Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing
Abstract
1. Introduction
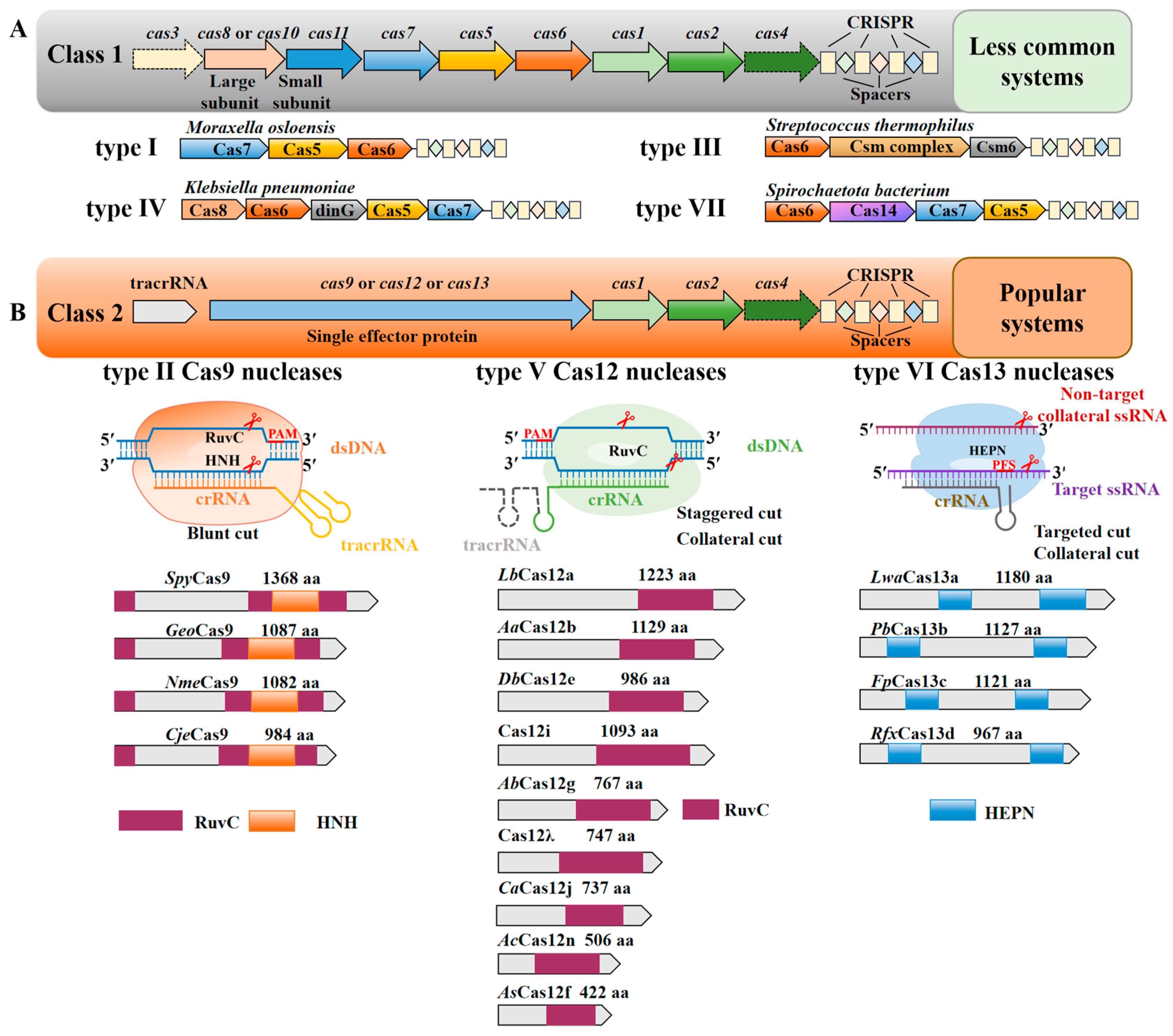
2. Classification of the CRISPR-Cas System
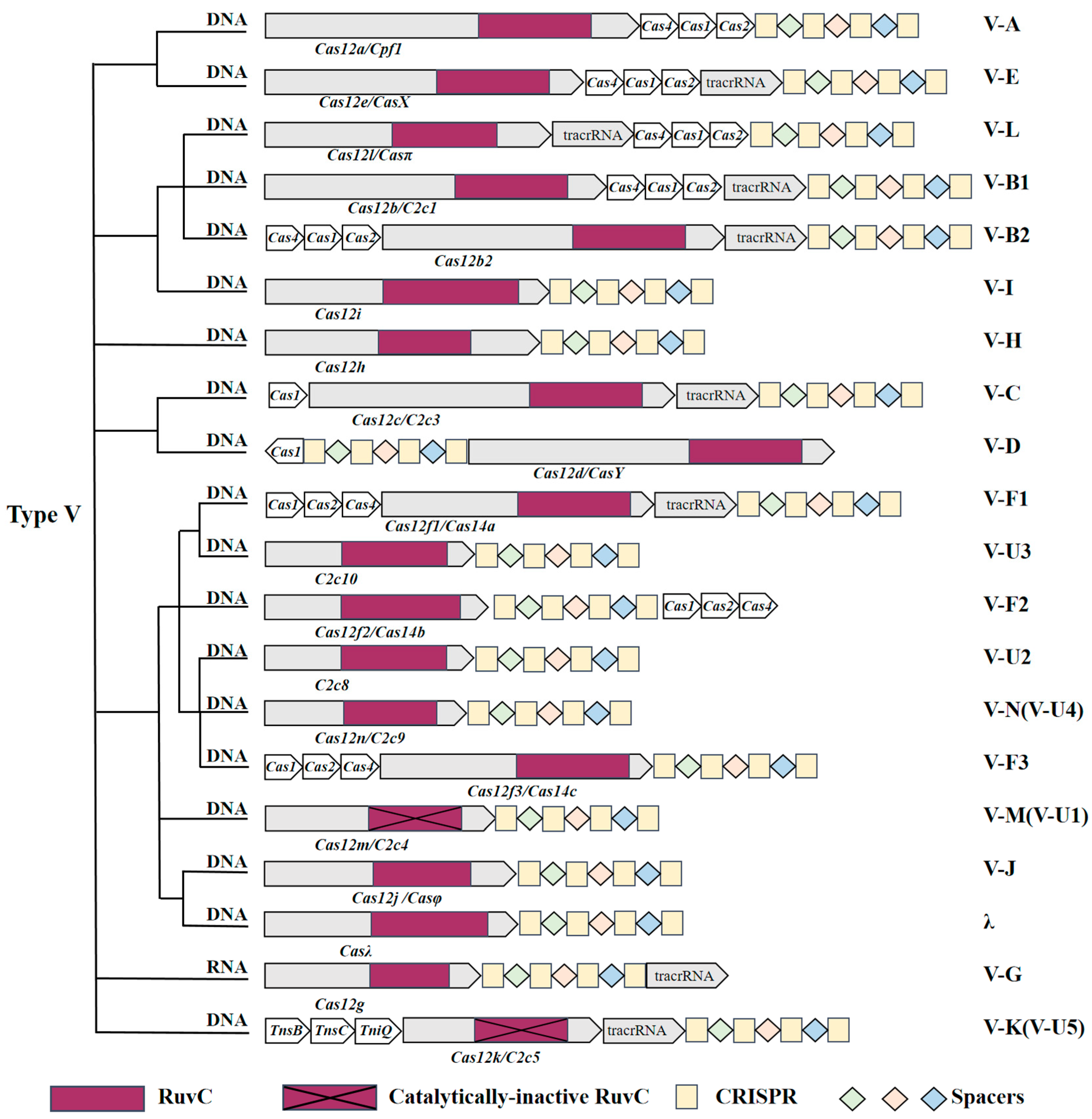
3. Characteristics of the Type V CRISPR-Cas System
| Type | Cas12 Protein | Size (aa) | PAM (5′-3′) | Processing precrRNA | tracrRNA | Target | Cleavage Activity | Classic Origin | References |
|---|---|---|---|---|---|---|---|---|---|
| V-A | Cas12a/Cpf1 | 1223 | TTTV | yes | no | dsDNA, ssDNA | cleavage | Lachnospiraceae bacterium | [33] |
| V-B | Cas12b/C2c1 | 1129 | TTN | no | yes | dsDNA, ssDNA | cleavage | Alicyclobacillus acidoterrestris | [34] |
| V-C | Cas12c/C2c3 | 1218 | TN | no | yes | dsDNA, ssDNA | no cleavage | uncultured archaeon | [35] |
| V-D | Cas12d/CasY | 1200 | TR | no | scoutRNA | dsDNA | cleavage | hot springs metagenome | [36] |
| V-E | Cas12e/CasX | 986 | TTCN | no | yes | dsDNA | cleavage | Deltaproteobacteria bacterium | [37] |
| V-F | Cas12f/Cas14 | 529 | TTN | no | yes | dsDNA, ssDNA | cleavage | uncultured archaeon | [38] |
| V-G | Cas12g | 767 | No PAM | no | yes | ssRNA | cleavage | Acidobacteriota bacterium | [12] |
| V-H | Cas12h | 871 | RTR | yes | no | dsDNA | cleavage | hypersaline lake sedimentmetagenome | [27] |
| V-I | Cas12i | 1093 | TTN | yes | no | dsDNA | cleavage | Metagenomic database | [32] |
| V-J | Cas12j/Casφ | 737 | TBN | yes | no | dsDNA | cleavage | Caudoviricetes | [13] |
| V-K (V-U5) | Cas12k/C2c5 | 639 | GTN | no | yes | dsDNA | no cleavage | Scytonema hofmannii | [39] |
| V-L | Cas12l/Casπ | 860 | CCN | no | yes | dsDNA | cleavage | Armatimonadetes bacterium | [40] |
| V-M (V-U1) | Cas12m/C2c4 | 607 | TTN | yes | no | dsDNA | no cleavage | Gordonia otitidis | [41] |
| V-N (V-U4) | Cas12n/C2c9 | 506 | AAN | no | yes | dsDNA | cleavage | Actinomadura craniellae | [15] |
| V-U2 | C2c8 | - | - | yes | no | dsDNA | cleavage | Cyanothece sp. Pcc sso | [27] |
| V-U3 | Cas12f1/C2c10 | 422 | TTN | no | yes | dsDNA | cleavage | Acidibacillus sulfuroxidans | [42] |
| - | Casλ | 747 | TTR | yes | no | dsDNA | cleavage | bacteriophage | [14] |
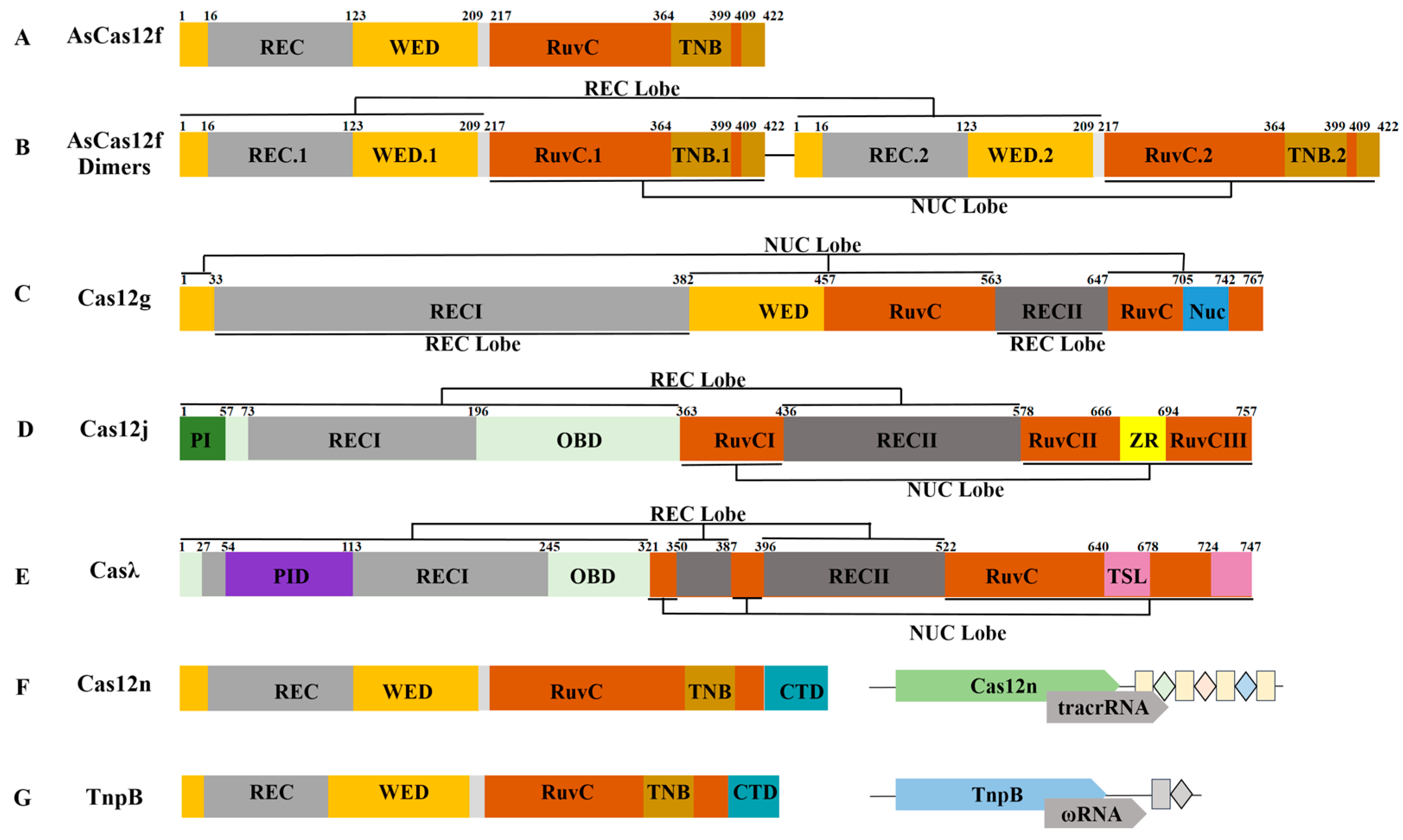
4. Structural Characteristics and Mechanism of the Miniature Cas12 Protein
4.1. Cas12f
4.2. Cas12g
4.3. Cas12j
4.4. Casλ
4.5. Cas12n
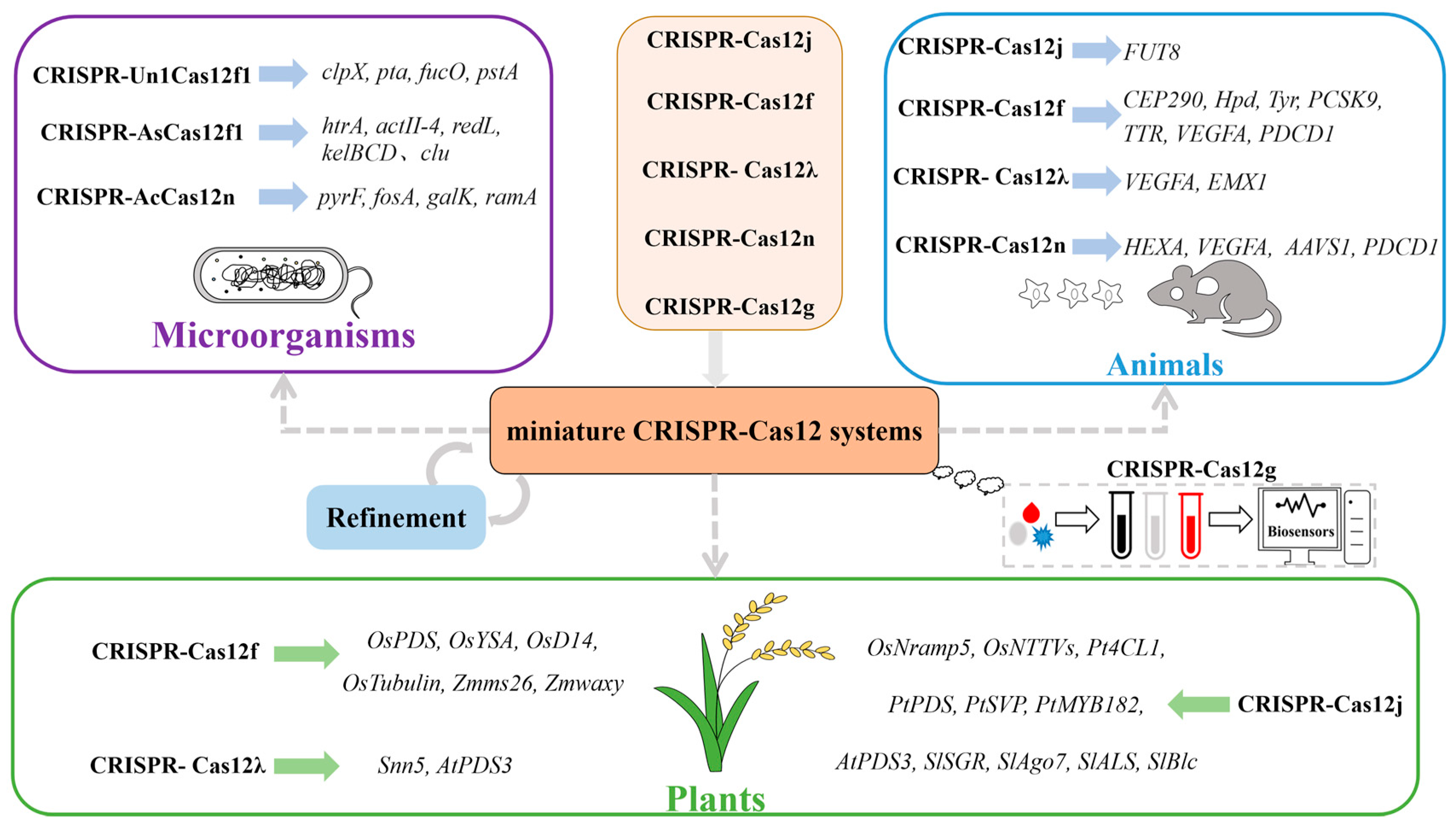
5. Application of the Miniature CRISPR-Cas12 System
5.1. Genome Editing of Microorganisms
5.2. Genome Editing of Humans and Animals
5.3. Genome Editing of Plants
6. Common Strategies to Improve the Editing Efficiency of Miniature CRISPR-Cas12 Systems
6.1. Optimization of gRNA Design
| Optimization Method | Gene Editing System | Processing | Efficiency | Activity | Species | References |
|---|---|---|---|---|---|---|
| gRNA engineering | Un1Cas12f1_ge4.1 | ge4.1 (MS2, MS3, MS4, MS5) | 867-fold | <1%→15.2% ± 12.0% | HEK293T | [58] |
| SpaCas12f1-gRNA_MS13 | MS13 (M10, AAGG, MS8) | Improvement | <1%→<21.95% | HEK293T | [84] | |
| CWCas12f1-TaRGET | TaRGET | 20-fold | <1%→20% | HEK293T | [83] | |
| Al2Cas12f1-tR3 | tR1-Stem1 truncation | 16-fold | 0.5%→8.2% | HEK293T | [85] | |
| Cas12Pro | sgRNA_v6 | 2-fold | 80% | HEK293T | [15] | |
| Cas12j2-TACG | crRNA-TACG | 2-fold | ~40% | Oryza sativa | [87] | |
| Cas12j2-STU.v2 | STU + crRNA-TACG | 5-fold | 78.80% | Oryza sativa | [87] | |
| Cas protein engineering | nCasΦ, vCasΦ | E159A, S160A, S164A, D167A, E168A, Δ155–176 (GSSG) | 20-fold | - | PCR product | [48] |
| nCas12j2 | E159A, S160A, S164A, D167A, E168A | Improvement | 50.00% | Oryza sativa | [62] | |
| Cas12j2-STU.v4-S511 | Npu split-S511 | Improvement | 6.5%→7.1% | Oryza sativa | [87] | |
| enRhCas12f1 | L270R, Rh-sg1.1 | 1.7-fold | 23.3 ± 26.8% | HEK293T | [60] | |
| enOsCas12f1 | T132R, D52R Os-sg2.6 | 3.9-fold | 54.7 ± 29.8% | HEK293T | [60] | |
| AsCas12f1-v5.1 | N70Q, K103R, A104R, S118A, D364R, sgRNA_T1 | 1.5–13.5-fold | - | HEK293T | [88] | |
| enAsCas12f | D196K, N199K, G276R, N328G, D364R, sgRNA-v2 | 11.3-fold | 69.80% | HEK293T | [46] | |
