Molecular Characterization of Subdomain Specification of Cochlear Duct Based on Foxg1 and Gata3
Abstract
1. Introduction
2. Results
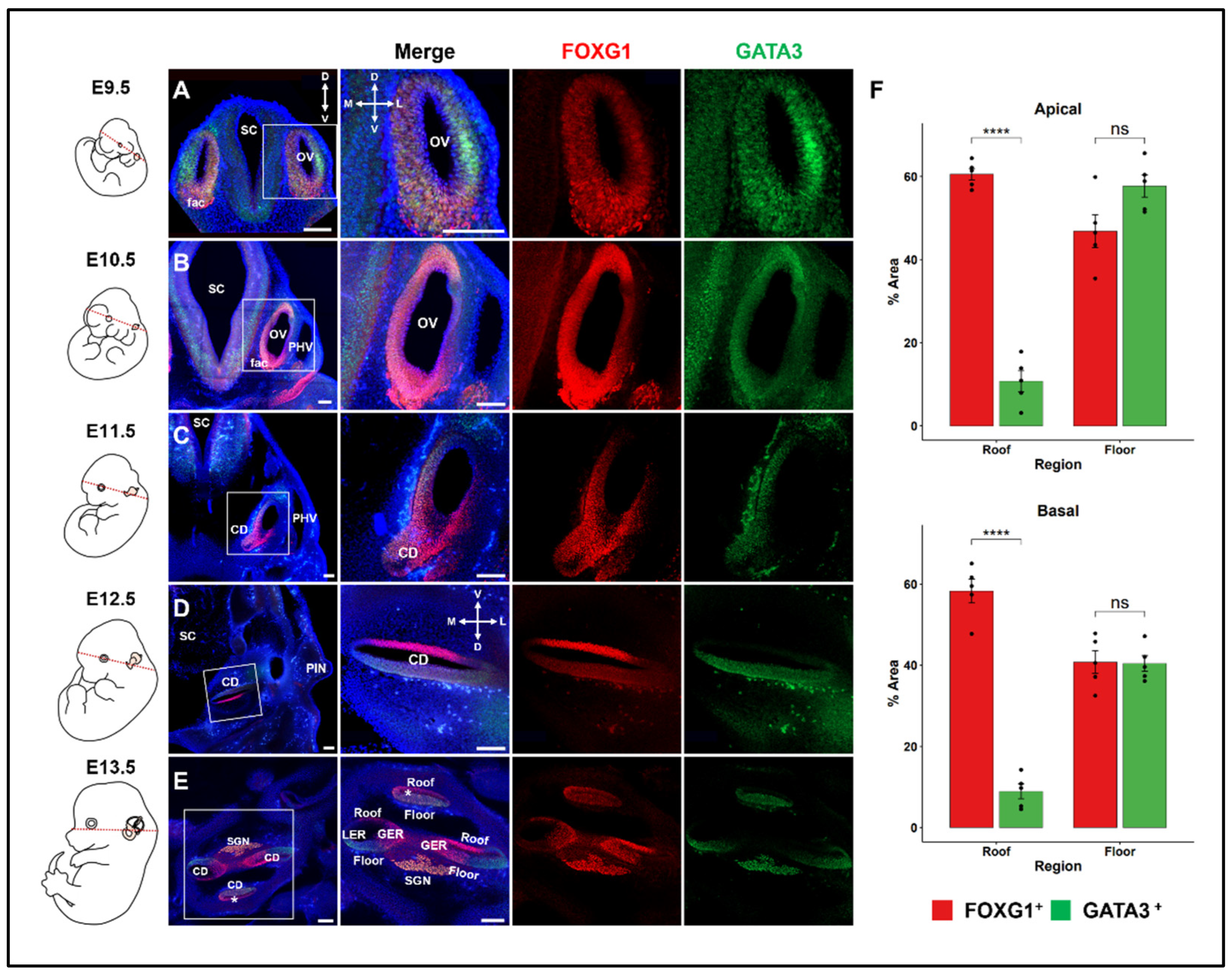
2.1. Dynamics of FOXG1 Expression Patterns in the Developing Cochlear Duct
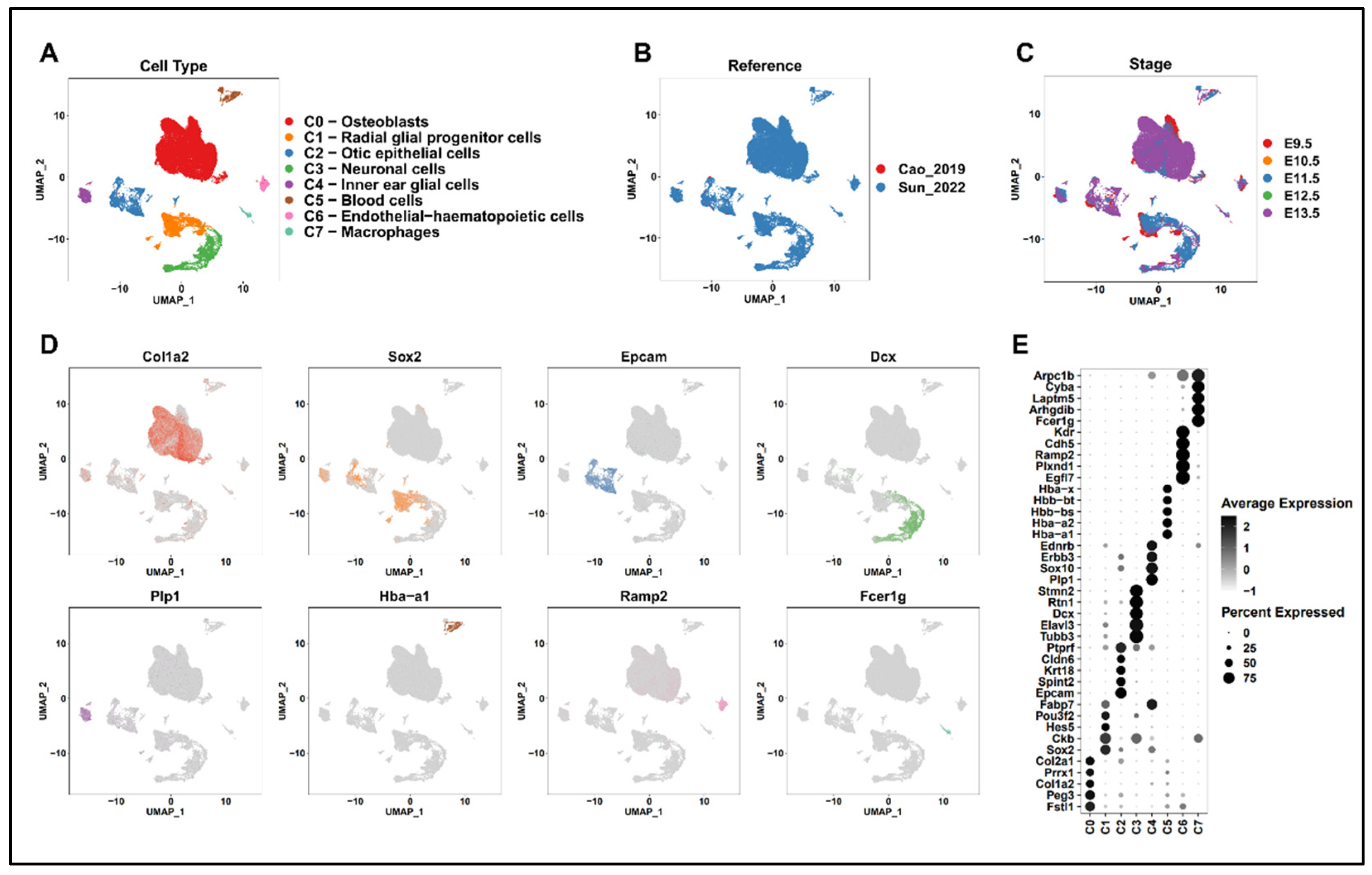
2.2. Identification of Inner-Ear Cell Types Using Public Data
2.3. Subclustering of Cochlear Epithelial Cell Subtypes
2.4. hdWGCNA of Cochlear Epithelial Cell Subtypes
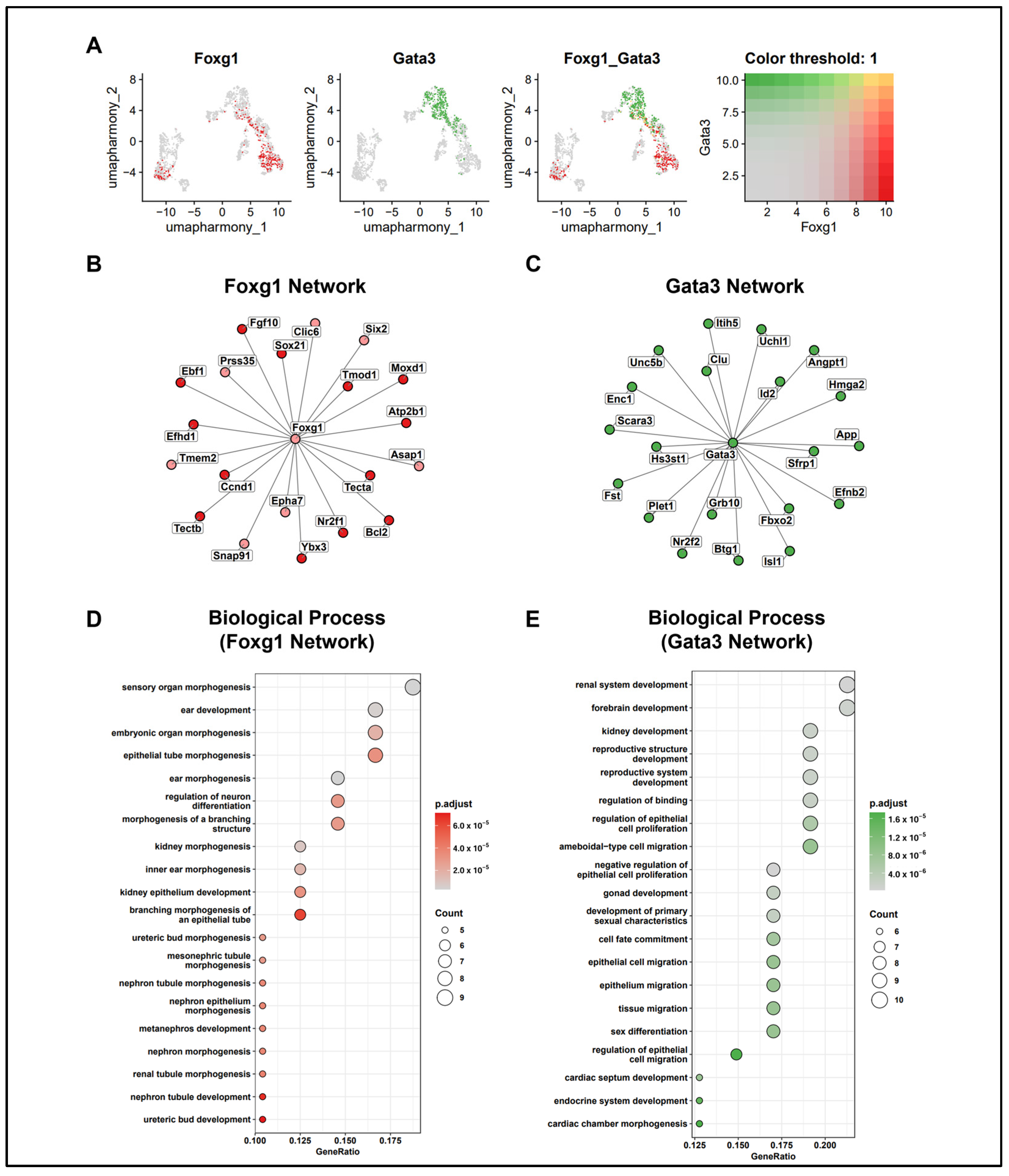
2.5. Network Analysis of FOXG1 and GATA3
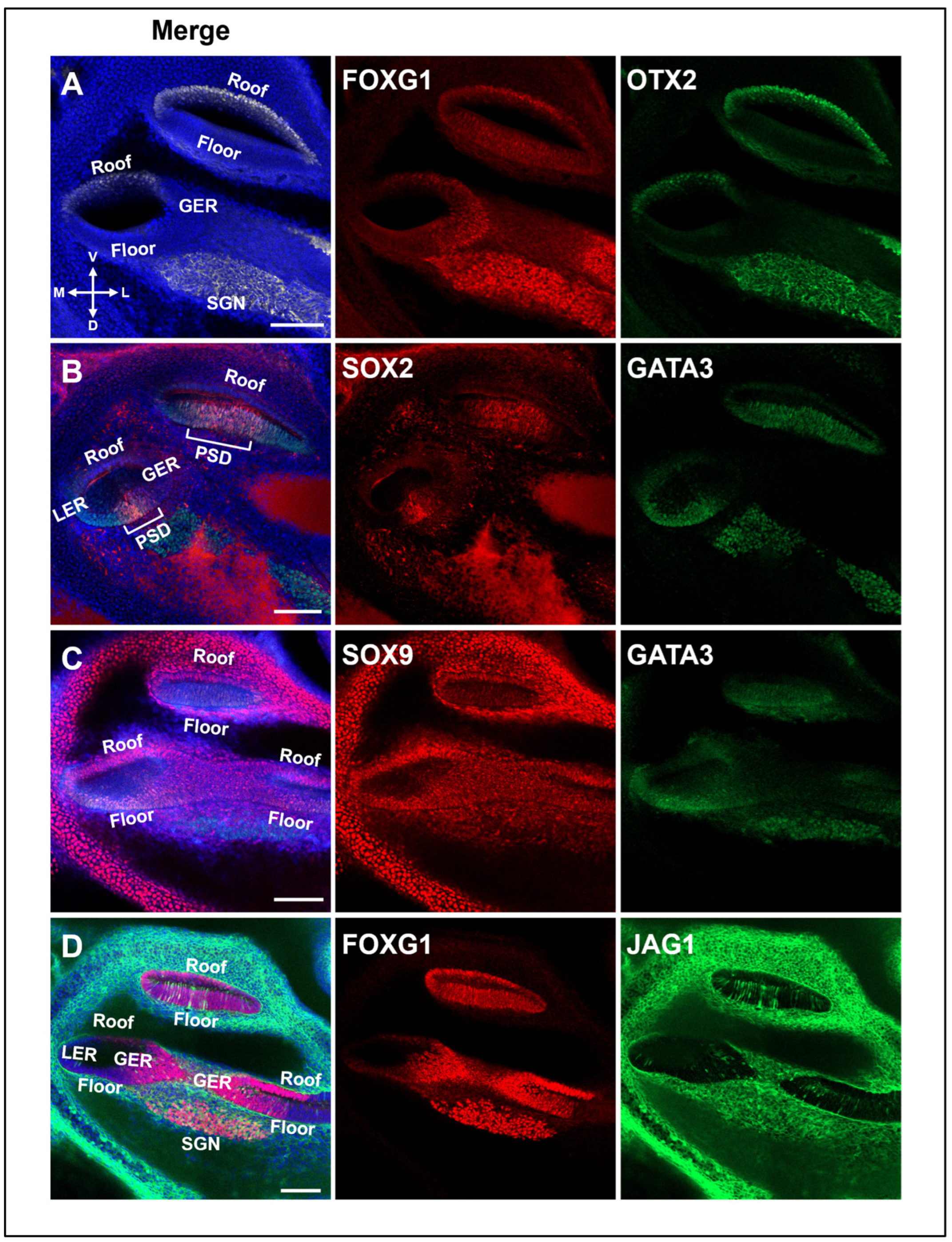
2.6. Expression of Various Marker Genes in the E13.5 Cochlear Duct
2.7. The Relationship Between Hearing Loss-Associated Disease and Modules
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Immunohistochemistry and Fluorescent Quantification Analysis
4.3. Dataset of Inner-Ear Samples
4.4. Preprocessing, Clustering, and Cell Type Identification
4.5. Subclustering of Otic Epithelial Cell Subtypes
4.6. High-Dimensional WGNCA Analysis
4.7. Network Analysis of Foxg1 and Gata3
4.8. Gene Ontology Enrichment Analysis
4.9. Hearing Loss-Associated Gene Enrichment and Correlation Analysis
4.10. Statistical Analysis and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kochhar, A.; Hildebrand, M.S.; Smith, R.J. Clinical aspects of hereditary hearing loss. Genet. Med. 2007, 9, 393–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kim, S.M.; Wang, W.; Cai, C.; Feng, Y.; Kong, W.; Lin, X. Cochlear Gene Therapy for Sensorineural Hearing Loss: Current Status and Major Remaining Hurdles for Translational Success. Front. Mol. Neurosci. 2018, 11, 221. [Google Scholar] [CrossRef] [PubMed]
