Supercomputer-Based Virtual Screening for Deoxyribonucleic Acid Methyltransferase 1 Inhibitors as Novel Anticancer Agents
Abstract
:1. Introduction
2. Results
2.1. Virtual Drug Screening and Molecular Docking
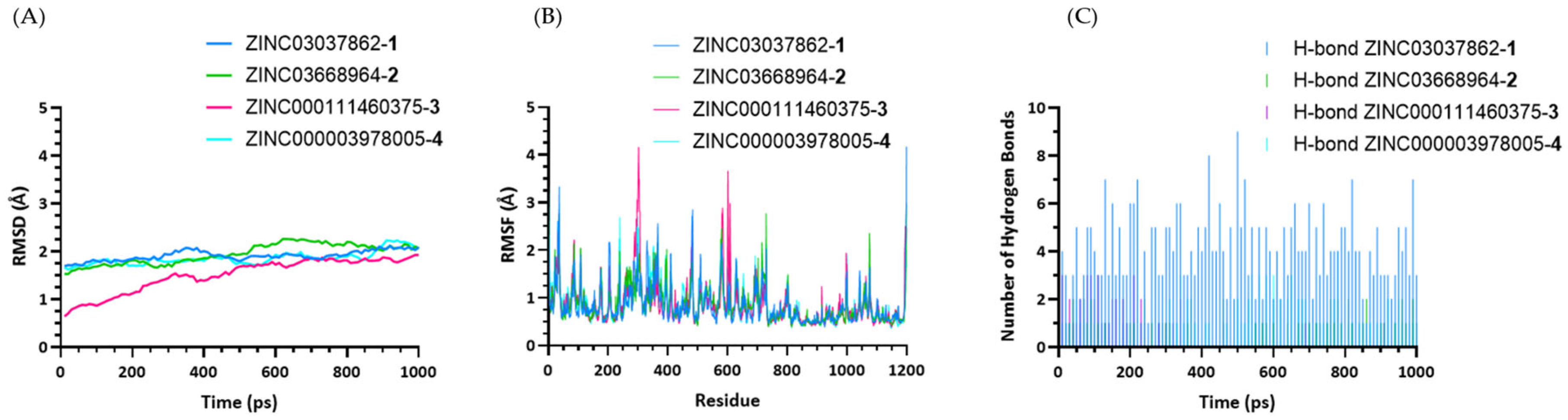
2.2. Molecular Dynamics Simulations for Protein–Ligand Complexes
2.3. Compounds 2 and 4 Inhibit DNMT1 Activity
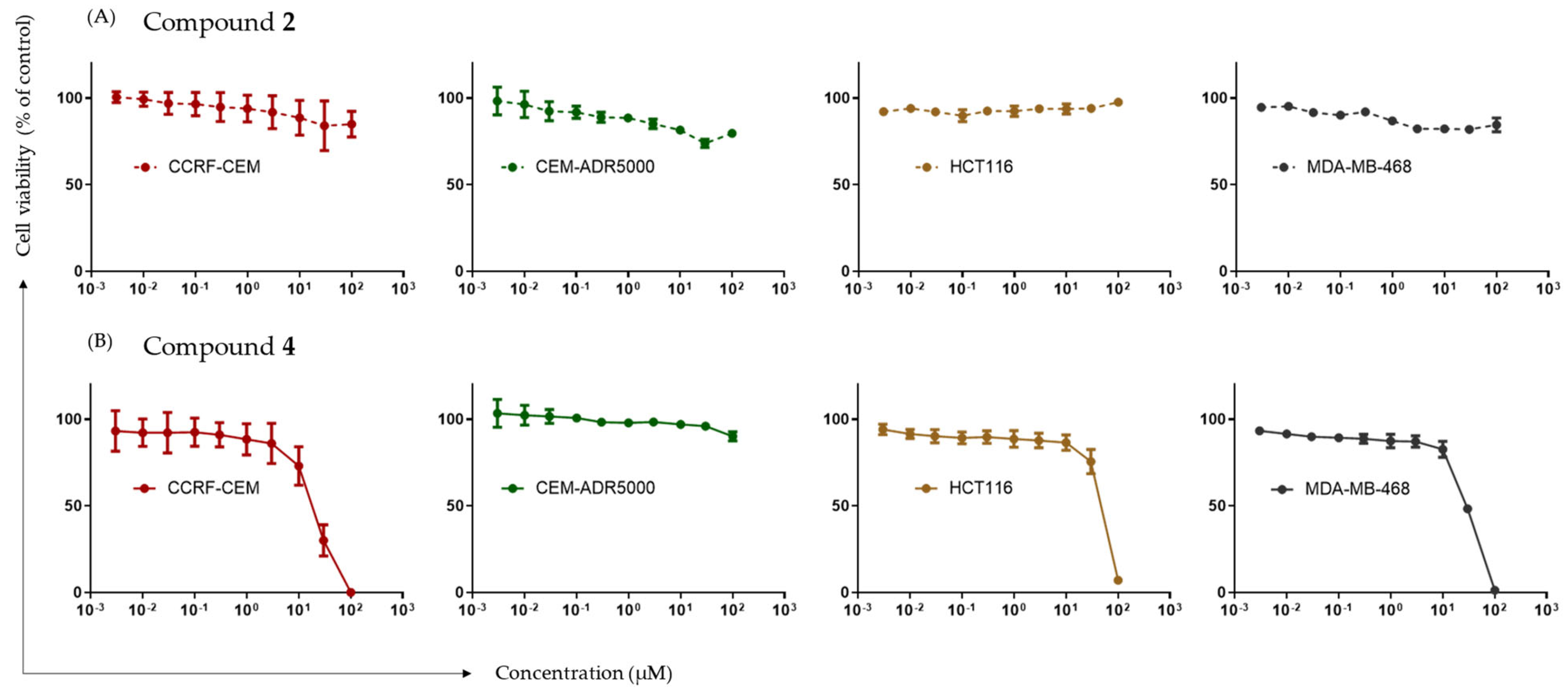
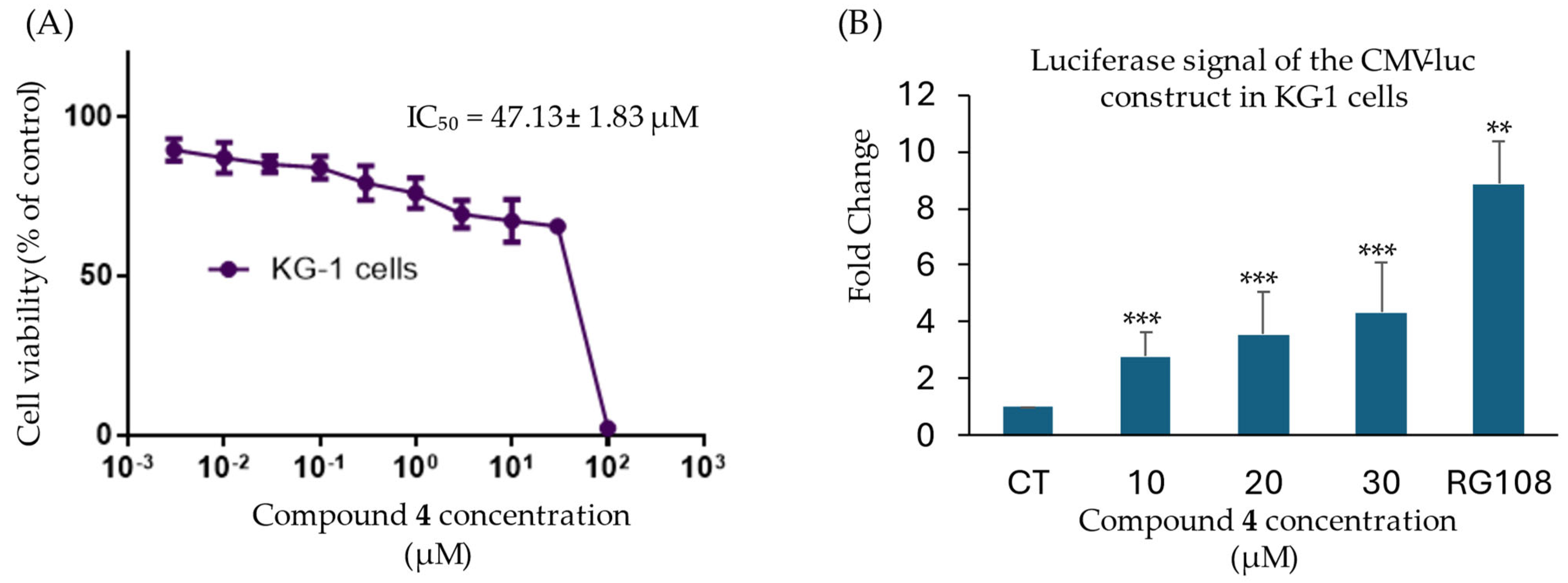
2.4. Cytotoxic Effects of Potential DNMT1 Inhibitors
2.5. Compound 4 Demethylates the Promotor of the CMV-Luciferase Gene Construct
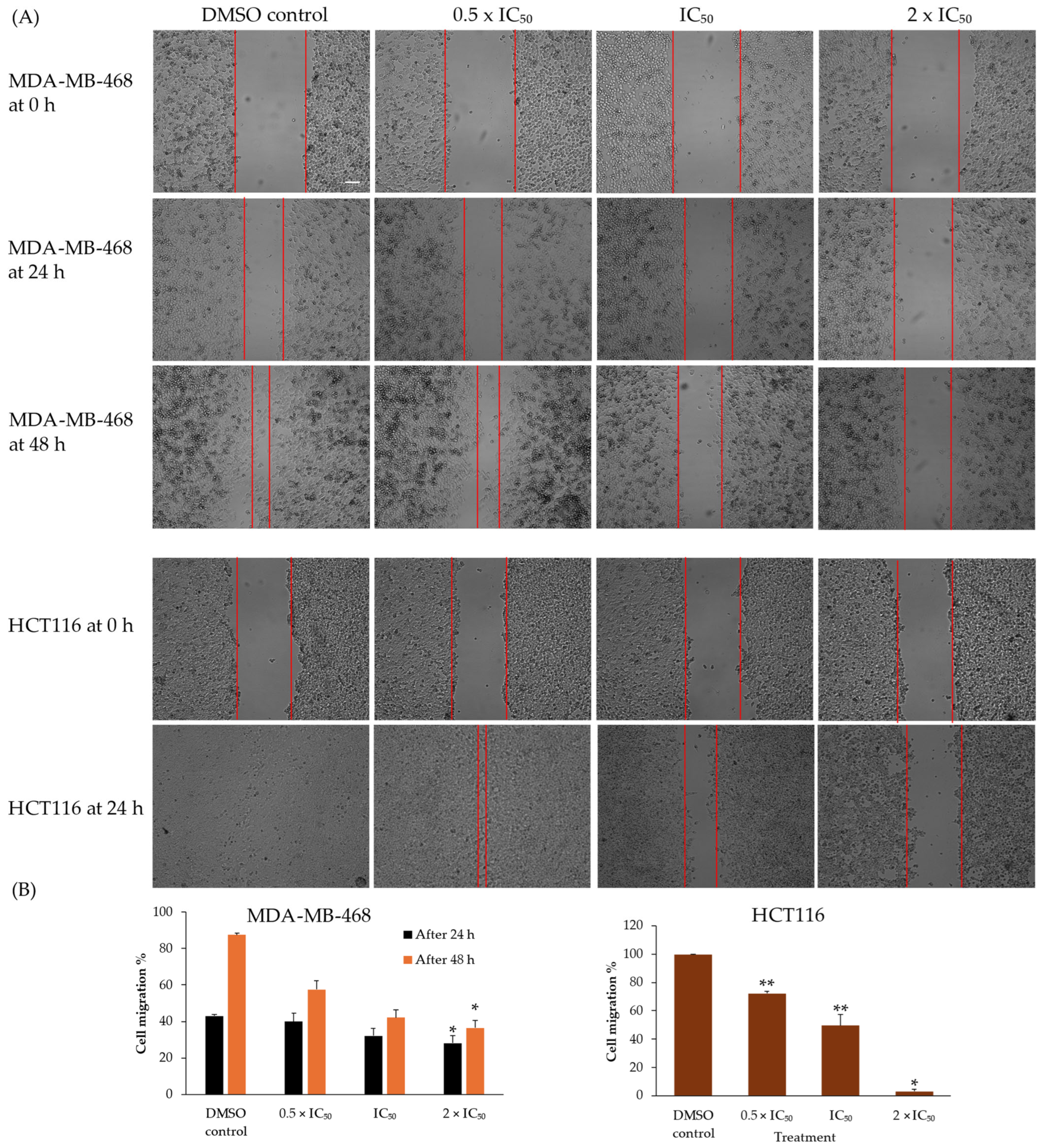
2.6. Compound 4 Inhibited 2D Migration of MDA-MB-468 Cells and HCT116 Cells
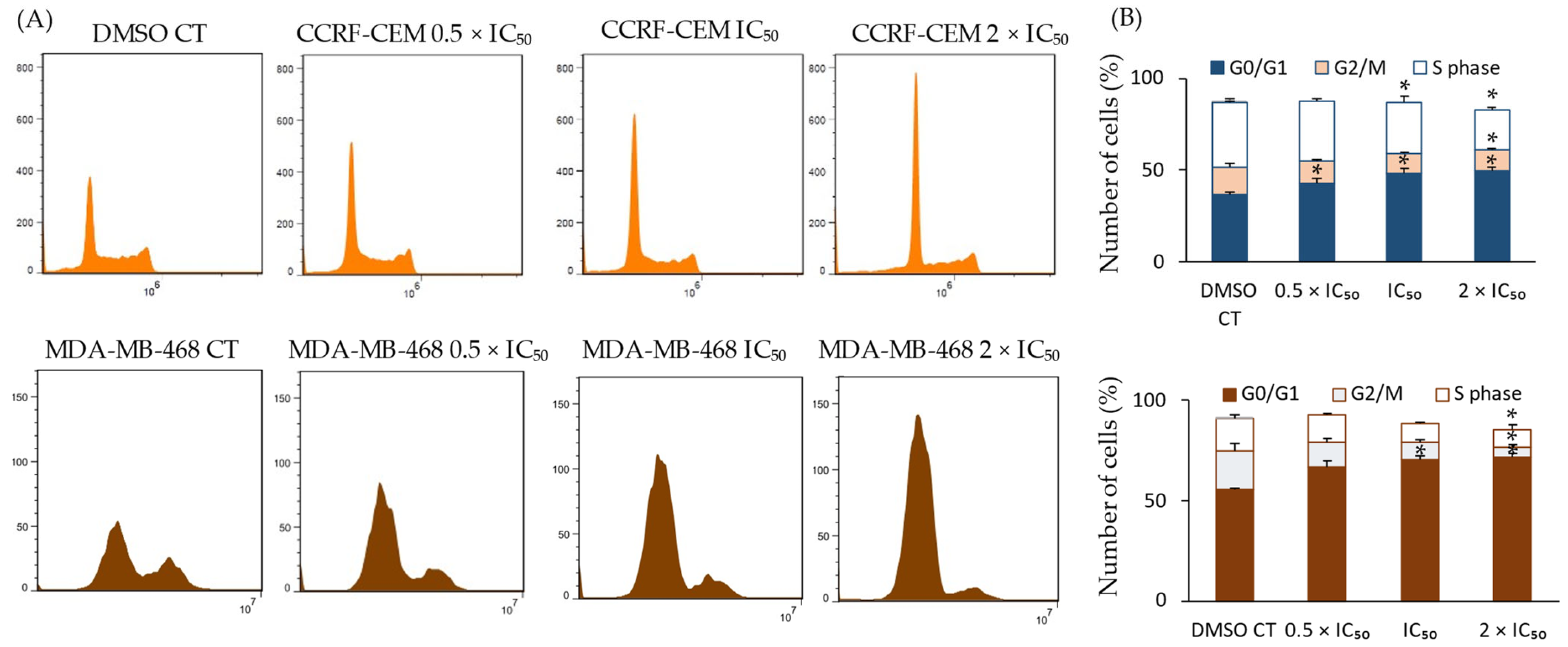
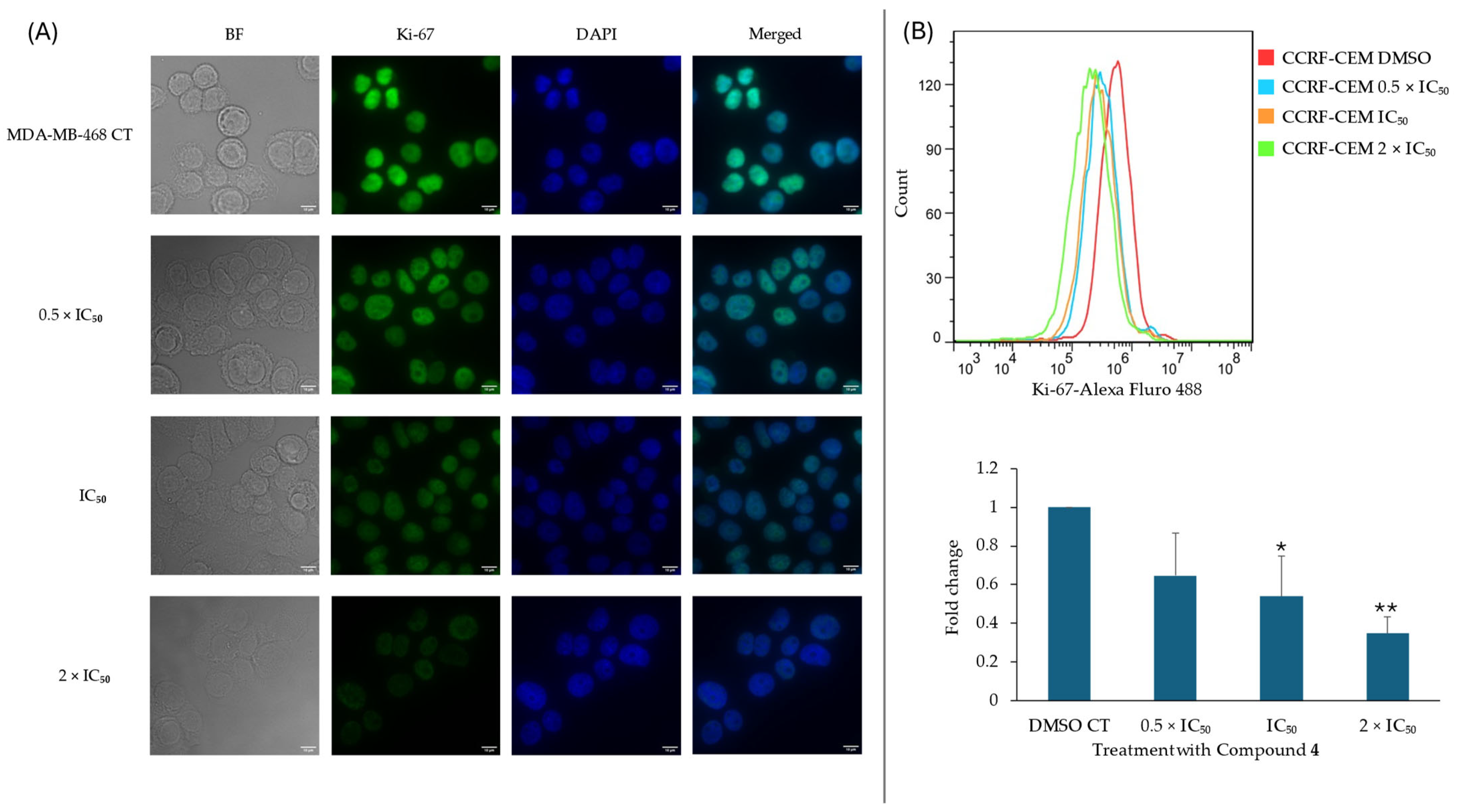
2.7. Effect of Compound 4 on the Cell Cycle and KI-67 Expression
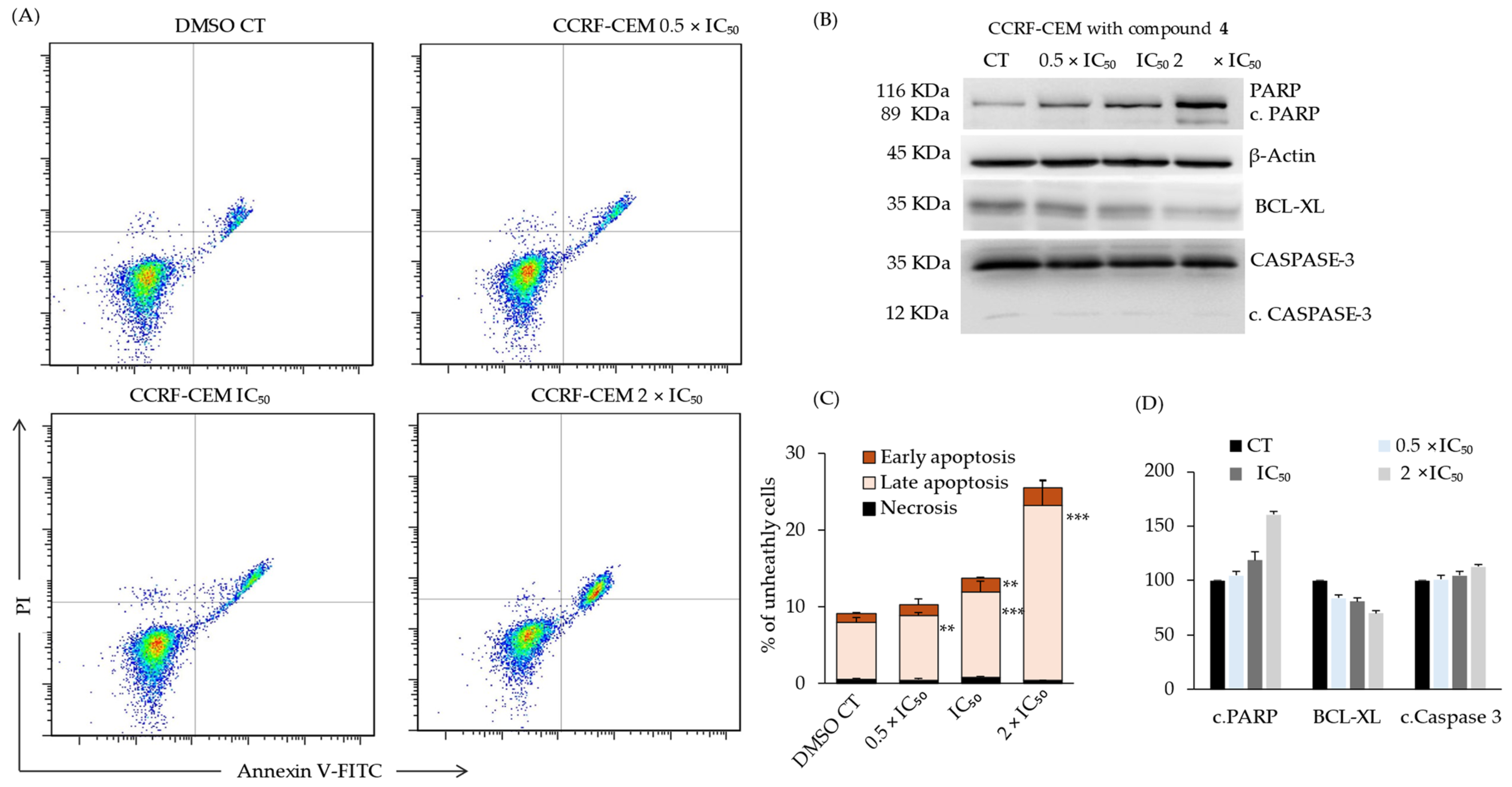
2.8. Compound 4-Induced Late Apoptosis of CCRF-CEM Cells via Deregulation of Apoptosis Markers
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Treatment Conditions
4.2. Virtual Drug Screening
4.3. Molecular Docking
4.4. MOE Docking
4.5. MD Simulations for Protein–Ligand Complexes
4.6. DNMT1 Activity Assay
4.7. Resazurin Cytotoxicity Assay
4.8. CMV-Luc Assay in KG-1 Cells
4.9. Migration Assay
4.10. Cell Cycle
4.11. Annexin V/PI Apoptosis Assay
4.12. Fluorescence Imaging
4.13. Flow Cytometry
4.14. SDS-Page and Western Blotting
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tronick, E.; Hunter, R.G. Waddington, Dynamic Systems, and Epigenetics. Front. Behav. Neurosci. 2016, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Peng, Q.; Jiang, X.; Tan, S.; Yang, Y.; Yang, W.; Han, Y.; Chen, Y.; Oyang, L.; Lin, J.; et al. Metabolic reprogramming and epigenetic modifications in cancer: From the impacts and mechanisms to the treatment potential. Exp. Mol. Med. 2023, 55, 1357–1370. [Google Scholar] [CrossRef]
- Nasrullah; Hussain, A.; Ahmed, S.; Rasool, M.; Shah, A.J. DNA methylation across the tree of life, from micro to macro-organism. Bioengineered 2022, 13, 1666–1685. [Google Scholar] [CrossRef] [PubMed]
