The Detailed Analysis of Polish Patients with Non-Small Cell Lung Cancer Through Insights from Molecular Testing (POL-MOL Study)
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
2.2. Molecular Testing
3. Discussion
4. Materials and Methods
- EGFR (possible use of afatinib, erlotinib, gefitinib as first line and osimertinib as second line in patients with mut T790 M) and criteria met:
- Presence of an activating mutation in the EGFR gene encoding the epidermal growth factor receptor (EGFR) confirmed using a validated test performed in a laboratory with a current European quality control program certificate for the given test;
- Presence of the T790M mutation in the EGFR gene confirmed using a validated test performed in a laboratory with a current European quality control program certificate for the given test;
- ALK (crizotinib, alectinib) and ROS1 (crizotinib)—presence of rearrangement in the ALK gene based on immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH) or new-generation sequencing (NGS) using a validated test performed in a laboratory with a current European quality control program certificate for the given test or presence of rearrangement in the ROS-1 gene based on fluorescence in situ hybridization (FISH) or new-generation sequencing (NGS) using a validated test performed in a laboratory with a current European quality control program certificate for the given test:
- Immunotherapy with pembrolizumab as the first line in monotherapy in patients with high PD-L1 expression;
- Presence of PDL1 expression in 50% or more of the tumor cells confirmed using the method indicated in the Product Characteristics or using DAKO 22C3 antibody concentrate or Ventana SP263 antibody;
- Exclusion of EGFR gene mutations and ALK gene rearrangement in the case of adenocarcinoma, large cell or non-small cell lung cancer NOS using a validated test performed in a laboratory with a current European quality control program certificate for the given test;
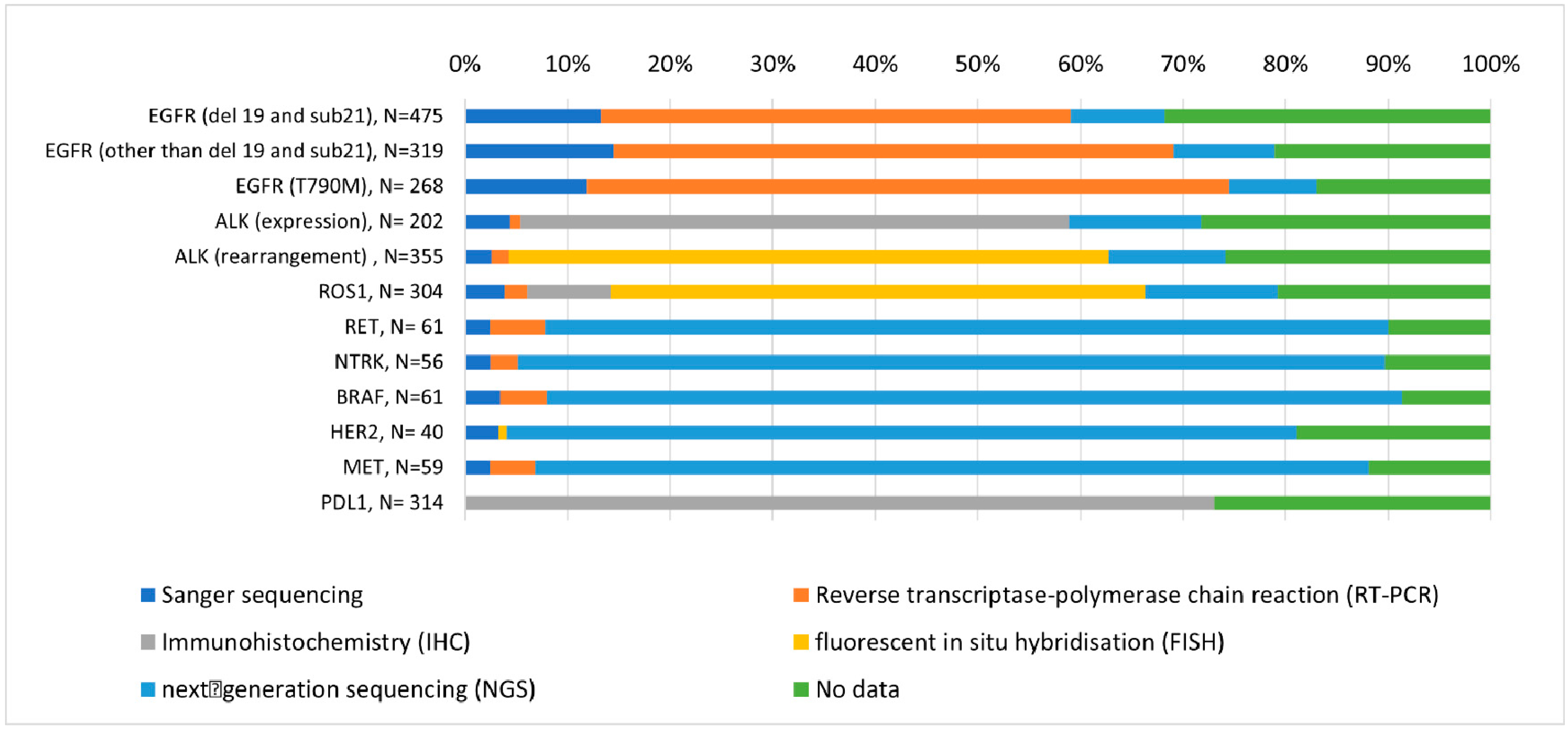
- NGSNext-generation sequencing (NGS) was used to identify mutations and gene fusions using the FusionPlex CTL Kit for Illumina, ArcherDx. The sequencing was performed using the MiniSeq (Illumina) instrument. The results were analyzed using Archer Analysis 5.1 and Archer Analysis 5.0 software. The scope of the analysis includes possible fusion variants of the following genes: ALK, ROS1, NTRK1/2/3, FGFR1/2/3, MET, NRG1, RET, and BRAF, and in the case of point mutations and deletions, insertions in the genes: ALK (T1151ins, L1152R, C1156Y, F1174L, L1196M, G1202R, S1206Y, 1269A); AKT (E17K); BRAF (G466V, G469, Y472, L597V, V600, D594G); DDR2 (S768R, T765P, G774); EGFR (variants in exons 18, 19, 20 and 21); HRAS (codons 12, 13 and 61); KRAS (codons 12, 13, 61, and 146); MAP2K1 (Q56P, K57N, D67N); MET (aberrant splice variant); NRAS (codons 12, 13, and 61); PIK3CA (E542K, E545, H1047); and ROS1 (G2032R). The average number of sequencing reads of the FusionPlex libraries was above 1,000,000 per sample. For DNA/RNA analysis, the average depth of coverage of the sequenced gene regions was not less than 500 reads; analytical sensitivity was 4% mutant DNA relative to normal DNA. The mutation detection rate was approximately 99.9% for mutations in the EGFR, KRAS, and BRAF genes in non-small cell lung cancer. For RNA analysis, the detection limit was not less than five fusion copies; analytical specificity was 99% for all known and new rearrangements of the ALK, RET, ROS1, NTRK1/2/3, FGFR1/2/3 genes. The indicated parameters are obtained when the neoplastic tissue constitutes no less than 20% in the preparation.