- Pyott, S.J.; Pavlinkova, G.; Yamoah, E.N.; Fritzsch, B. Harmony in the Molecular Orchestra of Hearing: Developmental Mechanisms from the Ear to the Brain. Annu. Rev. Neurosci. 2024, 47, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Egilmez, O.K.; Kalcioglu, M.T. Genetics of Nonsyndromic Congenital Hearing Loss. Scientifica 2016, 2016, 7576064. [Google Scholar] [CrossRef]
- Shearer, A.E.; Hildebrand, M.S.; Schaefer, A.M.; Smith, R.J. Genetic Hearing Loss Overview; GeneReviews®: Seattle, WA, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1434/ (accessed on 10 November 2024).
- Wang, L.; Kempton, J.B.; Brigande, J.V. Gene Therapy in Mouse Models of Deafness and Balance Dysfunction. Front. Mol. Neurosci. 2018, 11, 300. [Google Scholar] [CrossRef]
- Wu, D.K.; Kelley, M.W. Molecular mechanisms of inner ear development. Cold Spring Harb. Perspect. Biol. 2012, 4, a008409. [Google Scholar] [CrossRef]
- Zine, A.; Fritzsch, B. Early Steps towards Hearing: Placodes and Sensory Development. Int. J. Mol. Sci. 2023, 24, 6994. [Google Scholar] [CrossRef] [PubMed]
- Riccomagno, M.M.; Takada, S.; Epstein, D.J. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005, 19, 1612–1623. [Google Scholar] [CrossRef]
- Bok, J.; Raft, S.; Kong, K.A.; Koo, S.K.; Dräger, U.C.; Wu, D.K. Transient retinoic acid signaling confers anterior-posterior polarity to the inner ear. Proc. Natl. Acad. Sci. USA 2011, 108, 161–166. [Google Scholar] [CrossRef]
