Use of 3′ Rapid Amplification of cDNA Ends (3′ RACE)-Based Targeted RNA Sequencing for Profiling of Druggable Genetic Alterations in Urothelial Carcinomas
Abstract
1. Introduction
2. Results
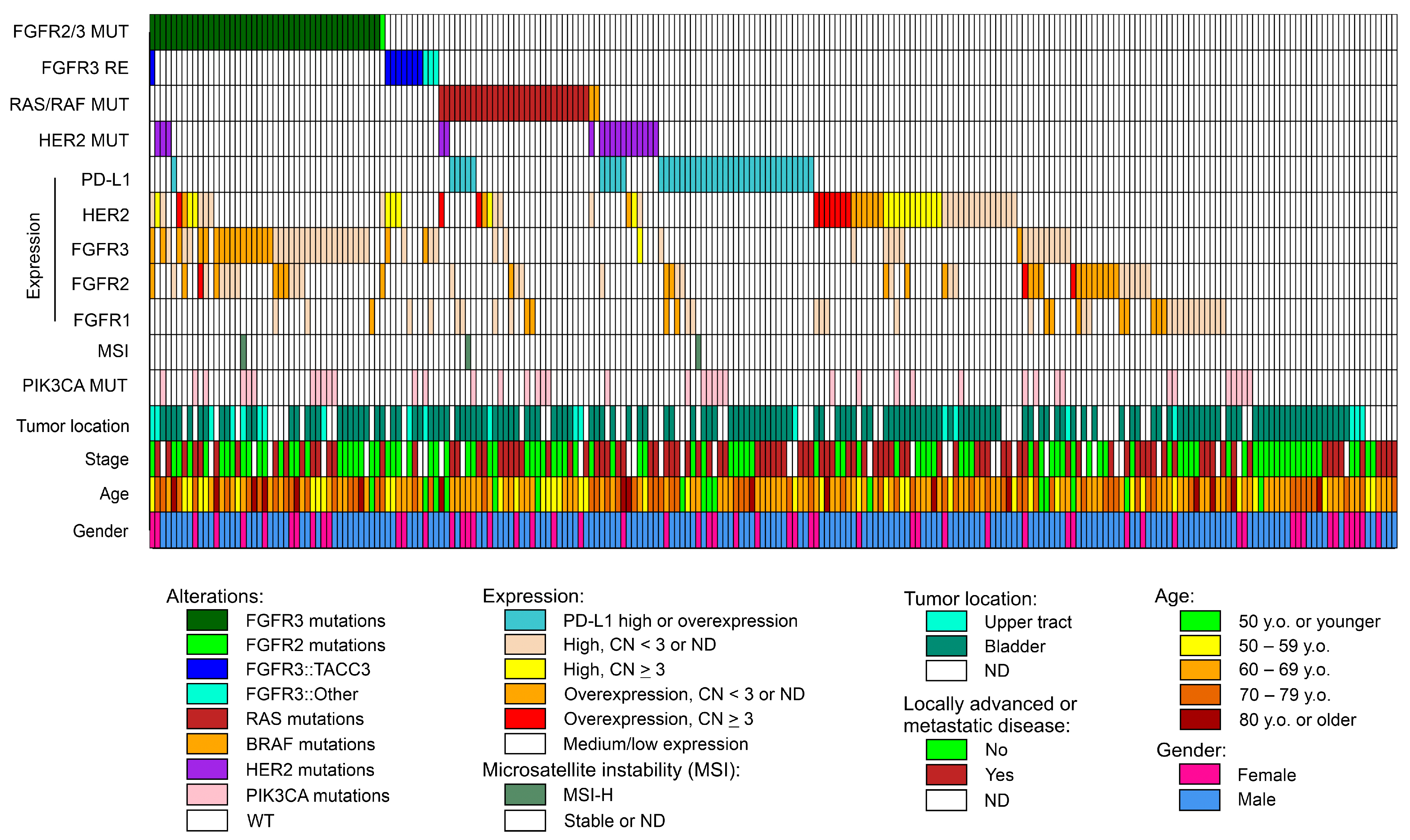
2.1. FGFR1-4 Gene Aberrations Identified by Targeted RNA Sequencing
2.2. Mutations in the Hot-Spot Regions of the RAS/RAF Genes
2.3. HER2 (ERBB2) Aberrations in UCs: Point Mutations, Amplifications and mRNA Overexpression
2.4. Predictive Markers for Treatment with Immune Checkpoint Inhibitors: PD-L1 Expression and Microsatellite Instability (MSI)
2.5. PIK3CA Mutations in UCs
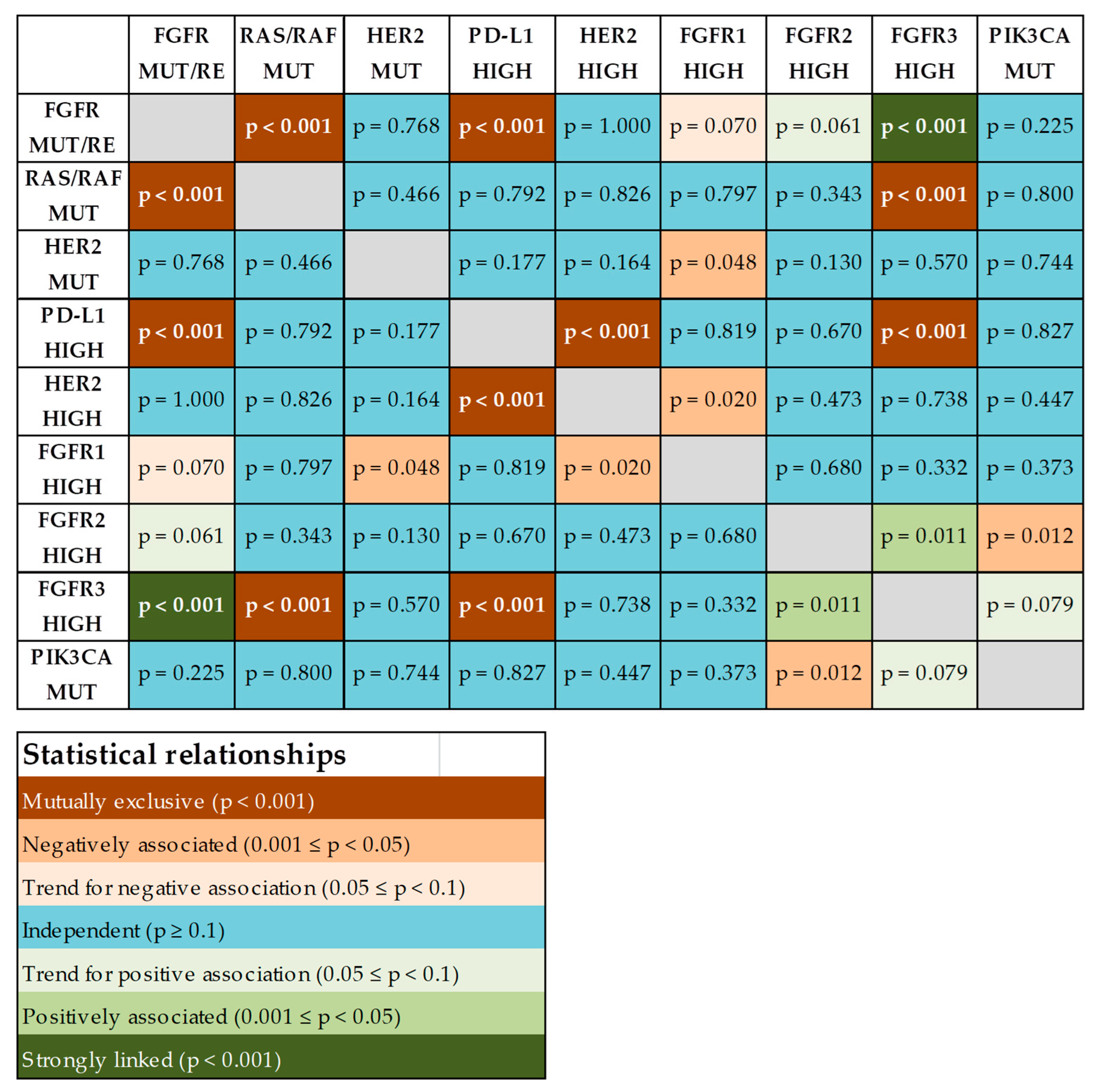
2.6. Co-Occurrence of Potentially Actionable Molecular Alterations
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
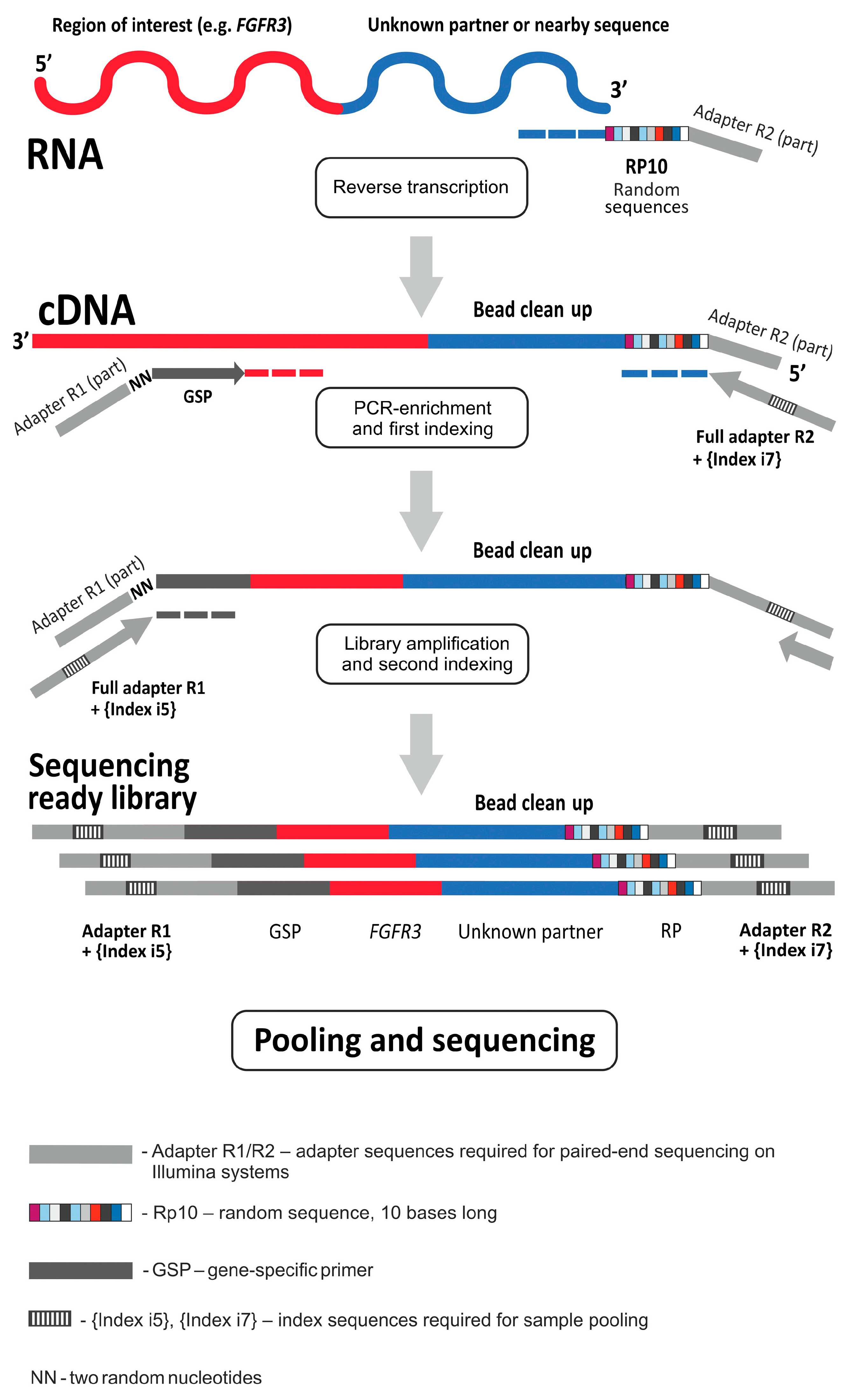
4.2. Targeted RNA Sequencing and Data Analysis
4.3. Other Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3′ RACE | 3′ Rapid Amplification of cDNA Ends |
| CN | Copy Number |
| ddPCR | digital droplet PCR |
| FDA | Food and Drug Administration |
| FFPE | Formalin-Fixed Paraffin-Embedded |
| FGFR | Fibroblast Growth Factor Receptor |
| ICI | Immune Checkpoint Inhibitor |