- qPCRDNA was isolated using the Agencourt FormaPure kit from Beckman Coulter. DNA concentration was assessed using the Quantus fluorimeter from Promega. EGFR gene status was assessed using the qPCR method using the commercial AmoyDx® EGFR 29 Mutations Detection Kit. Analyzed mutations: p.Gly719Cys/Ser/Ala, deletions in exon 19, insertions in exon 20, p.Thr790Met, p.Ser768Ile, p.Leu858Arg, p.Leu861Gln. Mutations were named according to the HGVS nomenclature. Reference sequence number: (EGFR:LRG_304). Mutation detection rate: approximately 99% of EGFR mutations occurring in lung cancer.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Padinharayil, H.; Varghese, J.; John, M.C.; Rajanikant, G.K.; Wilson, C.M.; Al-Yozbaki, M.; Renu, K.; Dewanjee, S.; Sanyal, R.; George, A.; et al. Non-small cell lung carcinoma (NSCLC): Implications on molecular pathology and advances in early diagnostics and therapeutics. Genes Dis. 2022, 10, 960–989. [Google Scholar] [CrossRef] [PubMed]
- Nalewaj, K.P.; Krawczyk, P.; Chmielewska, I.; Milanowski, J. Delays in the diagnosis of lung cancer patients in Poland. Oncol. Clin. Pract. 2023, 9, 22–27. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Global Cancer Facts & Figures, 4th ed.; American Cancer Society: Atlanta, GA, USA, 2018. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Adamek, M.; Biernat, W.; Chorostowska-Wynimko, J.; Didkowska, J.A.; Dziadziuszko, K.; Grodzki, T.; Jassem, J.; Kępka, L.; Kowalski, D.; Dziadziuszko, R.; et al. Lung Cancer in Poland. J. Thorac. Oncol. 2020, 15, 1271–1276. [Google Scholar] [CrossRef]
- European Union 2021. ECIS—European Cancer Information System. Available online: https://ecis.jrc.ec.europa.eu (accessed on 13 September 2024).
- Ferlay, J.; Laversanne, M.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Tomorrow; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/tomorrow (accessed on 20 January 2021).
- Didkowska, J.; Wojciechowska, U.; Śliwczyński, A. Report on the Stages of Advancement, Treatment, and Survival of Lung Cancer Patients Registered with the National Cancer Registry (KRN) from 2014 to 2016; National Cancer Registry; Maria Skłodowska-Curie National Research Institute of Oncology: Warsaw, Poland, 2019. [Google Scholar]
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report: Cancer Research for Cancer Prevention; International Agency for Research on Cancer, WHO: Lyon, France, 2020. [Google Scholar]
- Ryska, A.; Berzinec, P.; Brcic, L.; Cufer, T.; Dziadziuszko, R.; Gottfried, M.; Kovalszky, I.; Olszewski, W.; Oz, B.; Plank, L.; et al. NSCLC molecular testing in Central and Eastern European countries. BMC Cancer 2018, 18, 269. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef]
- American Cancer Society: About Non-Small Cell Lung Cancer. 2018. Available online: https://www.cancer.org/content/dam/CRC/PDF/Public/8703.00.pdf (accessed on 13 September 2024).
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The effect of advances in lung-cancer treatment on population mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B. The 2015 World Health Organization Classification of Lung Tumors. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar]
- Łaczmańska, I.; Dębicka, I.; Gil, J.; Michałowska, D.; Pawlak, I.; Sąsiadek, M.M. Personalised medicine in lung cancer. Nowotw. J. Oncol. 2021, 71, 122–128. [Google Scholar] [CrossRef]
- Kerr, K.M.; Bubendorf, L.; Edelman, M.J.; Marchetti, A.; Mok, T.; Novello, S.; O’Byrne, K.; Stahel, R.; Peters, S.; Felip, E. Second ESMO consensus conference on lung cancer: Pathology and molecular biomarkers for non-small-cell lung cancer. Ann. Oncol. 2014, 25, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung Adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed]
- Pikor, L.A.; Ramnarine, V.R.; Lam, S.; Lam, W.L. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 2013, 82, 179–189. [Google Scholar]
- Pujol, N.; Heeke, S.; Bontoux, C.; Boutros, J.; Ilié, M.; Hofman, V.; Marquette, C.H.; Hofman, P.; Benzaquen, J. Molecular profiling in non-squamous non-small cell lung carcinoma: Towards a switch to next-generation sequencing reflex testing. J. Pers. Med. 2022, 12, 1684. [Google Scholar] [CrossRef]
- Popat, S.; Navani, N.; Kerr, K.M.; Smit, E.F.; Batchelor, T.J.P.; Van Schil, P.; Senan, S.; McDonald, F. Navigating diagnostic and treatment decisions in non-small cell lung cancer: Expert commentary on the multidisciplinary team approach. Oncologist 2021, 26, e306–e315. [Google Scholar] [CrossRef] [PubMed]
- Kris, M.G.; Johnson, B.E.; Berry, L.D.; Kwiatkowski, D.J.; Iafrate, A.J.; Wistuba, I.I.; Varella-Garcia, M.; Franklin, W.A.; Aronson, S.L.; Su, P.F.; et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014, 311, 1998–2006. [Google Scholar] [CrossRef]
- Popper, H.H.; Tímár, J.; Ryska, A.; Olszewski, W. Minimal requirements for the molecular testing of lung cancer. Transl. Lung Cancer Res. 2014, 3, 301–304. [Google Scholar]
- Aisner, D.L.; Riely, G.J. Non–small cell lung cancer: Recommendations for biomarker testing and treatment. J. Natl. Compr. Cancer Netw. 2021, 19, 610–613. [Google Scholar] [CrossRef]
- Besse, B.; Adjei, A.; Baas, P.; Meldgaard, P.; Nicolson, M.; Paz-Ares, L.; Reck, M.; Smit, E.F.; Syrigos, K.; Stahel, R.; et al. 2nd ESMO consensus conference on lung cancer: Non-small-cell lung cancer first-line/second and further lines of treatment in advanced disease. Ann. Oncol. 2014, 25, 1475–1484. [Google Scholar] [CrossRef]
- Smolle, E.; Pichler, M. Non-smoking-associated lung cancer: A distinct entity in terms of tumor biology, patient characteristics and impact of hereditary cancer predisposition. Cancers 2019, 11, 204. [Google Scholar] [CrossRef]
- Travis, W.D.; Rekhtman, N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: Strategic management of tissue for molecular testing. Semin. Respir. Crit. Care Med. 2011, 32, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.L.F.; Lederer, D.J.; Ryerson, C.J.; Kolb, M.; Maher, T.M.; Nusser, R.; Poletti, V.; Richeldi, L.; Vancheri, C.; Wells, A.U.; et al. Diagnostic Likelihood Thresholds That Define a Working Diagnosis of Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 200, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Fukuoka, M.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef]