- Casadesús, J.; Low, D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. MMBR 2006, 70, 830–856. [Google Scholar] [CrossRef]
- Casadesús, J.; Sánchez-Romero, M.A. DNA Methylation in Prokaryotes. In DNA Methyltransferases—Role and Function; Jeltsch, A., Jurkowska, R.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 21–43. [Google Scholar] [CrossRef]
- Iyer, L.M.; Abhiman, S.; Aravind, L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011, 101, 25–104. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, I.S.; Kulakova, O.G.; Boyko, A.N.; Favorova, O.O. DNA Methylation As an Epigenetic Mechanism in the Development of Multiple Sclerosis. Acta Naturae 2021, 13, 45–57. [Google Scholar] [CrossRef]
- Kikuchi, A.; Onoda, H.; Yamaguchi, K.; Kori, S.; Matsuzawa, S.; Chiba, Y.; Tanimoto, S.; Yoshimi, S.; Sato, H.; Yamagata, A.; et al. Structural basis for activation of DNMT1. Nat. Commun. 2022, 13, 7130. [Google Scholar] [CrossRef]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. Adv. Exp. Med. Biol. 2016, 945, 151–172. [Google Scholar] [CrossRef]
- Lopez, M.; Gilbert, J.; Contreras, J.; Halby, L.; Arimondo, P.B. Inhibitors of DNA Methylation. Adv. Exp. Med. Biol. 2022, 1389, 471–513. [Google Scholar] [CrossRef]
- Yin, J. DNA Methyltransferase and its Clinical Applications. IOP Conf. Ser. Earth Environ. Sci. 2020, 512, 012082. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, G.; Li, Y.; Lei, D.; Xiang, J.; Ouyang, L.; Wang, Y.; Yang, J. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol. 2022, 13, 1072651. [Google Scholar] [CrossRef] [PubMed]
- Clements, E.G.; Mohammad, H.P.; Leadem, B.R.; Easwaran, H.; Cai, Y.; Van Neste, L.; Baylin, S.B. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012, 40, 4334–4346. [Google Scholar] [CrossRef]
- Schermelleh, L.; Spada, F.; Easwaran, H.P.; Zolghadr, K.; Margot, J.B.; Cardoso, M.C.; Leonhardt, H. Trapped in action: Direct visualization of DNA methyltransferase activity in living cells. Nat. Methods 2005, 2, 751–756. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Wyckoff, J.; Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Geng, Q.; Cao, Z.; Liu, B.; Li, L.; Lu, P.; Lin, L.; Wei, L.; Tan, Y.; He, X.; et al. Insights into DNMT1 and programmed cell death in diseases. Biomed. Pharmacother. 2023, 168, 115753. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Pisconti, S.; Della Vittoria Scarpati, G. P53 mutations and cancer: A tight linkage. Ann. Transl. Med. 2016, 4, 522. [Google Scholar] [CrossRef]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2016, 1865, 275–288. [Google Scholar] [CrossRef]
- Gnyszka, A.; JastrzĘBski, Z.; Flis, S. DNA Methyltransferase Inhibitors and Their Emerging Role in Epigenetic Therapy of Cancer. Anticancer Res. 2013, 33, 2989. [Google Scholar]
- Chen, T.; Mahdadi, S.; Vidal, M.; Desbène-Finck, S. Non-nucleoside inhibitors of DNMT1 and DNMT3 for targeted cancer therapy. Pharmacol. Res. 2024, 207, 107328. [Google Scholar] [CrossRef]
- Pappalardi, M.B.; Keenan, K.; Cockerill, M.; Kellner, W.A.; Stowell, A.; Sherk, C.; Wong, K.; Pathuri, S.; Briand, J.; Steidel, M.; et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2021, 2, 1002–1017. [Google Scholar] [CrossRef]
- Draškovič, T.; Hauptman, N. Discovery of novel DNA methylation biomarker panels for the diagnosis and differentiation between common adenocarcinomas and their liver metastases. Sci. Rep. 2024, 14, 3095. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, O.M.; Isakovic, L.; Llewellyn, D.B.; Zhan, L.; Bernstein, N.; Claridge, S.; Raeppel, F.; Vaisburg, A.; Elowe, N.; Petschner, A.J.; et al. SAR around (l)-S-adenosyl-l-homocysteine, an inhibitor of human DNA methyltransferase (DNMT) enzymes. Bioorg. Med. Chem. Lett. 2009, 19, 2747–2751. [Google Scholar] [CrossRef]
- Khader, A.; Bokhari, R.; Hakimelahi, R.; Scheirey, C.; Afnan, J.; Braschi-Amirfarzan, M.; Thomas, R. A radiologist’s guide to novel anticancer therapies in the era of precision medicine. Eur. J. Radiol. Open 2022, 9, 100406. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, B.; Gor, R.; Ramalingam, S.; Thiagarajan, A.; Sohn, H.; Madhavan, T. Repurposing FDA-approved compounds to target JAK2 for colon cancer treatment. Discov. Oncol. 2024, 15, 226. [Google Scholar] [CrossRef]