| AsCas12f-YHAM | F48Y, S188H, V232A, E316M, sgRNA_ΔS3-5_v7 | 12.6-fold | 3.0%→40.8% | HEK293T | [42] | |
| AsCas12f-HKRA | I123H, D195K, D208R, V232A, sgRNA_ΔS3-5_v7 | 13.9-fold | 3.0%→44.7% | HEK293T | [42] | |
| AsCas12f-YHAM | F48Y, S188H, V232A, E316M, sgRNA_ΔS3-5_v7 | Improvement | 4.0%, 0%, 25.0% | Oryza sativa (OsPDS, OsYSA, OsD14) | [63] | |
| AsCas12f-HKRA | I123H, D195K, D208R, V232A, sgRNA_ΔS3-5_v7 | Improvement | 27%, 24%, 53% | Oryza sativa (OsPDS, OsYSA, OsD14) | [63] | |
| Temperaturre treatment | Casλ | 23 °C, 28 °C, 32 °C | Improvement | 0%, ~2%, ~15% | Arabidopsis thaliana, Triticum aestivum | [14] |
| SpCas12f | 45 °C | Improvement | 20–50%, 1–1.5% | Zea mays L. (Zmms26, Zmwaxy) | [89] | |
| SpCas12f | 30 °C | Improvement | 28.8%, 55.6% | Oryza sativa (Tub-1, Tub-2) | [64] | |
| AsCas12f1 | 42, 27, 17 °C | Improvement | 1%, 46%, 86% | Escherichia coli (Double Target) | [90] | |
| AsCas12f1 | 37, 17 °C | Improvement | 3%, 35% | Escherichia coli (Triple Target) | [90] | |
| Other | Un1Cas12f1 | pOsU3 | 3-fold | 0.16–1.14%→0.34–3.70% | Oryza sativa | [78] |
| Cas12j2 | pAtU3 | Improvement | 0.29–2.66% | Solanum lycopersicum L. | [13] | |
| Cas12j2 | HH-HDV, Pol II | Improvement | 0–40%→15–50% | Arabidopsis thaliana, Oryza sativa | [62] | |
| eCWCas12f-VPR | ge4.1; D171R, T175R, E179A; D171R, T175R, K358R, E556R; NLS; FUS-IDR | 32.48-fold | - | HEK293T | [91] | |
| miniCRa | sgRNA (20nt), D143R, T147R, T203R | 4-fold | - | HEK293T, U2OS | [92] | |
| SminiCRa (Sso7d-dUn1Cas12f1-3R-VPR-C) | Sso7d, miniCRa | 29.65-fold | - | HEK293T, U2OS | [92] | |
| miniCRi | dUn1Cas12f1-3R-KRAB | improvement | - | HEK293T | [92] | |
| SminiCRi (Sso7d-dUn1Cas12f1-3R-KRAB) | Sso7d, miniCRi | improvement | - | HEK293T, Huh-7 | [92] | |
| STUminiABE | Sso7d, D143R, T147R, K330R, E528R | 4.4-fold | <10%→54% | HEK293T | [93] | |
| STUminiCBE | Sso7d | 3.5-fold | <10%→45% | HEK293T, Huh-7 | [93] |
6.2. Cas Protein Engineering
6.3. Selection of Treatment Temperatures
6.4. Other Optimization Methods
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Han, Y.; Jia, Z.; Xu, K.; Li, Y.; Lu, S.; Guan, L. CRISPR-Cpf1 system and its applications in animal genome editing. Mol. Genet. Genom. 2024, 299, 75. [Google Scholar] [CrossRef]
- Jain, D.; Kalia, A.; Sharma, S.; Manchanda, P. Genome editing tools based improved applications in macrofungi. Mol. Biol. Rep. 2024, 51, 873. [Google Scholar] [CrossRef]
- Pandey, S.; Divakar, S.; Singh, A. Genome editing prospects for heat stress tolerance in cereal crops. Plant Physiol. Biochem. 2024, 215, 108989. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Zhang, L.; Li, G.; Zhang, Y.; Cheng, Y.; Roberts, N.; Glenn, S.E.; DeZwaan-McCabe, D.; Rube, H.T.; Manthey, J.; Coleman, G.; et al. Boosting genome editing efficiency in human cells and plants with novel LbCas12a variants. Genome Biol. 2023, 24, 102. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfo, I.; et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar] [CrossRef]
- Xin, C.; Yin, J.; Yuan, S.; Ou, L.; Liu, M.; Zhang, W.; Hu, J. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption. Nat. Commun. 2022, 13, 5623. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H.; Xiao, R.; Han, R.; Chang, L. Cryo-EM structure of the RNA-guided ribonuclease Cas12g. Nat. Chem. Biol. 2021, 17, 387–393. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Al-Shayeb, B.; Skopintsev, P.; Soczek, K.M.; Stahl, E.C.; Li, Z.; Groover, E.; Smock, D.; Eggers, A.R.; Pausch, P.; Cress, B.F.; et al. Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell 2022, 185, 4574–4586. [Google Scholar] [CrossRef]
- Chen, W.; Ma, J.; Wu, Z.; Wang, Z.; Zhang, H.; Fu, W.; Pan, D.; Shi, J.; Ji, Q. Cas12n nucleases, early evolutionary intermediates of type V CRISPR, comprise a distinct family of miniature genome editors. Mol. Cell 2023, 83, 2768–2780. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gong, L.; Yu, H.; Li, M.; An, Q.; Liu, Z.; Fan, S.; Yang, C.; Zhao, D.; Han, J.; et al. Engineered minimal type I CRISPR-Cas system for transcriptional activation and base editing in human cells. Nat. Commun. 2024, 15, 7277. [Google Scholar] [CrossRef]