- Gregg, J.P.; Li, T.; Yoneda, K.Y. Molecular testing strategies in non-small cell lung cancer: Optimizing the diagnostic journey. Transl. Lung Cancer Res. 2019, 8, 286–301. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P.; Rouleau, E.; Sabourin, J.C.; Denis, M.; Deleuze, J.F.; Barlesi, F.; Laurent-Puig, P. Predictive molecular pathology in non-small cell lung cancer in France: The past, the present and the perspectives. Cancer Cytopathol. 2020, 128, 601–610. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; Hughes, M.; et al. NCCN Guidelines Insights: Non–Small Cell Lung Cancer, Version 2.2021: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Cancer Netw. 2021, 19, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. LCMC Investigators. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2015, 10, 768–777. [Google Scholar] [CrossRef]
- Dietel, M.; Bubendorf, L.; Dingemans, A.M.; Dooms, C.; Elmberger, G.; García, R.C.; Kerr, K.M.; Lim, E.; López-Ríos, F.; von Laffert, M.; et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): Recommendations of the European Expert Group. Thorax 2016, 71, 177–184. [Google Scholar] [CrossRef]
- Pązik, M.; Mirowski, M.; Balcerczak, E. Current Approach to Non-Small Cell Lung Cancer Diagnosis and Treatment—A Short Review; Wydawnictwo Uniwersytetu Medycznego w Łodzi: Łódź, Poland, 2022. [Google Scholar]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Sundaram, B.; et al. Molecular Testing Guideline for the Selection of Patients with Lung Cancer for Treatment with Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Yatabe, Y.; et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.S.; Ladanyi, M.; et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 2013, 8, 823–859. [Google Scholar] [PubMed]
- Aggarwal, C.; Bubendorf, L.; Cooper, W.A.; Illei, P.; Borralho Nunes, P.; Ong, B.H.; Tsao, M.S.; Yatabe, Y.; Kerr, K.M. Molecular testing in stage I-III non-small cell lung cancer: Approaches and challenges. Lung Cancer 2021, 162, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, P.; Ramlau, R.; Powrózek, T.; Wojas-Krawczyk, K.; Sura, S.; Jarosz, B.; Walczyna, B.; Pankowski, J.; Szumiło, J.; Milanowski, J.; et al. The detection of EGFR mutations in patients with non-small cell lung cancer in selected molecular diagnostics centers in Poland. Kardiochir. Torakochirurgia Pol. 2012, 9, 431–438. [Google Scholar] [CrossRef]
- Trembecki, Ł.; Sztuder, A.; Pawlak, I.; Matkowski, R.; Maciejczyk, A. Analysis of lung cancer measures of the National Cancer Network pilot study in Poland for potential improvement in the quality of advanced-stage lung cancer therapy. BMC Cancer 2021, 21, 1252. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Tsao, M.S.; Le, L.W.; Shepherd, F.A.; Feld, R.; Burkes, R.L.; Liu, G.; Kamel-Reid, S.; Hwang, D.; Leighl, N.B.; et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 2015, 26, 1415–1421. [Google Scholar] [CrossRef]
- Gutierrez, M.E.; Choi, K.; Lanman, R.B.; Licitra, E.J.; Skrzypczak, S.M.; Pe Benito, R.; Wu, T.; Arunajadai, S.; Kaur, S.; Goldberg, S.L.; et al. Genomic Profiling of Advanced Non-Small Cell Lung Cancer in Community Settings: Gaps and Opportunities. Clin. Lung Cancer 2017, 18, 651–659. [Google Scholar] [CrossRef]
- Vrijens, F.; De Gendt, C.; Verleye, L.; Robays, J.; Schillemans, V.; Camberlin, C.; Stordeur, S.; Dubois, C.; Van Eycken, E.; Van Meerbeeck, J.P.; et al. Quality of care and variability in lung cancer management across Belgian hospitals: A population-based study using routinely available data. Int. J. Qual. Health Care 2018, 30, 306–312. [Google Scholar] [CrossRef]
- Lee, D.H.; Tsao, M.S.; Kambartel, K.O.; Isobe, H.; Huang, M.S.; Barrios, C.H.; Khattak, A.; de Marinis, F.; Kothari, S.; de Castro, J.; et al. Molecular testing and treatment patterns for patients with advanced non-small cell lung cancer: PIvOTAL observational study. PLoS ONE 2018, 13, e0202865. [Google Scholar] [CrossRef]
- Ess, S.M.; Herrmann, C.; Frick, H.; Krapf, M.; Cerny, T.; Jochum, W.; Früh, M. Epidermal growth factor receptor and anaplastic lymphoma kinase testing and mutation prevalence in patients with advanced non-small cell lung cancer in Switzerland: A comprehensive evaluation of real world practices. Eur. J. Cancer Care 2017, 26, e12721. [Google Scholar] [CrossRef]
- Sandelin, M.; Berglund, A.; Sundström, M.; Micke, P.; Ekman, S.; Bergqvist, M.; Bergström, S.; Koyi, H.; Brandén, E.; Botling, J.; et al. Patients with Non-small Cell Lung Cancer Analyzed for EGFR: Adherence to Guidelines, Prevalence and Outcome. Anticancer Res. 2015, 35, 3979–3985. [Google Scholar]
- Kerr, K.M.; Bibeau, F.; Thunnissen, E.; Botling, J.; Ryška, A.; Wolf, J.; Öhrling, K.; Burdon, P.; Malapelle, U.; Büttner, R. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer 2021, 154, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Griesinger, F.; Eberhardt, W.; Nusch, A.; Reiser, M.; Zahn, M.O.; Maintz, C.; Bernhardt, C.; Losem, C.; Stenzinger, A.; Thomas, M.; et al. CRISP Registry Group: Biomarker testing in non-small cell lung cancer in routine care: Analysis of the first 3717 patients in the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315). Lung Cancer 2021, 152, 174–184. [Google Scholar] [CrossRef]
- Waterhouse, D.M.; Tseng, W.Y.; Espirito, J.L.; Robert, N.J. Understanding Contemporary Molecular Biomarker Testing Rates and Trends for Metastatic NSCLC Among Community Oncologists. Clin. Lung Cancer 2021, 22, e901–e910. [Google Scholar] [CrossRef]
- Gill, J.; Fontrier, A.-M.; Miracolo, A.; Kanavos, P. Access to Personalised Oncology in Europe; London School of Economics: London, UK, 2020. [Google Scholar]
- Reimbursement Announcement of the Polish Ministry of Health no. 67 (1 January 2023). Available online: https://www.gov.pl/attachment/420adc35-6fb1-4060-8c7a-f215886f7a23 (accessed on 13 September 2024).
