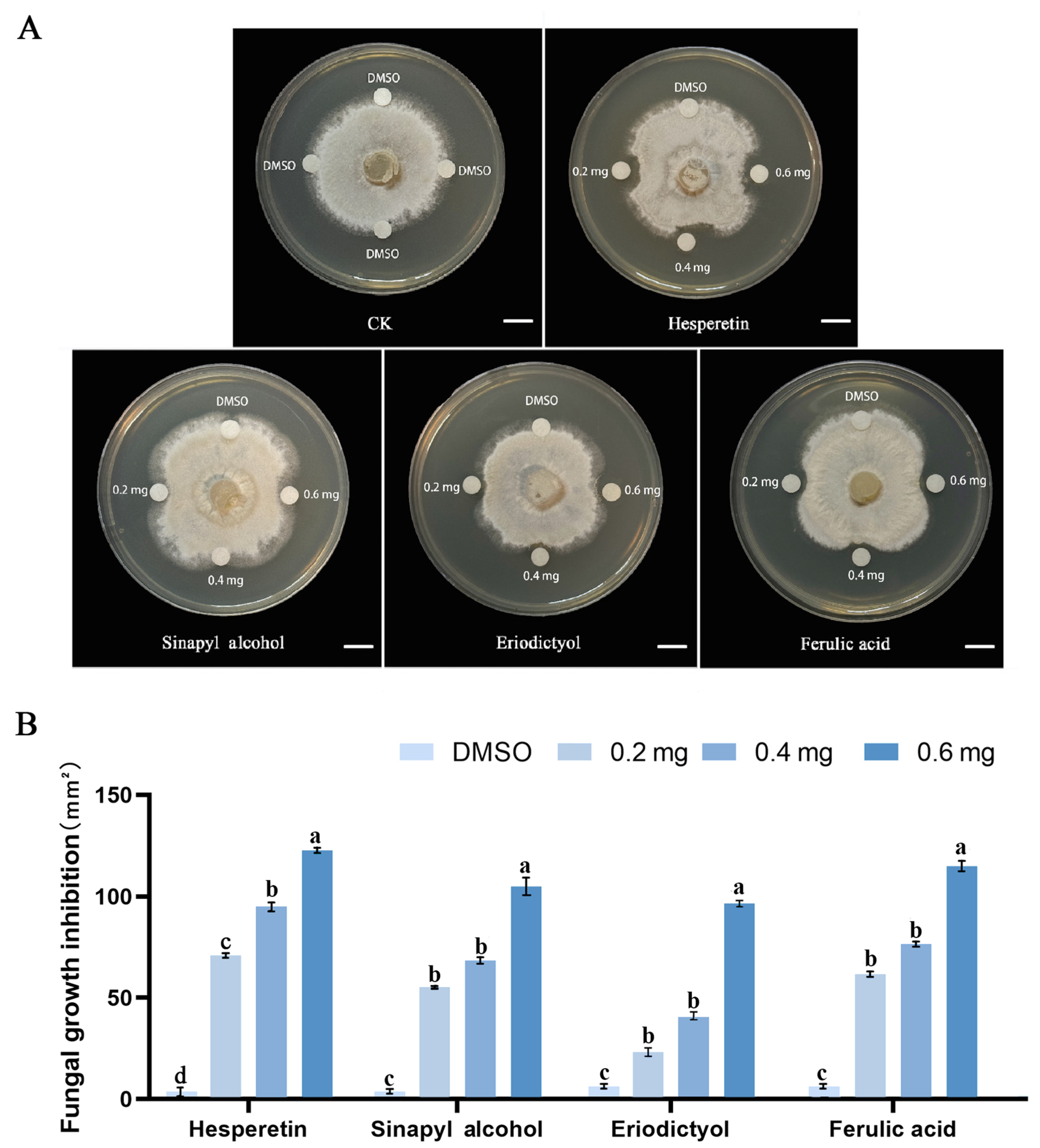
2.7. Phenylpropanes Inhibited B. elliptica Growth
3. Discussion
4. Materials and Methods
4.1. Lily Cultivation and B. elliptica Inoculation
4.2. Metabolomic Analysis
4.3. Transcriptomic Analysis
4.4. Transcriptional Regulatory Network Analysis
4.5. Validation of Gene Expression from Transcriptome Data
4.6. Plate Inhibition Assay of B. elliptica
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, J.D.G.; Jeffery, L.D. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Mullens, A.; Lipka, A.E.; Balint-Kurti, P.; Jamann, T. Exploring the relationship between pattern-triggered immunity and quantitative resistance to Xanthomonas vasicola pv. vasculorum in maize. Phytopathology 2023, 113, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Ökmen, B.; Zhu, W.; Sharon, A. Plant pathogenic fungi. In The Fungal Kingdom; Wiley: Hoboken, NJ, USA, 2017; pp. 701–726. [Google Scholar]
- Zhang, L.L.; Hou, M.W.; Zhang, X.R.; Cao, Y.Y.; Sun, S.L.; Zhu, Z.D.; Han, S.B.; Chen, Y.H.; Ku, L.X.; Duan, C.X. Integrative transcriptome and proteome analysis reveals maize responses to Fusarium verticillioides infection inside the stalks. Mol. Plant Pathol. 2023, 24, 693–710. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.S.; Wang, J.S.; Shaw, R.K.; Sheng, X.G.; Yu, H.F.; Branca, F.; Gu, H.H. Comparative transcriptome and targeted metabolome profiling unravel the key role of phenylpropanoid and glucosinolate pathways in defense against Alternaria brassicicola in broccoli. J. Agric. Food Chem. 2023, 71, 6499–6510. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Che, X.; Gu, Y.; Qu, Y.; Liu, D. Integrated multi-omics investigation revealed the importance of phenylpropanoid metabolism in the defense response of Lilium regale Wilson to Fusarium wilt. Hortic. Res. 2024, 11, uhae140. [Google Scholar] [CrossRef] [PubMed]
- Li, P.Q.; Ruan, Z.; Fei, Z.X.; Yan, J.J.; Tang, G.H. Integrated transcriptome and metabolome analysis revealed that flavonoid biosynthesis may dominate the resistance of Zanthoxylum bungeanum against stem canker. J. Agric. Food Chem. 2021, 69, 6360–6378. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.-Y.; Zhou, J.; Sun, Y.; Qiao, B.-X.; Wan, T.; Guo, R.-Q.; Zhang, J.; Shan, D.-Q.; Cai, Y.-L. Comparative transcriptome and metabolome analyses of cherry leaves spot disease caused by Alternaria alternata. Front. Plant Sci. 2023, 14, 1129515. [Google Scholar] [CrossRef]