- You, L.; Ma, J.; Wang, J.; Artamonova, D.; Wang, M.; Liu, L.; Xiang, H.; Severinov, K.; Zhang, X.; Wang, Y. Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 2019, 176, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Camara-Wilpert, S.; Russel, J.; Wandera, K.G.; Cepaite, R.; Ares-Arroyo, M.; Gomes, J.V.; Englert, F.; Kuehn, J.A.; Gloor, S.; et al. Type IV-A3 CRISPR-Cas systems drive inter-plasmid conflicts by acquiring spacers in trans. Cell Host Microbe 2024, 32, 875–886. [Google Scholar] [CrossRef]
- Altae-Tran, H.; Kannan, S.; Suberski, A.J.; Mears, K.S.; Demircioglu, F.E.; Moeller, L.; Kocalar, S.; Oshiro, R.; Makarova, K.S.; Macrae, R.K.; et al. Uncovering the functional diversity of rare CRISPR-Cas systems with deep terascale clustering. Science 2023, 382, eadi1910. [Google Scholar] [CrossRef]
- Yang, J.; Li, X.; He, Q.; Wang, X.; Tang, J.; Wang, T.; Zhang, Y.; Yu, F.; Zhang, S.; Liu, Z.; et al. Structural basis for the activity of the type VII CRISPR-Cas system. Nature 2024, 633, 465–472. [Google Scholar] [CrossRef]
- Carter, J.; Wiedenheft, B. SnapShot: CRISPR-RNA-guided adaptive immune systems. Cell 2015, 163, U260–U284. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Guo, M.; Wang, S.; Zhu, Y.; Wang, S.; Xiong, Z.; Yang, J.; Xu, Z.; Huang, Z. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 2017, 546, 436–439. [Google Scholar] [CrossRef]
- Swarts, D.C.; Jinek, M. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell 2019, 73, 589–600. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; Derby, M.D.L.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333. [Google Scholar] [CrossRef]
- Aquino-Jarquin, G. Genome and transcriptome engineering by compact and versatile CRISPR-Cas systems. Drug Discov. Today 2023, 28, 103793. [Google Scholar] [CrossRef]
- Yan, W.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88–91. [Google Scholar] [CrossRef]
- Wu, W.; Mohanraju, P.; Liao, C.; Adiego-Pérez, B.; Creutzburg, S.C.A.; Makarova, K.S.; Keessen, K.; Lindeboom, T.A.; Khan, T.S.; Prinsen, S.; et al. The miniature CRISPR-Cas12m effector binds DNA to block transcription. Mol. Cell 2022, 82, 4487–4502. [Google Scholar] [CrossRef]
- Saito, M.; Ladha, A.; Strecker, J.; Faure, G.; Neumann, E.; Altae-Tran, H.; Macrae, R.K.; Zhang, F. Dual modes of CRISPR-associated transposon homing. Cell 2021, 184, 2441–2453. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. CRISPR tools found in thousands of viruses could boost gene editing. Nature 2022, 612, 21. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, D.; Lü, P.; Ma, S.; Chen, K. Research progress on nucleic acid detection and genome editing of CRISPR/Cas12 system. Mol. Biol. Rep. 2023, 50, 3723–3738. [Google Scholar] [CrossRef] [PubMed]
- McGaw, C.; Garrity, A.J.; Munoz, G.Z.; Haswell, J.R.; Sengupta, S.; Keston-Smith, E.; Hunnewell, P.; Ornstein, A.; Bose, M.; Wessells, Q.; et al. Engineered Cas12i2 is a versatile high-efficiency platform for therapeutic genome editing. Nat. Commun. 2022, 13, 2833. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.; Chen, K.; Shi, H.; Stahl, E.C.; Adler, B.; Trinidad, M.; Liu, J.; Zhou, K.; Ye, J.; Doudna, J.A. Improved genome editing by an engineered CRISPR-Cas12a. Nucleic Acids Res. 2022, 50, 12689–12701. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.; Ren, Q.; Pan, C.; He, Y.; Zhang, Y.; Liu, S.; Zhong, Z.; Wang, J.; Malzahn, A.A.; Wu, J.; et al. CRISPR-Cas12b enables efficient plant genome engineering. Nat. Plants. 2020, 6, 202–208. [Google Scholar] [CrossRef]
- Huang, C.; Adler, B.A.; Doudna, J.A. A naturally DNase-free CRISPR-Cas12c enzyme silences gene expression. Mol. Cell 2022, 82, 2148–2160.e2144. [Google Scholar] [CrossRef]
- Chen, L.; Al-Shayeb, B.; Meheust, R.; Li, W.; Doudna, J.A.; Banfield, J.F. Candidate phyla radiation roizmanbacteria from hot springs have novel and unexpectedly abundant CRISPR-Cas systems. Front. Microbiol. 2019, 10, 928. [Google Scholar] [CrossRef]