- Mileham, K.F.; Schenkel, C.; Bruinooge, S.S.; Freeman-Daily, J.; Basu Roy, U.; Moore, A.; Smith, R.A.; Garrett-Mayer, E.; Rosenthal, L.; Silvestri, G.A.; et al. Defining comprehensive biomarker-related testing and treatment practices for advanced non-small-cell lung cancer: Results of a survey of U.S. oncologists. Cancer Med. 2022, 11, 530–538. [Google Scholar] [CrossRef]
- Association of Community Cancer Centers Operational Pathways for Biomarker Testing in NSCLC. Environmental Scan. 2021. Available online: https://www.accc-cancer.org/docs/projects/operational-pathways-in-nsclc/accc-operational-pathways_final-(2).pdf (accessed on 13 September 2024).
- Naidoo, J.; Drilon, A. Molecular Diagnostic Testing in Non-Small Cell Lung Cancer. Am. J. Hematol. Oncol. 2014, 10, 4–11. [Google Scholar]
- Guo, H.; Zhang, J.; Qin, C.; Yan, H.; Liu, T.; Hu, H.; Tang, S.; Tang, S.; Zhou, H. Biomarker-Targeted Therapies in Non-Small Cell Lung Cancer: Current Status and Perspectives. Cells 2022, 11, 3200. [Google Scholar] [CrossRef]
- Vanderpoel, J.; Stevens, A.L.; Emond, B.; Lafeuille, M.H.; Hilts, A.; Lefebvre, P.; Morrison, L. Total cost of testing for genomic alterations associated with next-generation sequencing versus polymerase chain reaction testing strategies among patients with metastatic non-small cell lung cancer. J. Med. Econ. 2022, 25, 457–468. [Google Scholar] [CrossRef]
- Zheng, Y.; Vioix, H.; Liu, F.X.; Singh, B.; Sharma, S.; Sharda, D. Diagnostic and economic value of biomarker testing for targetable mutations in non-small-cell lung cancer: A literature review. Future Oncol. 2022, 18, 505–518. [Google Scholar] [CrossRef]
- Isla, D.; Lozano, M.D.; Paz-Ares, L.; Salas, C.; de Castro, J.; Conde, E.; Felip, E.; Gómez-Román, J.; Garrido, P.; Enguita, A.B. New update to the guidelines on testing predictive biomarkers in non-small-cell lung cancer: A National Consensus of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin. Transl. Oncol. 2023, 25, 1252–1267. [Google Scholar] [CrossRef]



| Characteristic | All Patients (n = 1001) | Non-Squamous NSCLC (n = 542) | Squamous NSCLC (n = 378) | NOS (n = 79) | ||||
|---|---|---|---|---|---|---|---|---|
| Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | |
| Cancer Type | ||||||||
| Adenocarcinoma | 51 (508) | 48 | 94 (508) | |||||
| Large cell carcinoma | 3 (34) | 4 | 6 (34) | |||||
| NOS | 8 (79) | 9 | 100 (79) | |||||
| Squamouscell carcinoma | 36 (356) | 37 | 94 (356) | |||||
| Adenosquamous carcinoma | 2 (22) | 2 | 6 (22) | |||||
| No data | <1 (2) | 0 | ||||||
| Sex | ||||||||
| Male | 61 (615) | 62 | 56 (304) | 55 | 68 (257) | 70 | 66 (52) | 66 |