| IHC | Immunohistochemistry |
| MSI | Microsatellite Instability |
| NGS | Next Generation Sequencing |
| MIBC | Muscle-Invasive Bladder Carcinoma |
| NMIBC | Non-Muscle Invasive Bladder Carcinoma |
| OS | Overall Survival |
| PFS | Progression-Free Survival |
| PCR | Polymerase Chain Reaction |
| UC | Urothelial Carcinomas |
| UTR | Untranslated Region |
Appendix A
- The alignment of reads and removal of duplicates. The preprocessing of data was performed as described previously [17]. Pseudo-UMI sequences (representing the distal part of the random primer used in the reverse transcription reaction) were extracted from each read in *R2.fastq file with UMI-tools v. 1.1.4 [https://github.com/CGATOxford/UMI-tools (accessed on 9 September 2024)] [50]. Then, sequencing reads were aligned using the STAR program v2.7.8a [https://github.com/alexdobin/STAR (accessed on 9 September 2024)] [51]. Polymerase chain reaction (PCR) and optical duplicates were removed using UMI-tools. The resulting .bam file was sorted by queryname with the Picard toolkit v3.0.0 [https://broadinstitute.github.io/picard/ (accessed on 9 September 2024)]. Then, the custom python script Filter_RACEbam.py [https://github.com/MitiushkinaNV/RACE_NGS (accessed on 9 September 2024)] was applied to filter out improperly aligned/unspecific reads and create the catalogue of reads generated from each primer at the PCR-enrichment step. The filtered .bam file was coordinate-sorted and indexed using the Picard toolkit.
- Counting reads in selected regions. Analysis of gene expression was performed in a qPCR-like manner. A fragment containing an exon-exon junction was selected within a gene of interest. The primer for PCR enrichment was located 15–30 bp far from the 3′ end of the first exon (Figure A1). The coverage was counted in the second exon within 15 bp region located 75 bp away from the 5′ end position of the primer (Figure A1). The custom script Expression_count.py was used to calculate coverage in all selected regions [https://github.com/MitiushkinaNV/RACE_NGS (accessed on 9 September 2024)]. The following gene fragments were selected to assess expression in this study: exons 3–4 and 5–6 for the PD-L1 gene, exons 12–13 and 14–15 for the FGFR1 gene, exons 13–14 and 16–17 for the FGFR2 gene, exons 14–15 for the FGFR3 gene, exons 12–13 for the FGFR4 gene and exon 16–17 for the HER2 gene.