- Tsuchida, C.A.; Zhang, S.; Doost, M.S.; Zhao, Y.; Wang, J.; O’Brien, E.; Fang, H.; Li, C.; Li, D.; Hai, Z.; et al. Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity. Mol. Cell 2022, 82, 1199–1209. [Google Scholar] [CrossRef]
- Takeda, S.N.; Nakagawa, R.; Okazaki, S.; Hirano, H.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Nishimasu, H.; Nureki, O. Structure of the miniature type V-F CRISPR-Cas effector enzyme. Mol. Cell 2021, 81, 558–570. [Google Scholar] [CrossRef]
- Schmitz, M.; Querques, I.; Oberli, S.; Chanez, C.; Jinek, M. Structural basis for the assembly of the type V CRISPR-associated transposon complex. Cell 2022, 185, 4999–5010. [Google Scholar] [CrossRef]
- Sun, A.; Li, C.; Chen, Z.; Zhang, S.; Li, D.; Yang, Y.; Li, L.; Zhao, Y.; Wang, K.; Li, Z.; et al. The compact Casπ (Cas12l) ‘bracelet’ provides a unique structural platform for DNA manipulation. Cell Res. 2023, 33, 229–244. [Google Scholar] [CrossRef]
- Bigelyte, G.; Duchovska, B.; Zedaveinyte, R.; Sasnauskas, G.; Sinkunas, T.; Dalgediene, I.; Tamulaitiene, G.; Silanskas, A.; Kazlauskas, D.; Valancauskas, L.; et al. Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing. Nucleic Acids Res. 2024, 52, 3234–3248. [Google Scholar] [CrossRef] [PubMed]
- Hino, T.; Omura, S.N.; Nakagawa, R.; Togashi, T.; Takeda, S.N.; Hiramoto, T.; Tasaka, S.; Hirano, H.; Tokuyama, T.; Uosaki, H.; et al. An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis. Cell 2023, 186, 4920–4935. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, Y.; Yu, H.; Pan, D.; Wang, Y.; Wang, Y.; Li, F.; Liu, C.; Nan, H.; Chen, W.; et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 2021, 17, 1132–1138. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef]
- Su, M.; Li, F.; Wang, Y.; Gao, Y.; Lan, W.; Shao, Z.; Zhu, C.; Tang, N.; Gan, J.; Wu, Z.; et al. Molecular basis and engineering of miniature Cas12f with C-rich PAM specificity. Nat. Chem. Biol. 2024, 20, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Liu, C.; Zou, S.; Lyu, R.; Yang, B.; Yan, H.; Zhao, M.; Tang, W. An engineered hypercompact CRISPR-Cas12f system with boosted gene-editing activity. Nat. Chem. Biol. 2023, 19, 1384–1393. [Google Scholar] [CrossRef]
- Liu, M.; Li, Z.; Chen, J.; Lin, J.; Lu, Q.; Ye, Y.; Zhang, H.; Zhang, B.; Ouyang, S. Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage. PLoS Genet. 2023, 19, e1010930. [Google Scholar] [CrossRef]
- Pausch, P.; Soczek, K.M.; Herbst, D.A.; Tsuchida, C.A.; Al-Shayeb, B.; Banfield, J.F.; Nogales, E.; Doudna, J.A. DNA interference states of the hypercompact CRISPR-Casφ effector. Nat. Struct. Mol. Biol. 2021, 28, 652–661. [Google Scholar] [CrossRef]
- Carabias, A.; Fuglsang, A.; Temperini, P.; Pape, T.; Sofos, N.; Stella, S.; Erlendsson, S.; Montoya, G. Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3. Nat. Commun. 2021, 12, 4476. [Google Scholar] [CrossRef]
- Nguyen, G.T.; Dhingra, Y.; Sashital, D.G. Miniature CRISPR-Cas12 endonucleases-programmed DNA targeting in a smaller package. Curr. Opin. Struct. Biol. 2022, 77, 102466. [Google Scholar] [CrossRef]
- Duan, Z.; Zhang, X.; Zhang, J.; Li, S.; Liu, R.; Sun, J.; Zhao, Q.; Jia, N.; Jia, N.; Zhu, J. Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05. Cell Discov. 2023, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Al-Shayeb, B. Bogs, Bugs, Borgs, and Bacteriophages: Metagenomic and Biochemical Insights into the Enigmatic World of Extrachromosomal Genetic Elements. Ph.D. Thesis, University of California, Berkeley, CA, USA, 2022. [Google Scholar]
- Li, W.; Jiang, X.; Wang, W.; Hou, L.; Cai, R.; Li, Y.; Gu, Q.; Chuai, G.; Chen, Q.; Ma, P.; et al. Discovering CRISPR-Cas system with self-processing pre-crRNA capability by foundation models. Nat. Commun. 2024, 15, 10024. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Ji, Q. Miniature CRISPR-Cas12 systems: Mechanisms, engineering, and genome editing applications. ACS Chem. Biol. 2024, 19, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Okano, K.; Sato, Y.; Hizume, T.; Honda, K. Genome editing by miniature CRISPR/Cas12f1 enzyme in Escherichia coli. J. Biosci. Bioeng. 2021, 132, 120–124. [Google Scholar] [CrossRef]