- Lin, Z.; Cantos, R.; Patente, M.; Wu, D.K. Gbx2 is required for the morphogenesis of the mouse inner ear: A downstream candidate of hindbrain signaling. Development 2005, 132, 2309–2318. [Google Scholar] [CrossRef]
- Kopecky, B.; Johnson, S.; Schmitz, H.; Santi, P.; Fritzsch, B. Scanning thin-sheet laser imaging microscopy elucidates details on mouse ear development. Dev. Dyn. 2012, 241, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Muthu, V.; Rohacek, A.M.; Yao, Y.; Rakowiecki, S.M.; Brown, A.S.; Zhao, Y.T.; Meyers, J.; Won, K.J.; Ramdas, S.; Brown, C.D.; et al. Genomic architecture of Shh-dependent cochlear morphogenesis. Development 2019, 146, dev181339. [Google Scholar] [CrossRef]
- Morsli, H.; Tuorto, F.; Choo, D.; Postiglione, M.P.; Simeone, A.; Wu, D.K. Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development 1999, 126, 2335–2343. [Google Scholar] [CrossRef]
- Vendrell, V.; Lopez-Hernandez, I.; Duran Alonso, M.B.; Feijoo-Redondo, A.; Abello, G.; Galvez, H.; Giraldez, F.; Lamonerie, T.; Schimmang, T. Otx2 is a target of N-myc and acts as a suppressor of sensory development in the mammalian cochlea. Development 2015, 142, 2792–2800. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.W. Cochlear Development; New Tools and Approaches. Front. Cell Dev. Biol. 2022, 10, 884240. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, A.E.; Ahituv, N.; Fuchs, H.; Balling, R.; Avraham, K.B.; Steel, K.P.; Hrabé de Angelis, M. The Notch ligand Jagged1 is required for inner ear sensory development. Proc. Natl. Acad. Sci. USA 2001, 98, 3873–3878. [Google Scholar] [CrossRef]