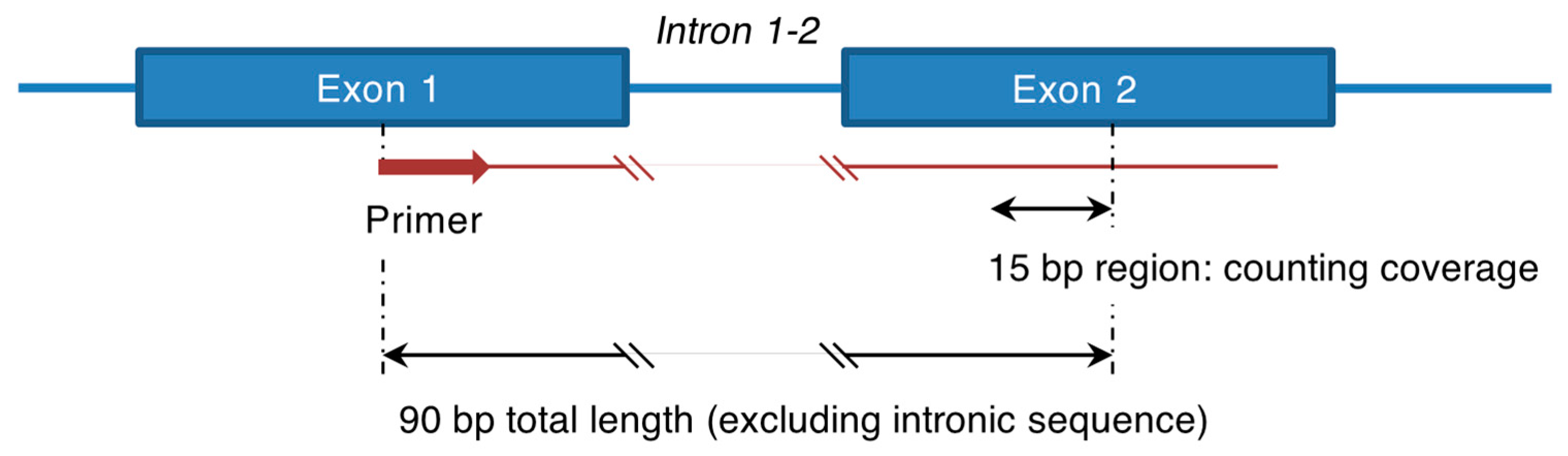
- 3.
- Normalization of data using a set of referee genes. In this study, three referee genes were used for normalization: DDX23 (exons 8–9), GOLGA5 (exons 5–6) and SEL1L (exons 13–14). These referee genes were selected based on the study of E. Eisenberg and E.Y. Levanon [57]. Normalization relied on the following assumptions: (1) the selected referee genes have stable levels of expression in the studied tissue samples and, thus, (2) the ratio of expression levels is also conserved for any pair of these genes. The following formula was used to estimate the normalization factor (NormFactor) for each sample:where RG1k, RG2k and RG3k are coverage values (numbers of sequencing reads) for the three referee genes, SEL1L, DDX23 and GOLGA5, respectively, in the sample k, and n is the total number of samples.
- 4.
- Definition of high expression or overexpression of a given gene. The normalized expression values for a single gene fragment were categorized as having “low”, “median” or “high” expression if they belonged to the first, second/third or the fourth quartile, respectively. However, if the value exceeded the third quartile plus 1.5 interquartile range, the respective fragment was considered to be “overexpressed”. If two fragments within a gene were used to assess its expression, the “high” expression of a gene meant that either both fragments had “high” expression or one of the fragments had “high” and the other had “overexpression” status. The “overexpression” status of a gene meant that both assessed fragments were found to be “overexpressed”.
References
- Nally, E.; Young, M.; Chauhan, V.; Wells, C.; Szabados, B.; Powles, T.; Jackson-Spence, F. Upper Tract Urothelial Carcinoma (UTUC): Prevalence; Impact and Management Challenge. Cancer Manag. Res. 2024, 16, 467–475. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Yu, E.M.; Belay, S.; Li, W.; Aragon-Ching, J.B. Non-urothelial and urothelial variants of bladder cancer. Cancer Treat. Res. Commun. 2022, 33, 100661. [Google Scholar] [CrossRef]
- Gill, E.; Perks, C.M. Mini-Review: Current Bladder Cancer Treatment-The Need for Improvement. Int. J. Mol. Sci. 2024, 25, 1557. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Cookson, M.S.; Guercio, B.J.; Cheng, L. Advances in diagnosis and treatment of bladder cancer. BMJ 2024, 384, e076743. [Google Scholar] [CrossRef]
- Ross, J.S.; Wang, K.; Khaira, D.; Ali, S.M.; Fisher, H.A.; Mian, B.; Nazeer, T.; Elvin, J.A.; Palma, N.; Yelensky, R.; et al. Comprehensive genomic profiling of 295 cases of clinically advanced urothelial carcinoma of the urinary bladder reveals a high frequency of clinically relevant genomic alterations. Cancer 2016, 122, 702–711. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Z.; Wang, X.; Bai, P.; Sun, A.; Shao, Z.; Luo, R.; Wu, Z.; Zhang, K.; Li, W.; et al. Identification of potential therapeutic targets in urothelial bladder carcinoma of Chinese population by targeted next-generation sequencing. Cancer Biol. Ther. 2020, 21, 709–716. [Google Scholar] [CrossRef]
- Tang, Q.; Zuo, W.; Wan, C.; Xiong, S.; Xu, C.; Yuan, C.; Sun, Q.; Zhou, L.; Li, X. Comprehensive genomic profiling of upper tract urothelial carcinoma and urothelial carcinoma of the bladder identifies distinct molecular characterizations with potential implications for targeted therapy & immunotherapy. Front. Immunol. 2023, 13, 1097730. [Google Scholar] [CrossRef]