- Wang, Y.; Sang, S.; Zhang, X.; Tao, H.; Guan, Q.; Liu, C. Efficient genome editing by a miniature CRISPR-AsCas12f1 nuclease in Bacillus anthracis. Front. Bioeng. Biotechnol. 2022, 9, 825493. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.; Xu, J.; Huang, X.; Zimin, A.; Wang, W.; Lu, Y. Low-toxicity and high-efficiency Streptomyces genome editing tool based on the miniature type V-F CRISPR/Cas nuclease AsCas12f1. J. Agric. Food. Chem. 2024, 72, 5358–5367. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lee, J.M.; Moon, S.B.; Chin, H.J.; Park, S.; Lim, Y.; Kim, D.; Koo, T.; Ko, J.H.; Kim, Y.S. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 2022, 40, 94–102. [Google Scholar] [CrossRef]
- Guo, R.; Li, Z.; Li, G.; Zhang, H.; Zhang, C.; Huo, X.; Zhang, X.; Yang, X.; Yang, R.; Liu, Y.; et al. In vivo treatment of tyrosinaemia with hypercompact Cas12f1. Cell Discov. 2023, 9, 73. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, H.; Li, G.; Wang, Z.; Kong, X.; Wang, L.; Xue, M.; Zhang, W.; Wang, Y.; Lin, J.; et al. Engineered CRISPR-OsCas12f1 and RhCas12f1 with robust activities and expanded target range for genome editing. Nat. Commun. 2023, 14, 2046. [Google Scholar] [CrossRef]
- Li, Z.; Zhong, Z.; Wu, Z.; Pausch, P.; Al-Shayeb, B.; Amerasekera, J.; Doudna, J.A.; Jacobsen, S.E. Genome editing in plants using the compact editor CasΦ. Proc. Natl. Acad. Sci. USA 2023, 120, e2216822120. [Google Scholar] [CrossRef]
- Liu, S.; Sretenovic, S.; Fan, T.; Cheng, Y.; Li, G.; Qi, A.; Tang, X.; Xu, Y.; Guo, W.; Zhong, Z.; et al. Hypercompact CRISPR-Cas12j2 (CasΦ) enables genome editing, gene activation, and epigenome editing in plants. Plant Commun. 2022, 3, 100453. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhang, Y.; He, S.; Li, S.; Luo, L.; Zhou, Y.; Tan, J.; Wan, J. Efficient genome editing in rice with miniature Cas12f variants. aBIOTECH 2024, 5, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Sukegawa, S.; Nureki, O.; Toki, S.; Saika, H. Genome editing in rice mediated by miniature size Cas nuclease SpCas12f. Front. Genome Ed. 2023, 5, 1138843. [Google Scholar] [CrossRef]
- Senthilnathan, R.; Ilangovan, I.; Kunale, M.; Easwaran, N.; Ramamoorthy, S.; Veeramuthu, A.; Muthukaliannan, G.K. An update on CRISPR-Cas12 as a versatile tool in genome editing. Mol. Biol. Rep. 2023, 50, 2865–2881. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef]
- Madigan, V.; Zhang, F.; Dahlman, J.E. Drug delivery systems for CRISPR-based genome editors. Nat. Rev. Drug Discov. 2023, 22, 875–894. [Google Scholar] [CrossRef]
- Stevanovic, M.; Piotter, E.; McClements, M.E.; MacLaren, R.E. CRISPR systems suitable for single AAV vector delivery. Curr. Gene Ther. 2022, 22, 1–14. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, F.; Gao, G. CRISPR-based therapeutic genome editing: Strategies and in vivo delivery by AAV vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Kong, X.; Li, T.; Yang, H. AAV-mediated gene therapies by miniature gene editing tools. Sci. China Life Sci. 2024. [Google Scholar] [CrossRef]
- Sharrar, A.; Meacham, Z.; Staples-Ager, J.; Arake, L.A.; Rabuka, D.; Collingwood, T.; Schelle, M. Viral delivery of compact CRISPR-Cas12f for gene editing applications. CRISPR J. 2024, 7, 150–155. [Google Scholar] [CrossRef]
- Li, Z.; Guo, R.; Sun, X.; Li, G.; Shao, Z.; Huo, X.; Yang, R.; Liu, X.; Cao, X.; Zhang, H.; et al. Engineering a transposon-associated TnpB-ωRNA system for efficient gene editing and phenotypic correction of a tyrosinaemia mouse model. Nat. Commun. 2024, 15, 831. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, T.; Liu, J.; Yang, Y.; Wang, Z.; Wang, Y.; Wang, T.; Li, M.; Li, M.; Lu, D.; et al. A highly specific CRISPR-Cas12j nuclease enables allele-specific genome editing. Sci. Adv. 2023, 9, eabo6405. [Google Scholar] [CrossRef] [PubMed]