- Kort, E.; Jovinge, S. Drug Repurposing: Claiming the Full Benefit from Drug Development. Curr. Cardiol. Rep. 2021, 23, 62. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014, 15, 49–63. [Google Scholar] [CrossRef]
- Zhang, X.; Nie, Y.; Zhang, R.; Yu, J.; Ge, J. Reduced DNMT1 levels induce cell apoptosis via upregulation of METTL3 in cardiac hypertrophy. Heliyon 2024, 10, e24572. [Google Scholar] [CrossRef]
- Chen, X.; Zheng, J.; Cai, J.; Li, H.; Li, S.; Wang, L.; Cheng, D.; Chen, H.; Yang, Y.; Chen, G.; et al. The cytoskeleton protein β-actin may mediate T cell apoptosis during acute rejection reaction after liver transplantation in a rat model. Am. J. Transl. Res. 2017, 9, 4888–4901. [Google Scholar]
- Dugina, V.; Khromova, N.; Rybko, V.; Blizniukov, O.; Shagieva, G.; Chaponnier, C.; Kopnin, B.; Kopnin, P. Tumor promotion by γ and suppression by β non-muscle actin isoforms. Oncotarget 2015, 6, 14556–14571. [Google Scholar] [CrossRef]
- Uxa, S.; Castillo-Binder, P.; Kohler, R.; Stangner, K.; Müller, G.A.; Engeland, K. Ki-67 gene expression. Cell Death Differ. 2021, 28, 3357–3370. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Kaufman, P.D. Ki-67: More than a proliferation marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Konkimalla, V.B.; Wang, Y.-F.; Sauerbrey, A.; Meinhardt, S.; Zintl, F.; Mattern, J.; Volm, M. Prediction of Broad Spectrum Resistance of Tumors towards Anticancer Drugs. Clin. Cancer Res. 2008, 14, 2405–2412. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Cao, J.; Kosyakova, N.; Mrasek, K.; Liehr, T.; Efferth, T. Genomic and transcriptomic profiling of resistant CEM/ADR-5000 and sensitive CCRF-CEM leukaemia cells for unravelling the full complexity of multi-factorial multidrug resistance. Sci. Rep. 2016, 6, 36754. [Google Scholar] [CrossRef]
- Efferth, T.; Verdorfer, I.; Miyachi, H.; Sauerbrey, A.; Drexler, H.G.; Chitambar, C.R.; Haber, M.; Gebhart, E. Genomic Imbalances in Drug-Resistant T-Cell Acute Lymphoblastic CEM Leukemia Cell Lines. Blood Cells Mol. Dis. 2002, 29, 1–13. [Google Scholar] [CrossRef]