- Murata, J.; Ohtsuka, T.; Tokunaga, A.; Nishiike, S.; Inohara, H.; Okano, H.; Kageyama, R. Notch-Hes1 pathway contributes to the cochlear prosensory formation potentially through the transcriptional down-regulation of p27Kip1. J. Neurosci. Res. 2009, 87, 3521–3534. [Google Scholar] [CrossRef]
- Lanford, P.J.; Lan, Y.; Jiang, R.; Lindsell, C.; Weinmaster, G.; Gridley, T.; Kelley, M.W. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat. Genet. 1999, 21, 289–292. [Google Scholar] [CrossRef]
- Dabdoub, A.; Puligilla, C.; Jones, J.M.; Fritzsch, B.; Cheah, K.S.; Pevny, L.H.; Kelley, M.W. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc. Natl. Acad. Sci. USA 2008, 105, 18396–18401. [Google Scholar] [CrossRef]
- Kempfle, J.S.; Turban, J.L.; Edge, A.S. Sox2 in the differentiation of cochlear progenitor cells. Sci. Rep. 2016, 6, 23293. [Google Scholar] [CrossRef]
- Kiernan, A.E.; Pelling, A.L.; Leung, K.K.; Tang, A.S.; Bell, D.M.; Tease, C.; Lovell-Badge, R.; Steel, K.P.; Cheah, K.S. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 2005, 434, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Lawoko-Kerali, G.; Rivolta, M.N.; Holley, M. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J. Comp. Neurol. 2002, 442, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.S.; Lim, K.C.; Engel, J.D.; Fritzsch, B. Limited inner ear morphogenesis and neurosensory development are possible in the absence of GATA3. Int. J. Dev. Biol. 2011, 55, 297–303. [Google Scholar] [CrossRef]