- Krępski, T.; Piasecka, A.; Święcicka, M.; Kańczurzewska, M.; Sawikowska, A.; Dmochowska-Boguta, M.; Rakoczy-Trojanowska, M.; Matuszkiewicz, M. Leaf rust (Puccinia recondita f. sp. secalis) triggers substantial changes in rye (Secale cereale L.) at the transcriptome and metabolome levels. BMC Plant Biol. 2024, 24, 107. [Google Scholar] [CrossRef]
- Lv, J.H.; Deng, M.H.; Li, Z.S.; Zhu, H.S.; Wang, Z.R.; Yue, Y.L.; Yang, Z.G.; Xu, J.Q.; Jiang, S.R.; Zhao, W.; et al. Integrative analysis of the transcriptome and metabolome reveals the response mechanism to tomato spotted wilt virus. Hortic. Plant J. 2023, 9, 958–970. [Google Scholar] [CrossRef]
- Qiao, S.C.; Ma, J.K.; Wang, Y.N.; Chen, J.W.; Kang, Z.H.; Bian, Q.Q.; Chen, J.J.; Yin, Y.M.; Cao, G.Z.; Zhao, G.R.; et al. Integrated transcriptome and metabolome analyses reveal details of the molecular regulation of resistance to stem nematode in sweet potato. Plants 2023, 12, 2052. [Google Scholar] [CrossRef]
- Lei, G.; Zhou, K.-H.; Chen, X.-J.; Huang, Y.-Q.; Yuan, X.-J.; Li, G.-G.; Xie, Y.-Y.; Fang, R. Transcriptome and metabolome analyses revealed the response mechanism of pepper roots to Phytophthora capsici infection. BMC Genom. 2023, 24, 626. [Google Scholar] [CrossRef]
- Adhikary, D.; Kisiala, A.; Sarkar, A.; Basu, U.; Rahman, H.; Emery, N.; Kav, N.N.V. Early-stage responses to Plasmodiophora brassicae at the transcriptome and metabolome levels in clubroot resistant and susceptible oilseed Brassica napus. Mol. Omics 2022, 18, 991–1014. [Google Scholar] [CrossRef]
- Terhem, R.B.; Staats, M.; van Kan, J.A. Mating type and sexual fruiting body of Botrytis elliptica, the causal agent of fire blight in lily. Eur. J. Plant Pathol. 2015, 142, 615–624. [Google Scholar] [CrossRef][Green Version]
- Malvestiti, M.C.; Immink, R.G.H.; Arens, P.; Monnens, T.Q.; van Kan, J.A.L. Fire blight susceptibility in Lilium spp. correlates to sensitivity to Botrytis elliptica secreted cell death inducing compounds. Front. Plant Sci. 2021, 12, 660337. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Cui, Q.; Cao, Q.-Z.; Liu, Q.; He, H.-B.; Zhang, D.-M.; Jia, G.-X. Transcriptome-wide analysis of Botrytis elliptica responsive microRNAs and their targets in Lilium regale Wilson by high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 753. [Google Scholar] [CrossRef]
- Hu, F.R.; Liu, G.X.; Hu, Y.M.; Guo, R.R.; Zhu, L.M.; Luo, F.X.; Wang, F. Authenticity identification and leaf blight resistance evaluation of the F1 hybrids from two Lilium cultivars ‘Sorbonne’ and ‘Francia’. Physiol. Mol. Plant Pathol. 2017, 100, 194–200. [Google Scholar] [CrossRef]