- Loriot, Y.; Kamal, M.; Syx, L.; Nicolle, R.; Dupain, C.; Mensourri, N.; Du-quesne, I.; Lavaud, P.; Nicotra, C.; Ngocamus, M.; et al. The genomic and transcriptomic landscape of metastastic urothelial cancer. Nat. Commun. 2024, 15, 8603. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Valderrama, B.P.; Gupta, S.; Bedke, J.; Kikuchi, E.; Hoffman-Censits, J.; Iyer, G.; Vulsteke, C.; Park, S.H.; Shin, S.J.; et al. Enfortumab Vedotin and Pembrolizumab in Untreated Advanced Urothelial Cancer. N. Engl. J. Med. 2024, 390, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Bellmunt, J.; Comperat, E.; De Santis, M.; Huddart, R.; Loriot, Y.; Necchi, A.; Valderrama, B.P.; Ravaud, A.; Shariat, S.F.; et al. ESMO Clinical Practice Guideline interim update on first-line therapy in advanced urothelial carcinoma. Ann. Oncol. 2024, 35, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Flaig, T.W.; Spiess, P.E.; Abern, M.; Agarwal, N.; Bangs, R.; Buyyounouski, M.K.; Chan, K.; Chang, S.S.; Chang, P.; Friedlander, T.; et al. NCCN Guidelines Insights: Bladder Cancer; Version 3.2024. J. Natl. Compr. Cancer Netw. 2024, 22, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Scotto-Lavino, E.; Du, G.; Frohman, M.A. 3’ end cDNA amplification using classic RACE. Nat. Protoc. 2006, 1, 2742–2745. [Google Scholar] [CrossRef]
- Kurose, K.; Mine, N.; Doi, D.; Ota, Y.; Yoneyama, K.; Konishi, H.; Araki, T.; Emi, M. Novel gene fusion of COX6C at 8q22-23 to HMGIC at 12q15 in a uterine leiomyoma. Genes Chromosomes Cancer 2000, 27, 303–307. [Google Scholar] [CrossRef]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef]
- Mitiushkina, N.V.; Tiurin, V.I.; Anuskina, A.A.; Bordovskaya, N.A.; Shestakova, A.D.; Martianov, A.S.; Bubnov, M.G.; Shishkina, A.S.; Semina, M.V.; Romanko, A.A.; et al. Molecular Analysis of Biliary Tract Cancers with the Custom 3’ RACE-Based NGS Panel. Diagnostics 2023, 13, 3168. [Google Scholar] [CrossRef]
- Loriot, Y.; Matsubara, N.; Park, S.H.; Huddart, R.A.; Burgess, E.F.; Houede, N.; Banek, S.; Guadalupi, V.; Ku, J.H.; Valderrama, B.P.; et al. Erdafitinib or Chemotherapy in Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2023, 389, 1961–1971. [Google Scholar] [CrossRef]
- Qin, A.; Johnson, A.; Ross, J.S.; Miller, V.A.; Ali, S.M.; Schrock, A.B.; Gadgeel, S.M. Detection of Known and Novel FGFR Fusions in Non-Small Cell Lung Cancer by Comprehensive Genomic Profiling. J. Thorac. Oncol. 2019, 14, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Gött, H.; Uhl, E. FGFR3-TACCs3 Fusions and Their Clinical Relevance in Human Glioblastoma. Int. J. Mol. Sci. 2022, 23, 8675. [Google Scholar] [CrossRef]
- Saridogan, T.; Akcakanat, A.; Zhao, M.; Evans, K.W.; Yuca, E.; Scott, S.; Kirby, B.P.; Zheng, X.; Ha, M.J.; Chen, H.; et al. Efficacy of futibatinib; an irreversible fibroblast growth factor receptor inhibitor; in FGFR-altered breast cancer. Sci. Rep. 2023, 13, 20223. [Google Scholar] [CrossRef] [PubMed]
- Brandi, G.; Relli, V.; Deserti, M.; Palloni, A.; Indio, V.; Astolfi, A.; Serravalle, S.; Mattiaccio, A.; Vasuri, F.; Malvi, D.; et al. Activated FGFR2 signalling as a biomarker for selection of intrahepatic cholangiocarcinoma patients candidate to FGFR targeted therapies. Sci. Rep. 2024, 14, 3136. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, A.; Govindan, R. Targeting KRAS G12C mutation in lung adenocarcinoma. Lung Cancer 2022, 165, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Bekaii-Saab, T.S.; Yaeger, R.; Spira, A.I.; Pelster, M.S.; Sabari, J.K.; Hafez, N.; Barve, M.; Velastegui, K.; Yan, X.; Shetty, A.; et al. Adagrasib in Advanced Solid Tumors Harboring a KRAS G12C Mutation. J. Clin. Oncol. 2023, 41, 4097–4106. [Google Scholar] [CrossRef] [PubMed]