- Steglich, B.; Mahringer, A.; Li, Y.; Posner, G.H.; Fricker, G.; Efferth, T. Inhibition of P-glycoprotein by two artemisinin derivatives. Nat. Prod. Bioprospect. 2012, 2, 59–64. [Google Scholar] [CrossRef]
- Kalinowsky, L.; Weber, J.; Balasupramaniam, S.; Baumann, K.; Proschak, E. A Diverse Benchmark Based on 3D Matched Molecular Pairs for Validating Scoring Functions. ACS Omega 2018, 3, 5704–5714. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, A.; Halby, L.; Fahy, J.; Arimondo, P.B. Targeting DNA methylation with small molecules: What’s next? J. Med. Chem. 2015, 58, 2569–2583. [Google Scholar] [CrossRef] [PubMed]
- Hossain, T.J. Methods for screening and evaluation of antimicrobial activity: A review of protocols, advantages, and limitations. Eur. J. Microbiol. Immunol. 2024, 14, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Rilova, E.; Erdmann, A.; Gros, C.; Masson, V.; Aussagues, Y.; Poughon-Cassabois, V.; Rajavelu, A.; Jeltsch, A.; Menon, Y.; Novosad, N.; et al. Design, synthesis and biological evaluation of 4-amino-N- (4-aminophenyl)benzamide analogues of quinoline-based SGI-1027 as inhibitors of DNA methylation. ChemMedChem 2014, 9, 590–601. [Google Scholar] [CrossRef]
- Damiescu, R.; Elbadawi, M.; Dawood, M.; Klauck, S.M.; Bringmann, G.; Efferth, T. Aniquinazoline B, a Fungal Natural Product, Activates the μ-Opioid Receptor. ChemMedChem 2024, 19, e202400213. [Google Scholar] [CrossRef]

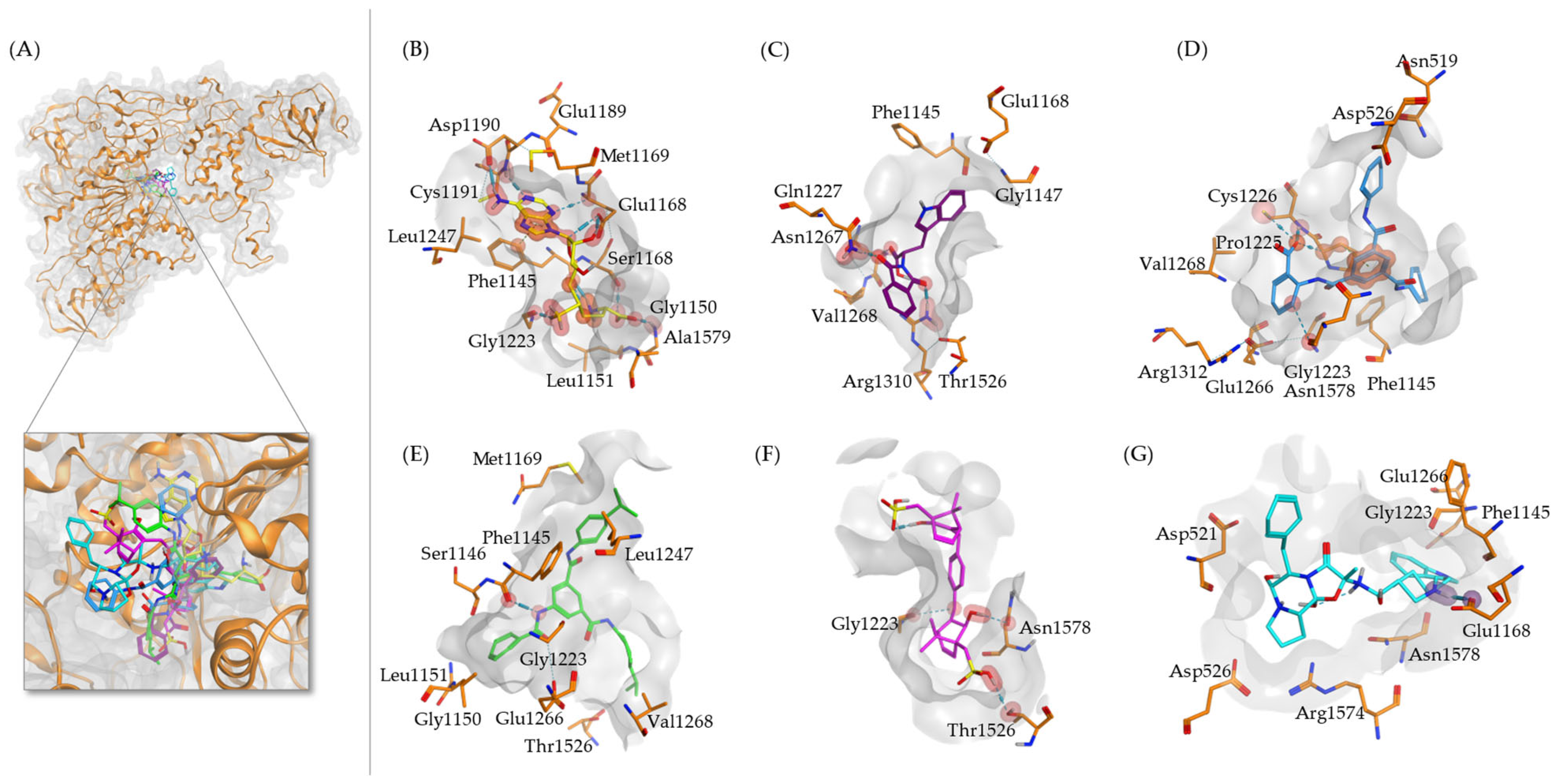
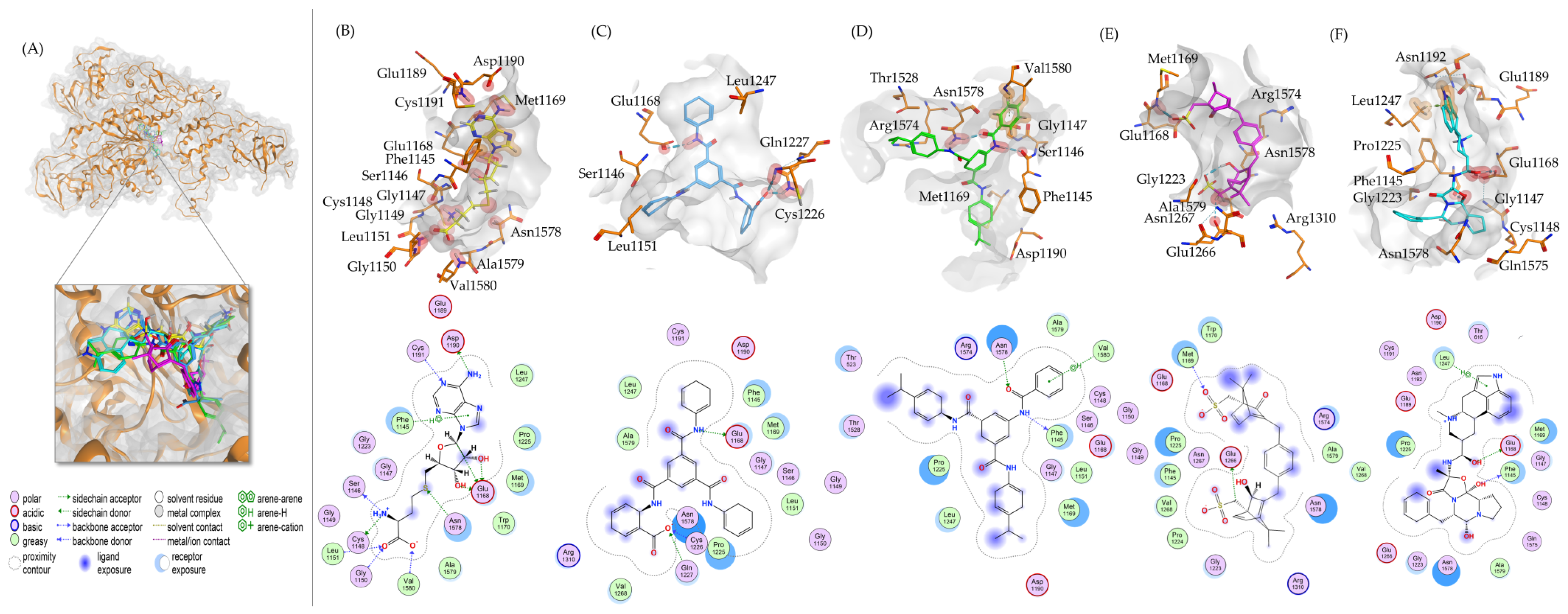

| No. | ZINC ID | Pyrx Binding Energy (kcal/mol) | Molecular Weight | logP |
|---|---|---|---|---|
| 1 | ZINC04050909 | −11.7 | 408.339 | 4.682 |
| 2 | ZINC03360933 | −11.3 | 487.61 | 4.407 |
| 3 | ZINC01038993 | −10.9 | 390.45 | 4.762 |
| 4 | ZINC02107822 | −10.9 | 423.516 | 4.416 |
| 5 | ZINC03037862 | −10.9 | 479.492 | 5.142 |
| 6 | ZINC01038994 | −10.8 | 394.413 | 4.592 |
| 7 | ZINC01071494 | −10.8 | 467.907 | 4.95 |
| 8 | ZINC02383479 | −10.8 | 430.508 | 4.962 |
| 9 | ZINC02971168 | −10.8 | 501.608 | 4.69 |
| 10 | ZINC02731106 | −10.7 | 484.467 | 3.756 |
| 11 | ZINC01038992 | −10.6 | 376.423 | 4.453 |