- Cui, Q.; Liu, Q.; Gao, X.; Yan, X.; Jia, G.X. Transcriptome-based identification of genes related to resistance against Botrytis elliptica in Lilium regale. Can. J. Plant Sci. 2018, 98, 1058–1071. [Google Scholar] [CrossRef]
- Chai, N.; Xu, J.; Zuo, R.; Sun, Z.; Cheng, Y.; Sui, S.; Li, M.; Liu, D. Metabolic and transcriptomic profiling of Lilium leaves infected with Botrytis elliptica reveals different stages of plant defense mechanisms. Front. Plant Sci. 2021, 12, 730620. [Google Scholar] [CrossRef]
- Xiang, J.; Lei, X.; Wu, Z.; Cao, X.; Zhang, D.; Teng, N. An efficient and novel method to screen Botrytis cinerea resistance genes based on TRV-induced gene silencing with lily petal discs. Physiol. Mol. Plant Pathol. 2022, 122, 101923. [Google Scholar] [CrossRef]
- Du, W.T.; Chai, N.; Sun, Z.Q.; Wang, H.R.; Liu, S.X.; Sui, S.Z.; Luo, L.; Liu, D.F. Full-length transcriptome characterization and functional analysis of pathogenesis-related proteins in Lilium oriental hybrid ‘Sorbonne’ infected with Botrytis elliptica. Int. J. Mol. Sci. 2023, 24, 425. [Google Scholar] [CrossRef]
- Lin, C.-H.; Liu, F.-W.; Pan, Y.-C.; Chen, C.-Y. Lilium gray mold suppression conferred by the host antimicrobial protein LsGRP1 involves main pathogen-targeted manipulation of the nonantimicrobial region LsGRP1N. J. Agric. Food Chem. 2023, 71, 12688–12699. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wu, Z.; Xiang, J.; Cao, X.; Xu, S.; Zhang, Y.; Zhang, D.; Teng, N. A LlWRKY33-LlHSFA4-LlCAT2 module confers resistance to Botrytis cinerea in lily. Hortic. Res. 2024, 11, uhad254. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Cui, Q.; Cao, Q.-Z.; Zhao, Y.-Q.; Liu, Q.; He, H.-B.; Jia, G.-X.; Zhang, D.-M. Evaluation of resistance to Botrytis elliptica in Lilium hybrid cultivars. Plant Physiol. Biochem. 2018, 123, 392–399. [Google Scholar] [CrossRef]
- Li, W.; Zhan, Q.; Guan, Y.; Wang, L.; Li, S.; Zheng, S.; Ma, H.; Liu, Y.; Ding, L.; Zhao, S.; et al. Heterografting enhances chrysanthemums resistance to Alternaria alternata via jasmonate-mediated increases in trichomes and terpenoids. J. Exp. Bot. 2024, 15, erae212. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.J.; Hao, Z.J.; Meng, J.S.; Liu, D.; Wei, M.R.; Tao, J. Digital gene expression analysis to screen disease resistance-relevant genes from leaves of herbaceous peony (Paeonia lactiflora Pall.) infected by Botrytis cinerea. PLoS ONE 2015, 10, e0133305. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, X.; Li, D.; Hong, B.; Gao, J.; Zhang, Z. Rose WRKY13 promotes disease protection to Botrytis by enhancing cytokinin content and reducing abscisic acid signaling. Plant Physiol. 2023, 191, 679–693. [Google Scholar] [CrossRef]