- Ros, J.; Vaghi, C.; Baraibar, I.; Saoudi González, N.; Rodríguez-Castells, M.; García, A.; Alcaraz, A.; Salva, F.; Tabernero, J.; Elez, E. Targeting KRAS G12C Mutation in Colorectal Cancer; A Review: New Arrows in the Quiver. Int. J. Mol. Sci. 2024, 25, 3304. [Google Scholar] [CrossRef]
- Gilardi, M.; Wang, Z.; Proietto, M.; Chillà, A.; Calleja-Valera, J.L.; Goto, Y.; Vanoni, M.; Janes, M.R.; Mikulski, Z.; Gualberto, A.; et al. Tipifarnib as a Precision Therapy for HRAS-Mutant Head and Neck Squamous Cell Carcinomas. Mol. Cancer Ther. 2020, 19, 1784–1796. [Google Scholar] [CrossRef]
- Lee, H.W.; Sa, J.K.; Gualberto, A.; Scholz, C.; Sung, H.H.; Jeong, B.C.; Choi, H.Y.; Kwon, G.Y.; Park, S.H. A Phase II Trial of Tipifarnib for Patients with Previously Treated; Metastatic Urothelial Carcinoma Harboring HRAS Mutations. Clin. Cancer Res. 2020, 26, 5113–5119. [Google Scholar] [CrossRef]
- Ho, A.L.; Brana, I.; Haddad, R.; Bauman, J.; Bible, K.; Oosting, S.; Wong, D.J.; Ahn, M.J.; Boni, V.; Even, C.; et al. Tipifarnib in Head and Neck Squamous Cell Carcinoma With HRAS Mutations. J. Clin. Oncol. 2021, 39, 1856–1864. [Google Scholar] [CrossRef]
- Odeniyide, P.; Yohe, M.E.; Pollard, K.; Vaseva, A.V.; Calizo, A.; Zhang, L.; Rodriguez, F.J.; Gross, J.M.; Allen, A.N.; Wan, X.; et al. Targeting farnesylation as a novel therapeutic approach in HRAS-mutant rhabdomyosarcoma. Oncogene 2022, 41, 2973–2983. [Google Scholar] [CrossRef]
- Dankner, M.; Rose, A.A.N.; Rajkumar, S.; Siegel, P.M.; Watson, I.R. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene 2018, 37, 3183–3199. [Google Scholar] [CrossRef]
- Turshudzhyan, A.; Vredenburgh, J. A Rare p.T599dup BRAF Mutant NSCLC in a Non-Smoker. Curr. Oncol. 2020, 28, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Miao, E.; Das, K.; Seetharamu, N. Clinical efficacy with dabrafenib and trametinib in a T599_V600insT poorly differentiated metastatic thyroid carcinoma. BMJ Case Rep. 2021, 14, e243264. [Google Scholar] [CrossRef]
- Qu, M.; Zhou, L.; Yan, X.; Li, S.; Wu, X.; Xu, H.; Li, J.; Guo, J.; Zhang, X.; Li, H.; et al. Advances in HER2-Targeted Treatment for Advanced/Metastatic Urothelial Carcinoma. Bladder 2023, 10, e21200012. [Google Scholar] [CrossRef]
- Roy-Chowdhuri, S.; Davies, K.D.; Ritterhouse, L.L.; Snow, A.N. ERBB2 (HER2) Alterations in Colorectal Cancer. J. Mol. Diagn. 2022, 24, 1064–1066. [Google Scholar] [CrossRef]
- Imyanitov, E.N.; Preobrazhenskaya, E.V.; Orlov, S.V. Current status of molecular diagnostics for lung cancer. Explor. Target. Anti-Tumor Ther. 2024, 5, 742–765. [Google Scholar] [CrossRef]
- Eturi, A.; Bhasin, A.; Zarrabi, K.K.; Tester, W.J. Predictive and Prognostic Biomarkers and Tumor Antigens for Targeted Therapy in Urothelial Carcinoma. Molecules 2024, 29, 1896. [Google Scholar] [CrossRef]
- Hainsworth, J.D.; Meric-Bernstam, F.; Swanton, C.; Hurwitz, H.; Spigel, D.R.; Sweeney, C.; Burris, H.; Bose, R.; Yoo, B.; Stein, A.; et al. Targeted Therapy for Advanced Solid Tumors on the Basis of Molecular Profiles: Results From MyPathway; an Open-Label; Phase IIa Multiple Basket Study. J. Clin. Oncol. 2018, 36, 536–542. [Google Scholar] [CrossRef]
- Sweeney, C.J.; Hainsworth, J.D.; Bose, R.; Burris, H.A.; Kurzrock, R.; Swanton, C.; Friedman, C.F.; Spigel, D.R.; Szado, T.; Schulze, K.; et al. MyPathway Human Epidermal Growth Factor Receptor 2 Basket Study: Pertuzumab + Trastuzumab Treatment of a Tissue-Agnostic Cohort of Patients With Human Epidermal Growth Factor Receptor 2-Altered Advanced Solid Tumors. J. Clin. Oncol. 2024, 42, 258–265. [Google Scholar] [CrossRef]