| 12 | ZINC02239620 | −10.6 | 465.509 | 5.937 |
| 13 | ZINC02241295 | −10.6 | 471.9 | 5.974 |
| 14 | ZINC02347371 | −10.6 | 469.472 | 5.768 |
| 15 | ZINC02690584 | −10.6 | 429.426 | 5.301 |
| 16 | ZINC02704495 | −10.6 | 419.383 | 4.522 |
| 17 | ZINC02860618 | −10.6 | 459.885 | 5.229 |
| 18 | ZINC03668964 | −10.6 | 519.645 | 7.69 |
| 19 | ZINC000008220909 | −12.2 | 665.733 | 0.12 |
| 20 | ZINC000111460375 | −11.8 | 562.706 | 4.24 |
| 21 | ZINC000003978005 | −11.7 | 583.689 | 2.081 |
| 22 | ZINC000169289767 | −11.4 | 872.894 | 6.67 |
| 23 | ZINC000052955754 | −11.3 | 581.673 | 1.991 |
| Compound | Lowest Binding Energy (kcal/mol) | pKi (nM) | Amino Acid Interactions | |
|---|---|---|---|---|
| 1 | ZINC03037862-1 | −10.27 ± 0.12 | 30.51 ± 6.25 | GLN1157, MET1077, SER1076, GLY1079, PRO1080, ASN1040, LEU1594, LYS1593, LEU1590, LYS1586 |
| 2 | ZINC03668964-2 | −10.25 ± 0.37 | 36.44 ± 19.20 | GLN594, VAL658, SER563, GLU562, GLU566, PRO574, ARG690, GLN687, GLN684, ARG1238, ASP571 |
| 3 | ZINC02731106 | −9.90 ± 0.11 | 56.24 ± 10.21 | GLN594 |
| 4 | ZINC03360933 | −9.74 ± 0.37 | 87.12 ± 51.28 | LEU1331, PHE1362, HIS1332, TRP1395, LEU1400, LYS1586, PRO1583, TYR1304, ALA1587, LEU1590, PHE1396, MET1077 |
| 5 | ZINC02239620 | −9.73 ± 0.46 | 97.42 ± 66.04 | ASP569, ASP565, GLU566, ASP571, GLN687, ALA669, PRO574, GLN684, GLU572, ARG690 |
| 6 | ZINC01038992 | −9.71 ± 0.09 | 77.09 ± 12.20 | GLY568, ASP569, GLN687, SER570, ARG690, GLU572, ASP565 |
| 7 | ZINC02241295 | −9.42 ± 0.20 | 131.36 ± 48.55 | ARG1453, PHE1492, PRO363, GLN 1491, LEU 365, TYR 359, GLN 358, ARG1490, ALA1488 |
| 8 | ZINC02347371 | −9.39 ± 0.33 | 154.90 ± 93.95 | CYS667, GLN687, ARG690, ASP571, ASP565, GLU566, GLU562, VAL658 |
| 9 | ZINC01038993 | −9.35 ± 0.05 | 141.0 ± 12.59 | SER1078, PRO1080, LEU1590, ASN1081, PHE1362, HIS1332, LEU1331, ASN1040, TRP1395 |
| 10 | ZINC01038994 | −8.97 ± 0.08 | 266.98 ± 32.49 | ALA838 |
| 11 | ZINC01071494 | −8.85 ± 0.10 | 331.18 ± 61.84 | GLY1147, ASN1576, GLU1168, GLU1256, MET1169, CYS1191, ILE1167, PHE1145, ASN1267, PRO1225 |
| 12 | ZINC02860618 | −8.83 ± 0.02 | 337.46 ± 10.41 | GLU1168, MET1169, GLY1223, PHE1145, PRO1225, LEU1247 |