- Csorba, A.B.; Kentelky, E.; Szabó, M.Ğ.E.; Jakab, M.; Nyárádi, I.I.; Bálint, J. Controlling grey mold (Botrytis cinerea) in flowering cyclamen production. Eur. J. Hortic. Sci. 2023, 88, 12. [Google Scholar] [CrossRef]
- Alam, M.; Alam, K.M.; Momotaz, R.; Arifunnahar, M.; Apu, M.R.B.; Siddique, S.S. Botrytis gray mold of Lilium in Bangladesh: Diagnosis, basic study and control. Heliyon 2024, 10, e33165. [Google Scholar] [CrossRef]
- Van Kan, J.L. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends. Plant Sci. 2006, 11, 247–253. [Google Scholar] [CrossRef]
- Valero-Jiménez, C.A.; Veloso, J.; Staats, M.; van Kan, J.A.L. Comparative genomics of plant pathogenic Botrytis species with distinct host specificity. BMC Genom. 2019, 20, 203. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.-F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.T.; Ding, Y.L. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Gupta, R.; Shokat, S.; Iqbal, N.; Kocsy, G.; Pérez-Pérez, J.M.; Riyazuddin, R. Ascorbate, plant hormones and their interactions during plant responses to biotic stress. Physiol. Plant. 2024, 176, e14388. [Google Scholar] [CrossRef]
- Al Aboody, M.S.; Mickymaray, S. Anti-fungal efficacy and mechanisms of flavonoids. Antibiotics 2020, 9, 45. [Google Scholar] [CrossRef]
- Zhang, L.X.; Bao, H.B.; Meng, F.L.; Ren, Y.; Tian, C.M. Transcriptome and metabolome reveal the role of flavonoids in poplar resistance to poplar anthracnose. Ind. Crop. Prod. 2023, 197, 116537. [Google Scholar] [CrossRef]
- Li, W.; Wang, K.; Chern, M.; Liu, Y.; Zhu, Z.; Liu, J.; Zhu, X.; Yin, J.; Ran, L.; Xiong, J.; et al. Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol. 2020, 226, 1850–1863. [Google Scholar] [CrossRef]
- Du, Y.W.; Jia, H.C.; Yang, Z.; Wang, S.H.; Liu, Y.Y.; Ma, H.Y.; Liang, X.F.; Wang, B.; Zhu, M.Q.; Meng, Y.A.; et al. Sufficient coumarin accumulation improves apple resistance to Cytospora mali under high-potassium status. Plant Physiol. 2023, 192, 1396–1419. [Google Scholar] [CrossRef]
- Wang, L.J.; Guo, D.Z.; Zhao, G.D.; Wang, J.Y.; Zhang, S.X.; Wang, C.; Guo, X.Q. Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytol. 2022, 236, 249–265. [Google Scholar] [CrossRef]