- Domb, C.; Garcia, J.A.; Barata, P.C.; Mendiratta, P.; Rao, S.; Brown, J.R. Systematic review of recent advancements in antibody-drug and bicycle toxin conjugates for the treatment of urothelial cancer. Ther. Adv. Urol. 2024, 16, 17562872241249073. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Makker, V.; Oaknin, A.; Oh, D.Y.; Banerjee, S.; González-Martín, A.; Jung, K.H.; Ługowska, I.; Manso, L.; Manzano, A.; et al. Efficacy and Safety of Trastuzumab Deruxtecan in Patients With HER2-Expressing Solid Tumors: Primary Results From the DESTINY-PanTumor02 Phase II Trial. J. Clin. Oncol. 2024, 42, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022, 387, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Al Nabhani, S.; Al Harthy, A.; Al Riyami, M.; Al Sinawi, S.; Al Rashdi, A.; Al Husseni, S.; Kumar, S. Programmed Death-ligand 1 (PD-L1) Expression in Bladder Cancer and its Correlation with Tumor Grade, Stage and Outcome. Oman Med. J. 2022, 37, e441. [Google Scholar] [CrossRef]
- Climent, M.Á.; Álvarez, C.; Morales, R.; Maroto, P.; Rodríguez-Vida, A.; Méndez-Vidal, M.J.; Del Muro, X.G.; Puente, J.; Láinez, N.; Vázquez, S.; et al. Exploratory analyses of treatment subgroup interaction by PD-L1 status and according to PD-L1 expression in the JAVELIN Bladder 100 trial. Clin. Transl. Oncol. 2024, 26, 1532–1538. [Google Scholar] [CrossRef]
- Lee, H.H.; Wang, Y.N.; Xia, W.; Chen, C.H.; Rau, K.M.; Ye, L.; Wei, Y.; Chou, C.K.; Wang, S.C.; Yan, M.; et al. Removal of N-Linked Glycosylation Enhances PD-L1 Detection and Predicts Anti-PD-1/PD-L1 Therapeutic Efficacy. Cancer Cell 2019, 36, 168–178.e4. [Google Scholar] [CrossRef]
- Germanà, E.; Pepe, L.; Pizzimenti, C.; Ballato, M.; Pierconti, F.; Tuccari, G.; Ieni, A.; Giuffrè, G.; Fadda, G.; Fiorentino, V.; et al. Programmed Cell Death Ligand 1 (PD-L1) Immunohistochemical Expression in Advanced Urothelial Bladder Carcinoma: An Updated Review with Clinical and Pathological Implications. Int. J. Mol. Sci. 2024, 25, 6750. [Google Scholar] [CrossRef]
- Plage, H.; Furlano, K.; Hofbauer, S.; Weinberger, S.; Ralla, B.; Franz, A.; Fendler, A.; de Martino, M.; Roßner, F.; Elezkurtaj, S.; et al. PD-L1 expression in tumor and inflammatory cells is associated with favorable tumor features and favorable prognosis in muscle-invasive urothelial carcinoma of the bladder not treated by immune checkpoint inhibitors. BMC Urol. 2024, 24, 96. [Google Scholar] [CrossRef]
- Maio, M.; Ascierto, P.A.; Manzyuk, L.; Motola-Kuba, D.; Penel, N.; Cassier, P.A.; Bariani, G.M.; De Jesus Acosta, A.; Doi, T.; Longo, F.; et al. Pembrolizumab in microsatellite instability high or mismatch repair deficient cancers: Updated analysis from the phase II KEYNOTE-158 study. Ann. Oncol. 2022, 33, 929–938. [Google Scholar] [CrossRef]
- Chandran, E.B.A.; Iannantuono, G.M.; Atiq, S.O.; Akbulut, D.; Sinaii, N.; Simon, N.I.; Banday, A.R.; Boudjadi, S.; Gurram, S.; Nassar, A.H.; et al. Mismatch repair deficiency and microsatellite instability in urothelial carcinoma: A systematic review and meta-analysis. BMJ Oncol. 2024, 3, e000335. [Google Scholar] [CrossRef]
- Sirico, M.; D’Angelo, A.; Gianni, C.; Casadei, C.; Merloni, F.; De Giorgi, U. Current State and Future Challenges for PI3K Inhibitors in Cancer Therapy. Cancers 2023, 15, 703. [Google Scholar] [CrossRef]
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Ensembl Canonical Transcript. Available online: https://www.ensembl.org/info/genome/genebuild/canonical.html (accessed on 8 August 2024).
- Haas, B.J.; Dobin, A.; Li, B.; Stransky, N.; Pochet, N.; Regev, A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019, 20, 213. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org (accessed on 8 August 2024).