- Cardi, T.; Murovec, J.; Bakhsh, A.; Boniecka, J.; Bruegmann, T.; Bull, S.E.; Eeckhaut, T.; Fladung, M.; Galovic, V.; Linkiewicz, A.; et al. CRISPR/Cas-mediated plant genome editing outstanding challenges a decade after implementation. Trends Plant Sci. 2023, 28, 1144–1165. [Google Scholar] [CrossRef]
- Li, B.; Sun, C.; Li, J.; Gao, C. Targeted genome-modification tools and their advanced applications in crop breeding. Nat. Rev. Genet. 2024, 25, 603–622. [Google Scholar] [CrossRef]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants. 2021, 7, 1166–1187. [Google Scholar] [CrossRef]
- Cai, Q.; Guo, D.; Cao, Y.; Li, Y.; Ma, R.; Liu, W. Application of CRISPR/CasΦ2 system for genome editing in plants. Int. J. Mol. Sci. 2022, 23, 5755. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Eid, A.; Zhang, R.; Cheng, Y.; Liu, A.; Chen, Y.; Chen, P.; Zhang, Y.; Qi, Y. Genome editing in rice and tomato with a small Un1Cas12f1 nuclease. Plant Genome 2024, 17, e20465. [Google Scholar] [CrossRef]
- Zhao, S.; Han, X.; Zhu, Y.; Han, Y.; Liu, H.; Chen, Z.; Li, H.; Wang, D.; Tian, C.; Yuan, Y.; et al. CRISPR/CasΦ2-mediated gene editing in wheat and rye. J. Integr. Plant Biol. 2024, 66, 638–641. [Google Scholar] [CrossRef]
- Liu, S.; Tang, X.; Qi, Y.; Zhang, Y. Optimizing rice genomics: Employing the hypercompact Cas12j2 system for targeted transcriptional regulation and epigenome modification. Methods Mol. Biol. 2024, 2844, 133–143. [Google Scholar] [CrossRef]
- Kawamata, M.; Suzuki, H.I.; Kimura, R.; Suzuki, A. Optimization of Cas9 activity through the addition of cytosine extensions to single-guide RNAs. Nat. Biomed. Eng. 2023, 7, 672–691. [Google Scholar] [CrossRef]
- Riesenberg, S.; Helmbrecht, N.; Kanis, P.; Maricic, T.; Paeaebo, S. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 2022, 13, 489. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Chung, Y.; Lee, Y.; Jeong, D.; Park, K.H.; Chin, H.J.; Lee, J.M.; Park, S.; Ko, S.; Ko, J.H.; et al. Hypercompact adenine base editors based on a Cas12f variant guided by engineered RNA. Nat. Chem. Biol. 2023, 19, 389. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Pan, D.; Yu, H.; Zhang, Y.; Chen, W.; Li, F.; Wu, Z.; Ji, Q. Guide RNA engineering enables efficient CRISPR editing with a miniature Syntrophomonas palmitatica Cas12f1 nuclease. Cell Rep. 2022, 40, 111418. [Google Scholar] [CrossRef]
- Sharrar, A.; de Tacca, L.A.; Collingwood, T.; Meacham, Z.; Rabuka, D.; Staples-Ager, J.; Schelle, M. Discovery and characterization of novel type V Cas12f nucleases with diverse protospacer adjacent motif preferences. CRISPR J. 2023, 6, 350–358. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Tang, N.; Wang, Z.; Pan, D.; Ji, Q. Characterization and engineering of a novel miniature Eubacterium siraeum CRISPR-Cas12f system. ACS Synth. Biol. 2024, 13, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, J.; Hu, Z.; Zhou, H.; Gao, Y.; Liu, Y.; Ji, Y.; Xu, G.; Guo, Y.; Zhang, Y.; et al. Engineer and split an efficient hypercompact CRISPR-CasΦ genome editor in plants. Plant Commun. 2024, 5, 100881. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, D.; Pan, D.; Yu, H.; Shi, J.; Ma, J.; Fu, W.; Wang, Z.; Zheng, Z.; Qu, Y.; et al. Structure and engineering of miniature Acidibacillus sulfuroxidans Cas12f1. Nat. Catal. 2023, 6, 695–709. [Google Scholar] [CrossRef]
- Bigelyte, G.; Young, J.K.; Karvelis, T.; Budre, K.; Zedaveinyte, R.; Djukanovic, V.; Van Ginkel, E.; Paulraj, S.; Gasior, S.; Jones, S.; et al. Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat. Commun. 2021, 12, 6191. [Google Scholar] [CrossRef]
- Lim, S.R.; Kim, H.J.; Lee, S.J. Efficient CRISPR-Cas12f1-mediated multiplex bacterial genome editing via low-temperature recovery. J. Microbiol. Biotechnol. 2024, 34, 1522–1529. [Google Scholar] [CrossRef]
- Oh, Y.; Gwon, L.W.; Lee, H.K.; Hur, J.K.; Park, K.H.; Kim, K.P.; Lee, S.H. Highly efficient and specific regulation of gene expression using enhanced CRISPR-Cas12f system. Gene Ther. 2024, 31, 358–365. [Google Scholar] [CrossRef]