| Female | 39 (386) | 38 | 44 (238) | 45 | 32 (121) | 30 | 34 (27) | 34 |
| Age (years) | ||||||||
| 18–54 | 12 (119) | 11 | 15 (84) | 15 | 7 (28) | 8 | 9 (7) | 8 |
| 55–65 | 29 (287) | 30 | 28 (151) | 30 | 29 (108) | 29 | 33 (26) | 38 |
| 66–75 | 30 (300) | 32 | 29 (158) | 31 | 31 (118) | 35 | 30 (24) | 29 |
| 76+ | 7 (71) | 8 | 6 (34) | 7 | 7 (28) | 9 | 11 (9) | 16 |
| No data | 22 (224) | 18 | 21 (115) | 18 | 25 (96) | 20 | 16 (13) | 9 |
| NSCLC stage at initiation of pharmacological treatment | ||||||||
| IIA | 2 (17) | 2 | 2 (10) | 2 | 2 (7) | 2 | <1 (1) | 0 |
| IIB | 6 (56) | 4 | 6 (35) | 5 | 5 (19) | 4 | 1 (1) | 0 |
| IIIA | 15 (150) | 16 | 12 (65) | 12 | 20 (76) | 21 | 10 (8) | 13 |
| IIIB | 18 (182) | 18 | 16 (86) | 16 | 21 (80) | 20 | 20 (16) | 22 |
| IV | 59 (588) | 59 | 63 (343) | 63 | 51 (191) | 51 | 68 (54) | 64 |
| No data | 1 (8) | 0 | 1 (3) | 0 | 1 (5) | 1 | 0 | 0 |
| ECOG Performance Status Scale grade at initiation of pharmacological treatment | ||||||||
| 0 | 8 (77) | 9 | 9 (48) | 10 | 7 (27) | 10 | 3 (2) | 2 |
| 1 | 72 (722) | 71 | 76 (412) | 76 | 70 (264) | 67 | 56 (44) | 54 |
| 2 | 17 (170) | 16 | 12 (65) | 11 | 21 (78) | 19 | 34 (27) | 33 |
| 3 | 2 (19) | 2 | 2 (11) | 2 | 2 (7) | 3 | 1 (1) | 2 |
| 4 | <1 (5) | 1 | <1 (1) | 0 | 0 | 0 | 5 (4) | 6 |
| No data | 1 (8) | 1 | 1 (5) | 1 | 1 (2) | 1 | 1 (1) | 3 |
| Tests Performed | All Patients (n = 1001) | Non-Squamous NSCLC (n = 542) | Squamous NSCLC (n = 378) | NOS (n = 79) | ||||
|---|---|---|---|---|---|---|---|---|
| Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | |
| Molecular tests and PD-L1 | 34 (337) | 33 | 51 (275) | 53 | 8 (29) | 6 | 42 (33) | 37 |
| Molecular tests without PD-L1 | 18 (184) | 18 | 27 (147) | 27 | 4 (15) | 3 | 28 (22) | 35 |
| PD-L1 only | 15 (151) | 16 | 1 (3) | 1 | 39 (147) | 41 | 1 (1) | 0 |
| None | 33 (329) | 32 | 22 (117) | 19 | 49 (187) | 50 | 29 (23) | 28 |
| All Patients (n = 1001) * | ||||||
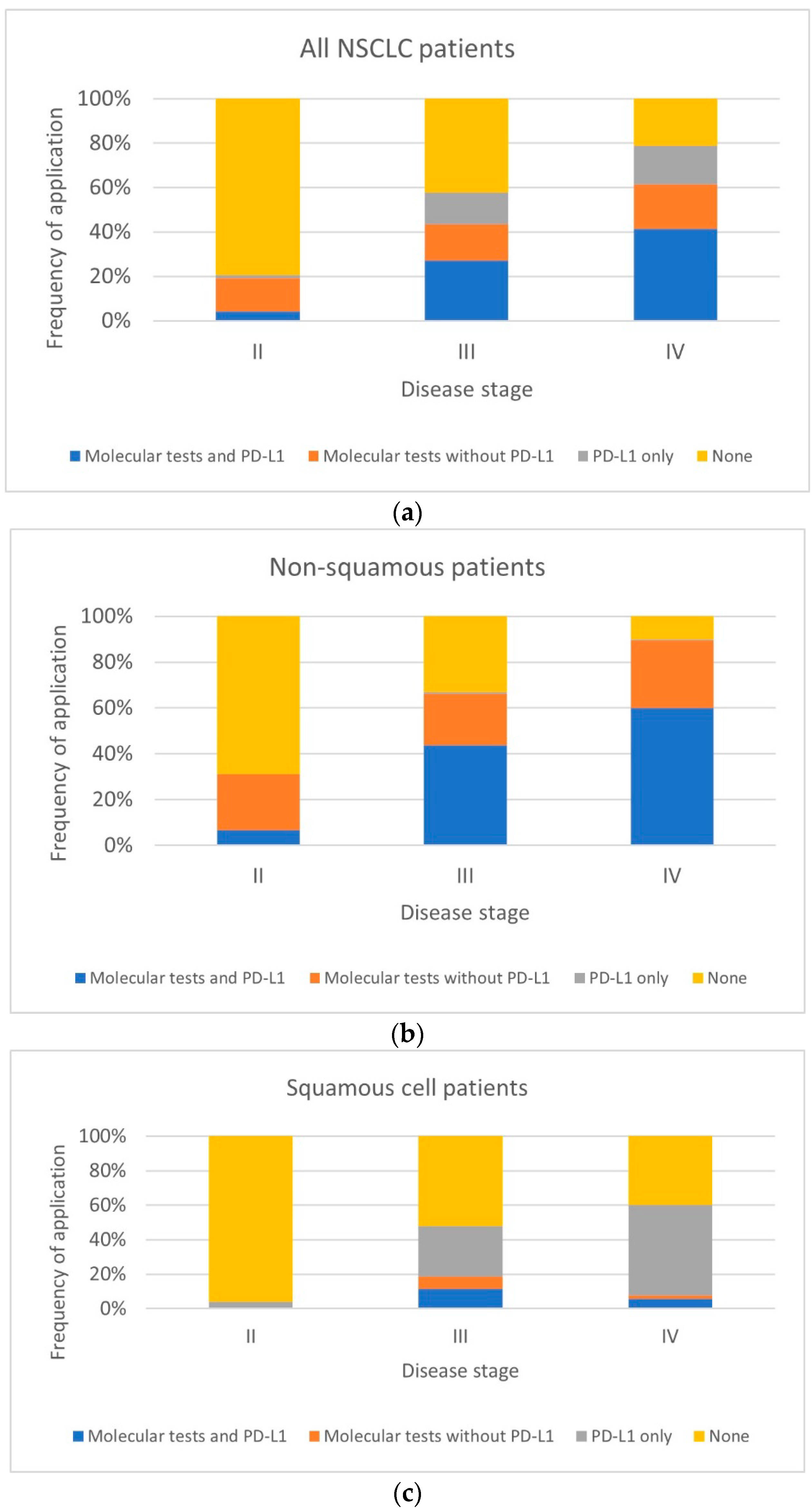
| Tests Performed | Stage II (n = 73) | Stage III (n = 332) | Stage IV (n = 588) | |||
| Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | |
| Molecular tests and PD-L1 | 4 (3) | 3 | 27 (90) | 24 | 41 (244) | 42 |
| Molecular tests without PD-L1 | 15 (11) | 25 | 17 (55) | 18 | 20 (117) | 18 |
| PD-L1 only | 1 (1) | 1 | 14 (47) | 16 | 18 (103) | 19 |
| None | 79 (58) | 71 | 42 (140) | 42 | 21 (124) | 22 |
| Non-squamous NSCLC (n = 542) | ||||||
| Tests performed | Stage II (n = 45) | Stage III (n = 151) | Stage IV (n = 343) | |||
| Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | |
| Molecular tests and PD-L1 | 7 (3) | 6 | 44 (66) | 41 | 60 (206) | 64 |
| Molecular tests without PD-L1 | 24 (11) | 42 | 23 (34) | 26 | 29 (101) | 25 |