| 13 | ZINC02690584 | −8.80 ± 0.22 | 379.17 ± 149.96 | CYS1191, PHE1146, PRO1225, MET1169, GLU1168, GLY1147, ASN1578, CYS1148 |
| 14 | ZINC02107822 | −8.65 ± 0.41 | 593.29 ± 443.15 | PRO363, ASP364, GLN358, GLN1491, ARG1490, PHE1492, ARG552 |
| 15 | ZINC02704495 | −8.54 ± 0.20 | 443.35 ± 276.67 | GLU1168, PHE1145, GLY1147, GLY1223, SER1146, ASN1578, ASN1267, VAL1268, PRO1225 |
| 16 | ZINC02971168 | −8.52 ± 0.55 | 823.7 ± 611.19 | ASP364, PRO363, THR424, PHE1492, ARG1453, ALA1488, ARG1490 |
| 17 | ZINC02383479 | −8.46 ± 0.25 | 681.75 ± 237.69 | GLY568, GLU566, ASP565, GLN687, ALA669, SER570, ARG690, GLU572 |
| 18 | ZINC04050909 | −8.02 ± 0.005 | 1330 ± 16.33 | VAL658, GLU566, GLU562, ASP565, PRO574, GLU572, ASP571, SER570, ARG690 |
| ZINC-15 FDA | ||||
| 19 | ZINC000111460375-3 | −11.09 ± 0.02 | 7.40 ± 0.34 | ARG1603, THR1602, VAL1604, LYS1323, LYS881, PRO880, ARG898 |
| 20 | ZINC000003978005-4 | −9.67 ± 0.22 | 86.65 ± 27.28 | GLN358, LEU365, ASP364, PHE1492, PRO363, ARG1453, LYS505, ASN1493, ARG1490, ALA1488, MET1451 |
| 21 | ZINC000052955754 | −8.88 ± 0.58 | 437.91 ± 23.24 | RG1574, TRP1170, MET1169, PHE1145, LEU1247, SER1246, PRO1225, GLY1228, PHE1229 |
| 22 | ZINC000169289767 | −8.72 ± 0.35 | 473.68 ±246.88 | ARG1310, ASN1578, GLU1168, PRO1225, ASP521, LYS1242, ASN519, LYS1242, SER520 |
| 23 | ZINC000008220909 | −7.99 ± 0.03 | 1390.0 ± 80 | SER570, GLU572, ASP571, ASP565, GLU566, VAL658, PRO574, ARG 690 |
| Compound | Docking Score (kcal/mol) | Amino Acid Interactions |
|---|---|---|
| SAH | −8.23 ± 0.23 | Phe1145, Ser1146, Gly1147, Cys1148, Gly1149, Gly1150, Leu1151, Ile1167, Glu1168, Met1169, Trp1170, Ala1173, Glu1189, Asp1190, Cys1191, Gly1223, Pro1225, Leu1247, Asn1578, Ala1579, Val1580 |
| Compound 1 | −10.03 ± 0.16 | Asp1143, Phe1145, Ser1146, Gly1147, Cys1148, Gly1149, Gly1150, Leu1151, Ile1167, Glu1168, Met1169, Glu1189, Asp1190, Cys1191, Gly1223, Pro1224, Pro1225, Cys1226, Gln1227, Leu1247, Asn1267, Val1268, Arg1310, Thr1528, Gly1577, Asn1578, Ala1579, Val1580 |