- Martianov, A.S.; Mitiushkina, N.V.; Ershova, A.N.; Martynenko, D.E.; Bubnov, M.G.; Amankwah, P.; Yanus, G.A.; Aleksakhina, S.N.; Tiurin, V.I.; Venina, A.R.; et al. KRAS, NRAS, BRAF, HER2 and MSI Status in a Large Consecutive Series of Colorectal Carcinomas. Int. J. Mol. Sci. 2023, 24, 4868. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes; revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef]

| Characteristic | Patients, n = 233 | |
|---|---|---|
| Age | Years, median (range) | 66 (20–92) |
| Gender | Male, n (%) | 176 (75.5%) |
| Female, n (%) | 57 (24.5%) | |
| Tumor location | Upper tract, n (%) | 21 (9.0%) |
| Bladder, n (%) | 147 (63.1%) | |
| ND, n (%) | 65 (27.9%) | |
| Locally advanced or metastatic disease (stage IIIB or IV) | Yes, n (%) | 99 (42.5%) |
| No, n (%) | 99 (42.5%) | |
| ND, n (%) | 35 (15.0%) |
| Patient | #Alternative Transcript | 5′ Partner Gene | 5′ Ensembl Transcript ID | 5′ Partner Exon | 5′ Insertion of the Intronic Sequence | 3′ Partner Gene | 3′ Ensembl Transcript ID | 3′ Partner Exon | 3′ Insertion of the Intronic Sequence | 3′ Gene Reading Frame | Number of Supporting NGS Reads | FGFR3 mRNA Expression Level α |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #77 | 1 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 4 | −1591_−1512 β | INFRAME | 192 | medium | |
| #147 | 1 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 5 | FRAMESHIFT | 6 | high | ||
| 2 | FGFR3 | ENST00000440486 | 17 | +1_+49 | TACC3 | ENST00000313288 | 5 | −1535_−1481 β | INFRAME | 43 | ||
| #58 | 1 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 8 | INFRAME | 186 | medium | ||
| #211 | 1 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 11 | INFRAME | 20,387 | overexpression | ||
| #30 | 1 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 11 | INFRAME | 1 γ | medium | ||
| #52 | 1 | FGFR3 | ENST00000440486 | 17 | +1_+3 | TACC3 | ENST00000313288 | 11 | −742_? δ | ? | 2247 | overexpression |
| 2 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 11 | INFRAME | 722 | ||||
| #9 | 1 | FGFR3 | ENST00000440486 | 17 | +1_+32 | TACC3 | ENST00000313288 | 12 | −39_−1 | INFRAME | 129 | medium |
| 2 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 12 | FRAMESHIFT | 2 | ||||
| #2 | 1 | FGFR3 | ENST00000440486 | 17 | TACC3 | ENST00000313288 | 14 | INFRAME | 48 | medium | ||
| 2 | FGFR3 | ENST00000440486 | 17 | +1_+25 | TACC3 | ENST00000313288 | 14 | −2987_? δ | ? | 5 | ||
| #106 | 1 | FGFR3 | ENST00000440486 | 17 | ADD1 | ENST00000683351 | 3 | INFRAME | 5306 | high | ||
| #130 | 1 | FGFR3 | ENST00000440486 | 17 | SMIM14 | ENST00000295958 | 3 | INFRAME | 4411 | overexpression | ||
| #6 | 1 | FGFR3 | ENST00000440486 | 17 | UACA | ENST00000322954 | 14 | INFRAME | 70 | high |
| HER2 Gene Amplification | HER2 mRNA Expression Level α | Total | ||
|---|---|---|---|---|
| Low or Medium | High | Overexpression | ||
| High-level (CN ≥ 10) | 0 | 1 (12.5%) | 7 (87.5%) | 8 |
| Medium-level (5 ≤ CN < 10) | 10 (58.8%) | 6 (35.3%) | 1 (5.9%) | 17 |
| Low-level (3 ≤ CN < 5) | 31 (68.8%) | 12 (26.7%) | 2 (4.4%) | 45 |
| No amplification (CN ≤ 3) | 123 (80.4%) | 23 (15.0%) | 7 (4.6%) | 153 |
| ND | 7 (70.0%) | 1 (10.0%) | 2 (20.0%) | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitiushkina, N.V.; Tiurin, V.I.; Anuskina, A.A.; Bordovskaya, N.A.; Nalivalkina, E.A.; Terina, D.M.; Berkut, M.V.; Shestakova, A.D.; Syomina, M.V.; Kuligina, E.S.; et al. Use of 3′ Rapid Amplification of cDNA Ends (3′ RACE)-Based Targeted RNA Sequencing for Profiling of Druggable Genetic Alterations in Urothelial Carcinomas. Int. J. Mol. Sci. 2024, 25, 12126. https://doi.org/10.3390/ijms252212126
Mitiushkina NV, Tiurin VI, Anuskina AA, Bordovskaya NA, Nalivalkina EA, Terina DM, Berkut MV, Shestakova AD, Syomina MV, Kuligina ES, et al. Use of 3′ Rapid Amplification of cDNA Ends (3′ RACE)-Based Targeted RNA Sequencing for Profiling of Druggable Genetic Alterations in Urothelial Carcinomas. International Journal of Molecular Sciences. 2024; 25(22):12126. https://doi.org/10.3390/ijms252212126
Chicago/Turabian StyleMitiushkina, Natalia V., Vladislav I. Tiurin, Aleksandra A. Anuskina, Natalia A. Bordovskaya, Ekaterina A. Nalivalkina, Darya M. Terina, Mariya V. Berkut, Anna D. Shestakova, Maria V. Syomina, Ekaterina Sh. Kuligina, and et al. 2024. "Use of 3′ Rapid Amplification of cDNA Ends (3′ RACE)-Based Targeted RNA Sequencing for Profiling of Druggable Genetic Alterations in Urothelial Carcinomas" International Journal of Molecular Sciences 25, no. 22: 12126. https://doi.org/10.3390/ijms252212126
APA StyleMitiushkina, N. V., Tiurin, V. I., Anuskina, A. A., Bordovskaya, N. A., Nalivalkina, E. A., Terina, D. M., Berkut, M. V., Shestakova, A. D., Syomina, M. V., Kuligina, E. S., Togo, A. V., & Imyanitov, E. N. (2024). Use of 3′ Rapid Amplification of cDNA Ends (3′ RACE)-Based Targeted RNA Sequencing for Profiling of Druggable Genetic Alterations in Urothelial Carcinomas. International Journal of Molecular Sciences, 25(22), 12126. https://doi.org/10.3390/ijms252212126