- Wang, X.; Li, L.; Guo, L.; Feng, Y.; Du, Z.; Jiang, W.; Wu, X.; Zheng, J.; Xiao, X.; Zheng, H.; et al. Robust miniature Cas-based transcriptional modulation by engineering Un1Cas12f1 and tethering Sso7d. Mol. Ther. 2024, 32, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, L.; Mo, Q.; Du, Z.; Jiang, W.; Wu, X.; Zheng, J.; Xiao, X.; Sun, Y.; Ma, H. Engineering miniature CRISPR-Cas Un1Cas12f1 for efficient base editing. Mol. Ther. Nucleic Acids 2024, 35, 102201. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xie, W.; Bao, Z. Broadening the targetable space: Engineering and discovery of proteins. Trends Microbiol. 2024, 32, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, M.A.; Davletshin, A.I.; Karpov, D.S. Engineering Cas9: Next generation of genomic editors. Appl. Microbiol. Biotechnol. 2024, 108, 209. [Google Scholar] [CrossRef] [PubMed]
- Eggers, A.R.; Chen, K.; Soczek, K.M.; Tuck, O.T.; Doherty, E.E.; Xu, B.; Trinidad, M.I.; Thornton, B.W.; Yoon, P.H.; Doudna, J.A. Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9. Cell 2024, 187, 3249–3261. [Google Scholar] [CrossRef]
- Zhang, G.; Song, Z.; Huang, S.; Wang, Y.; Sun, J.; Qiao, L.; Li, G.; Feng, Y.; Han, W.; Tang, J.; et al. nCas9 engineering for improved target interaction presents an effective strategy to enhance base editing. Adv. Sci. 2024, 11, e2405426. [Google Scholar] [CrossRef]
- Banakar, R.; Schubert, M.; Kurgan, G.; Rai, K.M.; Beaudoin, S.F.; Collingwood, M.A.; Vakulskas, C.A.; Wang, K.; Zhang, F. Efficiency, specificity and temperature sensitivity of Cas9 and Cas12a RNPs for DNA-free genome editing in plants. Front. Genome Ed. 2021, 3, 760820. [Google Scholar] [CrossRef]
- Blomme, J.; Develtere, W.; Kose, A.; Ribera, J.A.; Brugmans, C.; Jaraba-Wallace, J.; Decaestecker, W.; Rombaut, D.; Baekelandt, A.; Fernandez, A.D.F.; et al. The heat is on: A simple method to increase genome editing efficiency in plants. BMC Plant Biol. 2022, 22, 142. [Google Scholar] [CrossRef]
- Illa-Berenguer, E.; LaFayette, P.R.; Parrott, W.A. Editing efficiencies with Cas9 orthologs, Cas12a endonucleases, and temperature in rice. Front. Genome Ed. 2023, 5, 1074641. [Google Scholar] [CrossRef]
- Xu, X.; Chemparathy, A.; Zeng, L.; Kempton, H.R.; Shang, S.; Nakamura, M.; Qi, L.S. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell 2021, 81, 4333–4345. [Google Scholar] [CrossRef]
- Long, T.; Zhou, S.; Huang, Z.; Li, G.; Zhong, Q.; Zhang, X.; Li, Y.; Chen, C.; Xia, L.; Wei, R.; et al. Innovative delivery system combining CRISPR-Cas12f for combatting antimicrobial resistance in gram-negative bacteria. ACS Synth. Biol. 2024, 13, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Badon, I.W.; Oh, Y.; Kim, H.J.; Lee, S.H. Recent application of CRISPR-Cas12 and OMEGA system for genome editing. Mol. Ther. 2024, 32, 32–43. [Google Scholar] [CrossRef]
- Gunitseva, N.; Evteeva, M.; Korzhenkov, A.; Patrushev, M. A new RNA-dependent Cas12g nuclease. Int. J. Mol. Sci. 2023, 24, 17105. [Google Scholar] [CrossRef]
- Hui, F.; Tang, X.; Li, B.; Alariqi, M.; Xu, Z.; Meng, Q.; Hu, Y.; Wang, G.; Zhang, Y.; Zhang, X.; et al. Robust CRISPR/Mb2Cas12a genome editing tools in cotton plants. iMeta 2024, 3, e209. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Y.; Dailey, M.; Qi, Y. Hs1Cas12a and Ev1Cas12a confer efficient genome editing in plants. Front. Genome Ed. 2023, 5, 1251903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chai, N.; Liu, T.; Zheng, Z.; Lin, Q.; Xie, X.; Wen, J.; Yang, Z.; Liu, Y.; Zhu, Q. The type V effectors for CRISPR/Cas-mediated genome engineering in plants. Biotechnol. Adv. 2024, 74, 108382. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, B. A review on CRISPR/Cas: A versatile tool for cancer screening, diagnosis, and clinic treatment. Funct. Integr. Genom. 2023, 23, 182. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Hu, X.; Weng, X.; Wang, H.; Yu, J.; Jiang, T.; Zou, L.; Zhou, X.; Lyu, Z.; Liu, J.; et al. Efficient, specific and direct detection of double-stranded DNA targets using Cas12f1 nucleases and engineered guide RNAs. Biosens. Bioelectron. 2024, 260, 116428. [Google Scholar] [CrossRef]