- Duncan, J.S.; Fritzsch, B. Continued expression of GATA3 is necessary for cochlear neurosensory development. PLoS ONE 2013, 8, e62046. [Google Scholar] [CrossRef] [PubMed]
- Morsli, H.; Choo, D.; Ryan, A.; Johnson, R.; Wu, D.K. Development of the mouse inner ear and origin of its sensory organs. J. Neurosci. 1998, 18, 3327–3335. [Google Scholar] [CrossRef]
- Kil, S.H.; Collazo, A. Origins of inner ear sensory organs revealed by fate map and time-lapse analyses. Dev. Biol. 2001, 233, 365–379. [Google Scholar] [CrossRef][Green Version]
- Ohyama, T.; Basch, M.L.; Mishina, Y.; Lyons, K.M.; Segil, N.; Groves, A.K. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J. Neurosci. 2010, 30, 15044–15051. [Google Scholar] [CrossRef]
- Blauwkamp, M.N.; Beyer, L.A.; Kabara, L.; Takemura, K.; Buck, T.; King, W.M.; Dolan, D.F.; Barald, K.F.; Raphael, Y.; Koenig, R.J. The role of bone morphogenetic protein 4 in inner ear development and function. Hear. Res. 2007, 225, 71–79. [Google Scholar] [CrossRef][Green Version]
- Hébert, J.M.; McConnell, S.K. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 2000, 222, 296–306. [Google Scholar] [CrossRef]
- Yang, H.; Ryu, J.; Lim, C.; Choi, J.W.; Park, Y.J.; Jang, S.W.; Shim, S. SOXE group transcription factors regulates the expression of FoxG1 during inner ear development. Biochem. Biophys. Res. Commun. 2022, 623, 96–103. [Google Scholar] [CrossRef]