- Abbruscato, P.; Tosi, S.; Crispino, L.; Biazzi, E.; Menin, B.; Picco, A.M.; Pecetti, L.; Avato, P.; Tava, A. Triterpenoid glycosides from Medicago sativa as antifungal agents against Pyricularia oryzae. J. Agric. Food Chem. 2014, 62, 11030–11036. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid with anolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Wei, X.Y.; Mo, C.J.; Wei, M.H.; Li, Y.X.; Fan, Y.X.; Gu, X.J.; Zhang, X.J.; Zhang, Y.B.; Kong, Q.S. Integrated full-length transcriptome and metabolome analysis reveals the defence response of melon to gummy stem blight. Plant Cell Environ. 2024, 47, 1997–2010. [Google Scholar] [CrossRef] [PubMed]
- Bakhshaie, M.; Khosravi, S.; Azadi, P.; Bagheri, H.; van Tuyl, J.M. Biotechnological advances in Lilium. Plant Cell Rep. 2016, 35, 1799–1826. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, L.-J.; Sun, X.-F.; Zhao, L.-L.; Wang, P.-C. Transcriptomic and metabolomic analyses reveal key metabolites, pathways and candidate genes in Sophora davidii (Franch.) skeels seedlings under drought stress. Front. Plant Sci. 2022, 13, 785702. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2012, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Mei, Z.L.; Zeng, C.W.; Liu, S.Q. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D822–D827. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Q.; Li, X.; Hu, S.; Yang, D.; Abozeid, A.; Yang, Z.; Jiang, J.; Ren, Z.; Li, D.; Li, D.; et al. The Critical Role of Phenylpropanoid Biosynthesis Pathway in Lily Resistance Against Gray Mold. Int. J. Mol. Sci. 2024, 25, 11068. https://doi.org/10.3390/ijms252011068
Cui Q, Li X, Hu S, Yang D, Abozeid A, Yang Z, Jiang J, Ren Z, Li D, Li D, et al. The Critical Role of Phenylpropanoid Biosynthesis Pathway in Lily Resistance Against Gray Mold. International Journal of Molecular Sciences. 2024; 25(20):11068. https://doi.org/10.3390/ijms252011068
Chicago/Turabian StyleCui, Qi, Xinran Li, Shanshan Hu, Dongfeng Yang, Ann Abozeid, Zongqi Yang, Junhao Jiang, Ziming Ren, Danqing Li, Dongze Li, and et al. 2024. "The Critical Role of Phenylpropanoid Biosynthesis Pathway in Lily Resistance Against Gray Mold" International Journal of Molecular Sciences 25, no. 20: 11068. https://doi.org/10.3390/ijms252011068
APA StyleCui, Q., Li, X., Hu, S., Yang, D., Abozeid, A., Yang, Z., Jiang, J., Ren, Z., Li, D., Li, D., Zheng, L., & Qin, A. (2024). The Critical Role of Phenylpropanoid Biosynthesis Pathway in Lily Resistance Against Gray Mold. International Journal of Molecular Sciences, 25(20), 11068. https://doi.org/10.3390/ijms252011068