- Jiao, C.; Peeck, N.L.; Yu, J.; Ghaem Maghami, M.; Kono, S.; Collias, D.; Martinez Diaz, S.L.; Larose, R.; Beisel, C.L. TracrRNA reprogramming enables direct PAM-independent detection of RNA with diverse DNA-targeting Cas12 nucleases. Nat. Commun. 2024, 15, 5909. [Google Scholar] [CrossRef]
- Awan, M.J.A.; Amin, I.; Mansoor, S. Mini CRISPR-Cas12f1: A new genome editing tool. Trends Plant Sci. 2022, 27, 110–112. [Google Scholar] [CrossRef]
- Gong, Z.; Previtera, D.A.; Wang, Y.; Botella, J.R. Geminiviral-induced genome editing using miniature CRISPR/Cas12j (CasΦ) and Cas12f variants in plants. Plant Cell Rep. 2024, 43, 71. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zheng, Q.; Yao, R.; Wang, M. Lung-specific supramolecular nanoparticles for efficient delivery of therapeutic proteins and genome editing nucleases. Proc. Natl. Acad. Sci. USA 2024, 121, e2406654121. [Google Scholar] [CrossRef] [PubMed]

| Biological | Miniature Cas12 System | Gene | Efficiency | Species | References |
|---|---|---|---|---|---|
| Microorganisms | Un1Cas12f1 | clpX, pta, fucO, pstA | 63–100% | Escherichia coli | [55] |
| AsCas12f1 | htrA | 100.00% | Bacillus anthracis | [56] | |
| AsCas12f1 | (actII-4 + redL), (act + red) | 46.7%, 40% | Streptomyces hygroscopicus M145 | [57] | |
| AsCas12f1 | kelBCD, clu | 70%, 30% | Streptomyces hygroscopicus SIPI-054 | [57] | |
| AcCas12n | pyrF, fosA, galK, ramA | 99.97%, 99.84%, 99.71%, 99.85% | Klebsiella pneumoniae | [15] | |
| Animals | Un1Cas12f1 | CEP290 | 46.00% | HEK293T | [58] |
| Un1Cas12f1 | Hpd, Tyr | 20.00% | mouse zygotic embryos | [59] | |
| enRhCas12f1 | PCSK9, TTR, VEGFA | 23.3 ± 26.8% | HEK293T | [60] | |
| enOsCas12f1 | PCSK9, TTR, VEGFA | 54.7 ± 29.8% | HEK293T | [60] | |
| enOsCas12f1 | exon51 | 22.7 ± 9.2% | whole muscle | [60] | |
| AsCas12f1 | VEGFA, PDCD1 | 11.50% | HEK293, U-2 OS, and Huh-7 | [43] | |
| AsCas12f-HKRA | TTR | 66.30% | mouse liver | [42] | |
| CasΦ−2 (Cas12j) | EGFP | 33% | HEK293 | [13] | |
| Cas12j-SF05 | FUT8 | ~12% | CHO cell | [51] | |
| AcCas12n | 35 different human genomic sites | 2.2–73.7% | HEK293T | [15] | |
| Casλ | VEGFA, EMX1 | 20–50%, 1–1.5% | HEK293T | [14] | |
| Plants | CasΦ−2 (Cas12j) | AtPDS3 | 0.85% | Arabidopsis thaliana | [61] |
| vCas12j, nCas12j | AtPDS3 | 6.12%, 6.07% | Arabidopsis thaliana | [61] | |
| Casλ | AtPDS3 | 18% | Arabidopsis thaliana | [14] | |
| Casλ | Snn5 | <4% | Triticum aestivum L. | [14] | |
| Cas12j-SF05 | OsNramp5 | 20% | Oryza sativa | [51] | |
| Cas12j2 | Pt4CL1, PtPDS, PtSVP, PtMYB182 | 1.20% | Populus tomentosa | [62] | |
| Cas12j2 | OsNTTVs | 15–60% | Oryza sativa | [62] | |
| Cas12j2 | SlSGR, SlAgo7, SlALS, SlBlc | <15% | Solanum lycopersicum L. | [62] | |
| AsCas12f-HKRA | OsPDS, OsYSA, OsD14 | 27.1%, 24%, 53.1% | Oryza sativa | [63] | |
| SpCas12f | OsTubulin (Tub-1, Tub-2) | 28.8%, 55.6% | Oryza sativa | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xuan, Q.; Wang, J.; Nie, Y.; Fang, C.; Liang, W. Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing. Int. J. Mol. Sci. 2024, 25, 12686. https://doi.org/10.3390/ijms252312686
Xuan Q, Wang J, Nie Y, Fang C, Liang W. Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing. International Journal of Molecular Sciences. 2024; 25(23):12686. https://doi.org/10.3390/ijms252312686
Chicago/Turabian StyleXuan, Qiangbing, Junjie Wang, Yuanqing Nie, Chaowei Fang, and Weihong Liang. 2024. "Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing" International Journal of Molecular Sciences 25, no. 23: 12686. https://doi.org/10.3390/ijms252312686
APA StyleXuan, Q., Wang, J., Nie, Y., Fang, C., & Liang, W. (2024). Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing. International Journal of Molecular Sciences, 25(23), 12686. https://doi.org/10.3390/ijms252312686