| Compound 2 | −9.76 ± 0.15 | Thr523, Thr616, Phe1145, Ser1146, Gly1147, Cys1148, Gly1149, Gly1150, Leu1151, Glu1168, Met1169, Asp1190, Asn1192, Gly1223, Pro1225, Gln1227, Leu1247, Thr1528, Gln1536, Arg1574, Gln1575, Asn1578, Ala1579, Val1580 |
| Compound 3 | −8.08 ± 0.29 | Phe1145, Glu1168, Met1169, Trp1170, Gly1223, Pro1224, Pro1225, Gln1227, Leu1247, Glu1266, Asn1267, Val1268, Arg1310, Arg1574, Asn1578, Ala1579, Val1580 |
| Compound 4 | −9.85 ± 0.14 | Thr616, Phe1145, Ser1146, Gly1147, Cys1148, Ile1167, Glu1168, Met1169, Trp1170, Ala1173, Glu1189, Asp1190, Cys1191, Asn1192, Gly1223, Pro1224, Pro1225, Cys1226, Leu1247, Glu1266, Asn1267, Val1268, Arg1574, Gln1575, Asn1578, Ala1579 |
| Cell Lines | IC50 Values of Compound 2 (µM) | IC50 Values of Compound 4 (µM) |
|---|---|---|
| CCRF-CEM | >100 | 18.25 ± 4.37 |
| CEM-ADR5000 | >100 | >100 |
| HCT116 | >100 | 46.82 ± 3.04 |
| MDA-MB-468 | >100 | 29.42 ± 2–37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Friedrich, L.J.; Guthart, A.; Zhou, M.; Arimondo, P.B.; Efferth, T.; Dawood, M. Supercomputer-Based Virtual Screening for Deoxyribonucleic Acid Methyltransferase 1 Inhibitors as Novel Anticancer Agents. Int. J. Mol. Sci. 2024, 25, 11870. https://doi.org/10.3390/ijms252211870
Friedrich LJ, Guthart A, Zhou M, Arimondo PB, Efferth T, Dawood M. Supercomputer-Based Virtual Screening for Deoxyribonucleic Acid Methyltransferase 1 Inhibitors as Novel Anticancer Agents. International Journal of Molecular Sciences. 2024; 25(22):11870. https://doi.org/10.3390/ijms252211870
Chicago/Turabian StyleFriedrich, Lara Johanna, Axel Guthart, Min Zhou, Paola B. Arimondo, Thomas Efferth, and Mona Dawood. 2024. "Supercomputer-Based Virtual Screening for Deoxyribonucleic Acid Methyltransferase 1 Inhibitors as Novel Anticancer Agents" International Journal of Molecular Sciences 25, no. 22: 11870. https://doi.org/10.3390/ijms252211870
APA StyleFriedrich, L. J., Guthart, A., Zhou, M., Arimondo, P. B., Efferth, T., & Dawood, M. (2024). Supercomputer-Based Virtual Screening for Deoxyribonucleic Acid Methyltransferase 1 Inhibitors as Novel Anticancer Agents. International Journal of Molecular Sciences, 25(22), 11870. https://doi.org/10.3390/ijms252211870