| PD-L1 only | 0 | 0 | 1 (1) | 1 | 1 (2) | 1 |
| None | 69 (31) | 52 | 33 (50) | 31 | 10 (34) | 10 |
| Squamous NSCLC (n = 378) | ||||||
| Tests performed | Stage II (n = 26) | Stage III (n = 156) | Stage IV | |||
| Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | |
| Molecular tests and PD-L1 | 0 | 0 | 12 (18) | 8 | 6 (11) | 5 |
| Molecular tests without PD-L1 | 0 | 0 | 7 (11) | 5 | 2 (4) | 1 |
| PD-L1 only | 4 (1) | 1 | 29 (46) | 33 | 52 (100) | 54 |
| None | 96 (25) | 99 | 52 (81) | 55 | 40 (76) | 40 |
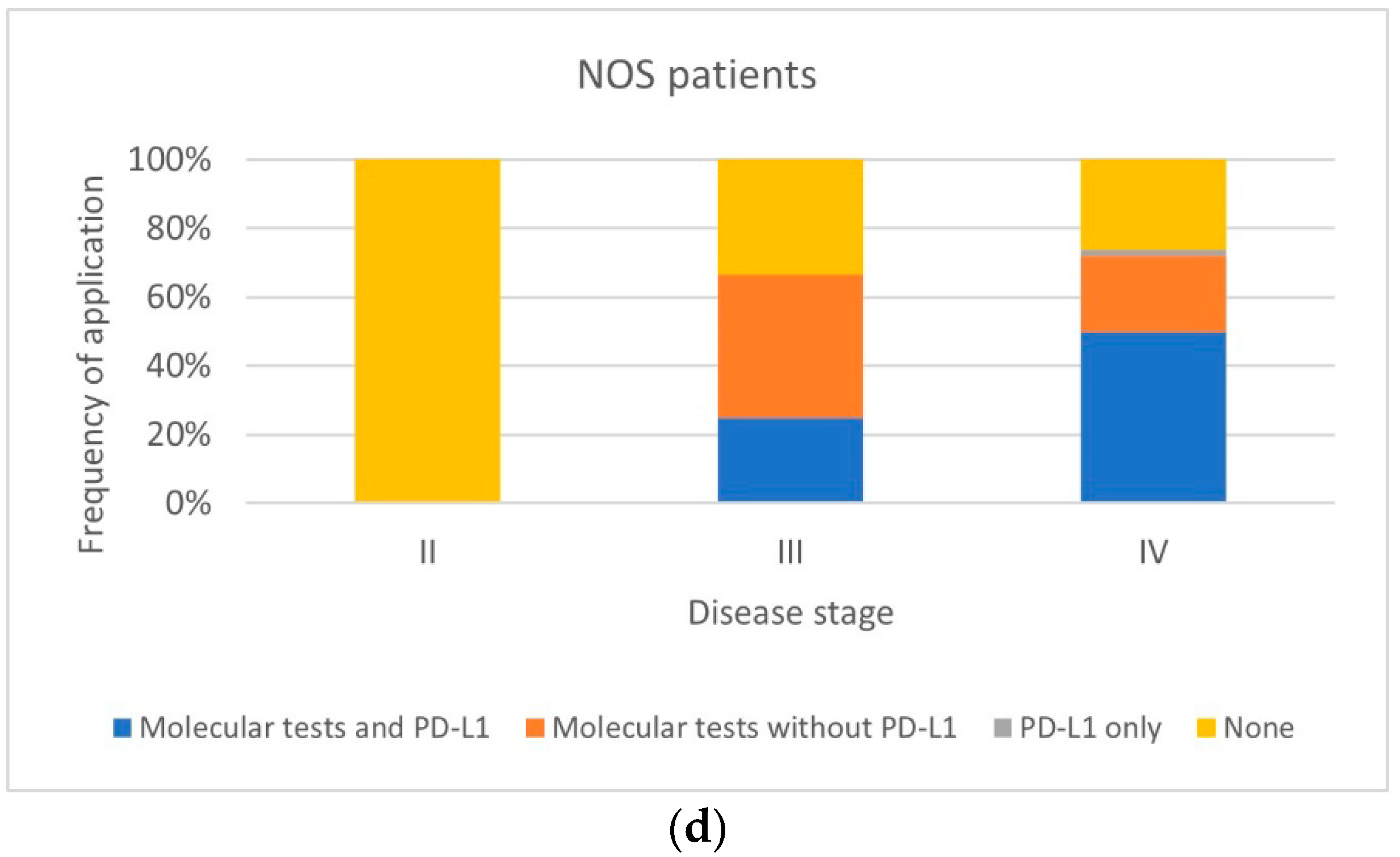
| NOS patients (n = 79) | ||||||
| Tests performed | Stage II (n = 1) | Stage III (n = 24) | Stage IV (n = 54) | |||
| Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | |
| Molecular tests and PD-L1 | 0 | 0 | 25 (6) | 26 | 50 (27) | 43 |
| Molecular tests without PD-L1 | 0 | 0 | 42 (10) | 49 | 22 (12) | 27 |
| PD-L1 only | 0 | 0 | 0 | 0 | 2 (1) | 1 |
| None | 100 (1) | 100 | 33 (8) | 25 | 26 (14) | 29 |
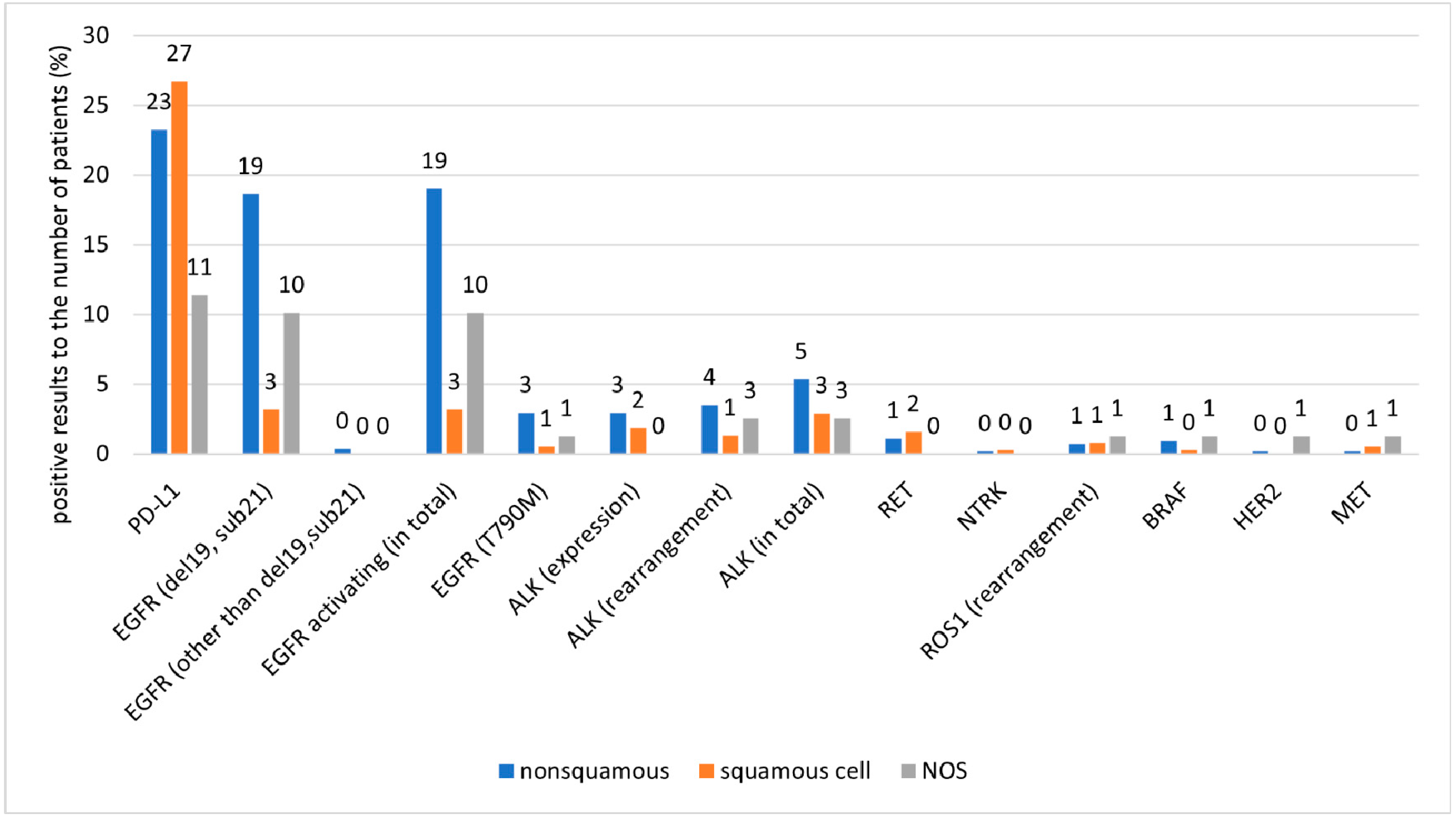
| All Patients (n = 1001) | ||||||
| Test | Tests Performed | Positive Results Related to the Number of Patients | Positive Results Related to the Number of Tests Performed | |||
| Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | Unweighted Data, % (n) | Weighted Data, % | |
| PD-L1 | 49 (488) | 50 | 24 (236) | 23 | 48 (236) | 46 |
| EGFR (del19, sub21) | 51 (508) | 50 | 12 (121) | 11 | 24 (121) | 21 |
| EGFR (other than del19, sub21) | 35 (345) | 35 | <1 (2) | 0 | 1 (2) | 0 |
| EGFR activating (in total) | 51 (513) | 51 | 12 (123) | 11 | 24 (123) | 21 |
| EGFR (T790M) | 28 (282) | 26 | 2 (19) | 2 | 7 (19) | 6 |
| ALK (expression) | 22 (216) | 28 | 2 (23) | 2 | 11 (23) | 8 |
| ALK (rearrangement) | 37 (368) | 38 | 3 (26) | 2 | 7 (26) | 6 |
| ALK (in total) | 43 (431) | 44 | 4 (42) | 3 | 10 (42) | 8 |
| RET | 8 (82) | 8 | 1 (12) | 1 | 15 (12) | 10 |
| NTRK | 8 (77) | 8 | <1 (2) | 0 | 3 (2) | 2 |
| ROS1 (rearrangement) | 33 (327) | 36 | 1 (8) | 0 | 2 (8) | 1 |
| BRAF | 8 (77) | 8 | 1 (7) | 0 | 9 (7) | 6 |
| HER2 | 5 (52) | 5 | <1 (2) | 0 | 4 (2) | 2 |
| MET | 8 (83) | 8 | <1 (5) | 0 | 6 (5) | 4 |
| Test | Tests performed | Positive results related to the number of patients | Positive results related to the number of tests performed | |||
| Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | |
| PD-L1 | 51 (278) | 54 | 23 (126) | 21 | 45 (126) | 40 |
| EGFR (del19, sub21) | 76 (414) | 78 | 19 (101) | 17 | 24 (101) | 22 |
| EGFR (other than del19, sub21) | 52 (284) | 56 | <1 (2) | 0 | 1 (2) | 1 |
| EGFR activating (in total) | 77 (417) | 79 | 19 (103) | 17 | 25 (103) | 22 |
| EGFR (T790M) | 42 (226) | 40 | 3 (16) | 3 | 7 (16) | 6 |