- Kumamoto, T.; Hanashima, C. Evolutionary conservation and conversion of Foxg1 function in brain development. Dev. Growth Differ. 2017, 59, 258–269. [Google Scholar] [CrossRef]
- Pauley, S.; Lai, E.; Fritzsch, B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 2006, 235, 2470–2482. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.H.; Simeone, A.; Lai, E.; Wu, D.K. Foxg1 is required for proper separation and formation of sensory cristae during inner ear development. Dev. Dyn. 2009, 238, 2725–2734. [Google Scholar] [CrossRef]
- He, Z.H.; Li, M.; Fang, Q.J.; Liao, F.L.; Zou, S.Y.; Wu, X.; Sun, H.Y.; Zhao, X.Y.; Hu, Y.J.; Xu, X.X.; et al. FOXG1 promotes aging inner ear hair cell survival through activation of the autophagy pathway. Autophagy 2021, 17, 4341–4362. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Fang, Q.; Li, H.; Shao, B.; Zhang, Y.; Zhang, Y.; Han, X.; Guo, R.; Cheng, C.; Guo, L.; et al. The role of FOXG1 in the postnatal development and survival of mouse cochlear hair cells. Neuropharmacology 2019, 144, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Y.; Dong, Y.; Guo, L.; Zhang, Z.; Shao, B.; Qi, J.; Zhou, H.; Zhu, W.; Yan, X.; et al. Knockdown of Foxg1 in supporting cells increases the trans-differentiation of supporting cells into hair cells in the neonatal mouse cochlea. Cell Mol. Life Sci. 2020, 77, 1401–1419. [Google Scholar] [CrossRef]