| ALK (expression) | 30 (165) | 43 | 3 (16) | 3 | 10 (16) | 8 |
| ALK (rearrangement) | 55 (299) | 61 | 4 (19) | 3 | 6 (19) | 5 |
| ALK (in total) | 65 (352) | 70 | 5 (29) | 5 | 8 (29) | 7 |
| RET | 8 (43) | 9 | 1 (6) | 1 | 14 (6) | 8 |
| NTRK | 8 (41) | 9 | <1 (1) | 0 | 2 (1) | 1 |
| ROS1 (rearrangement) | 47 (257) | 56 | 1 (4) | 0 | 2 (4) | 1 |
| BRAF | 8 (44) | 9 | 1 (5) | 1 | 11 (5) | 6 |
| HER2 | 5 (27) | 6 | <1 (1) | 0 | 4 (1) | 2 |
| MET | 5 (27) | 9 | <1 (1) | 0 | 4 (1) | 3 |
| Test | Tests performed | Positive results related to the number of patients | Positive results to tests performed | |||
| Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | |
| PD-L1 | 47 (176) | 47 | 27 (101) | 29 | 57 (101) | 62 |
| EGFR (del19, sub21) | 11 (40) | 8 | 3 (12) | 2 | 30 (12) | 26 |
| EGFR (other than del19, sub21) | 7 (26) | 5 | 0 | 0 | 0 | 0 |
| EGFR activating (in total) | 11 (42) | 8 | 3 (12) | 2 | 29 (12) | 25 |
| EGFR (T790M) | 7 (28) | 5 | 1 (2) | 0 | 7 (2) | 7 |
| ALK (expression) | 6 (23) | 4 | 2 (7) | 1 | 30 (7) | 27 |
| ALK (rearrangement) | 8 (30) | 6 | 1 (5) | 1 | 17 (5) | 15 |
| ALK (in total) | 9 (33) | 6 | 3 (11) | 2 | 33 (11) | 29 |
| RET | 7 (26 | 4 | 2 (6) | 1 | 23 (6) | 23 |
| NTRK | 6 (24) | 4 | <1 (1) | 0 | 4 (1) | 4 |
| ROS1 (rearrangement) | 10 (36) | 7 | 1 (3) | 1 | 8 (3) | 7 |
| BRAF | 5 (20) | 3 | <1 (1) | 0 | 5 (1) | 5 |
| HER2 | 4 (15) | 3 | 0 | 0 | 0 | 0 |
| MET | 7 (26) | 4 | 1 (2) | 0 | 8 (2) | 8 |
| Test | Tests performed | Positive results related to the number of patients | Positive results related to the number of tests performed | |||
| Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | Unweighted data, % (n) | Weighted data, % | |
| PD-L1 | 43 (34) | 37 | 11 (9) | 5 | 26 (9) | 14 |
| EGFR (del19, sub21) | 68 (54) | 71 | 10 (8) | 10 | 15 (8) | 15 |
| EGFR (other than del19, sub21) | 44 (35) | 48 | 0 | 0 | 0 | 0 |
| EGFR activating (in total) | 68 (54) | 71 | 10 (8) | 10 | 15 (8) | 15 |
| EGFR (T790M) | 35 (28) | 37 | 1 (1) | 1 | 4 (1) | 2 |
| ALK (expression) | 35 (28) | 41 | 0 | 0 | 0 | 0 |
| ALK (rearrangement) | 49 (39) | 50 | 3 (2) | 3 | 5 (2) | 6 |
| ALK (in total) | 58 (46) | 59 | 3 (2) | 3 | 4 (2) | 6 |
| RET | 29 (23) | 17 | 0 | 0 | 0 | 0 |
| NTRK | 15 (12) | 16 | 0 | 0 | 0 | 0 |
| ROS1 (rearrangement) | 43 (34) | 45 | 1 (1) | 0 | 3 (1) | 1 |
| BRAF | 16 (13) | 17 | 1 (1) | 1 | 8 (1) | 4 |
| HER2 | 13 (10) | 13 | 1 (1) | 1 | 10 (1) | 5 |
| MET | 16 (13) | 17 | 1 (1) | 1 | 8 (1) | 4 |
| Size of Drug Contract (PLN) | No. of Centers in Poland | % of the Total Contract Value in 2020 | % of Contract Value after Exclusion of the Centers with <1 Million PLN Contracts | No. of Included Centers | Max. Number of Patients |
|---|---|---|---|---|---|
| >3 million | 21 | 61 | 68 | 12 | 68 |
| >1.5–3 million | 20 | 23 | 25 | 7 | 36 |
| >1–1.5 million | 10 | 6 | 7 | 2 | 28 |
| <1 million | 42 | 10 | - | - | - |
| Basic Questionnaire | Extended Questionnaire | |
|---|---|---|
| Data collection | Retrospective | |
| Data collector | Physicians providing NSCLC treatment | |
| Data source | Medical records | |
| Population | Patients admitted to the center in the second half of 2019 and treated for NSCLC | Patients with adenocarcinoma or NOS |
| Aim | To collect data regarding the entire population of NSCLC patients receiving systemic treatment and to assess the frequency of molecular testing in this population | To collect more detailed data on molecular diagnostics and therapy in patients with adenocarcinoma or NOS |
| Topic/questions | Basic questions regarding NSCLC patients and molecular tests applied | Detailed questions regarding molecular diagnosis and treatment in patients with adenocarcinoma or NOS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalski, D.M.; Zaborowska-Szmit, M.; Bryl, M.; Byszek, A.; Dziedzic, D.A.; Jaśkiewicz, P.; Langfort, R.; Krzakowski, M.; Orłowski, T.; Ramlau, R.; et al. The Detailed Analysis of Polish Patients with Non-Small Cell Lung Cancer Through Insights from Molecular Testing (POL-MOL Study). Int. J. Mol. Sci. 2024, 25, 11354. https://doi.org/10.3390/ijms252111354
Kowalski DM, Zaborowska-Szmit M, Bryl M, Byszek A, Dziedzic DA, Jaśkiewicz P, Langfort R, Krzakowski M, Orłowski T, Ramlau R, et al. The Detailed Analysis of Polish Patients with Non-Small Cell Lung Cancer Through Insights from Molecular Testing (POL-MOL Study). International Journal of Molecular Sciences. 2024; 25(21):11354. https://doi.org/10.3390/ijms252111354
Chicago/Turabian StyleKowalski, Dariusz M., Magdalena Zaborowska-Szmit, Maciej Bryl, Agnieszka Byszek, Dariusz Adam Dziedzic, Piotr Jaśkiewicz, Renata Langfort, Maciej Krzakowski, Tadeusz Orłowski, Rodryg Ramlau, and et al. 2024. "The Detailed Analysis of Polish Patients with Non-Small Cell Lung Cancer Through Insights from Molecular Testing (POL-MOL Study)" International Journal of Molecular Sciences 25, no. 21: 11354. https://doi.org/10.3390/ijms252111354
APA StyleKowalski, D. M., Zaborowska-Szmit, M., Bryl, M., Byszek, A., Dziedzic, D. A., Jaśkiewicz, P., Langfort, R., Krzakowski, M., Orłowski, T., Ramlau, R., & Szmit, S. (2024). The Detailed Analysis of Polish Patients with Non-Small Cell Lung Cancer Through Insights from Molecular Testing (POL-MOL Study). International Journal of Molecular Sciences, 25(21), 11354. https://doi.org/10.3390/ijms252111354