- Driver, E.C.; Kelley, M.W. Development of the cochlea. Development 2020, 147, dev162263. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, L.; Zhu, T.; Wu, B.; Wang, G.; Luo, Z.; Li, C.; Wei, W.; Liu, Z. Single-cell transcriptomic landscapes of the otic neuronal lineage at multiple early embryonic ages. Cell Rep. 2022, 38, 110542. [Google Scholar] [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Li, W.; Alahdal, M.; Deng, Z.; Liu, J.; Zhao, Z.; Cheng, X.; Chen, X.; Li, J.; Yin, J.; Li, Y.; et al. Molecular functions of FSTL1 in the osteoarthritis. Int. Immunopharmacol. 2020, 83, 106465. [Google Scholar] [CrossRef] [PubMed]
- Edri, R.; Yaffe, Y.; Ziller, M.J.; Mutukula, N.; Volkman, R.; David, E.; Jacob-Hirsch, J.; Malcov, H.; Levy, C.; Rechavi, G.; et al. Analysing human neural stem cell ontogeny by consecutive isolation of Notch active neural progenitors. Nat. Commun. 2015, 6, 6500. [Google Scholar] [CrossRef]
- Hertzano, R.; Puligilla, C.; Chan, S.L.; Timothy, C.; Depireux, D.A.; Ahmed, Z.; Wolf, J.; Eisenman, D.J.; Friedman, T.B.; Riazuddin, S.; et al. CD44 is a marker for the outer pillar cells in the early postnatal mouse inner ear. J. Assoc. Res. Otolaryngol. 2010, 11, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Tischfield, M.A.; Baris, H.N.; Wu, C.; Rudolph, G.; Van Maldergem, L.; He, W.; Chan, W.M.; Andrews, C.; Demer, J.L.; Robertson, R.L.; et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 2010, 140, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.S.; Shetty, A.K. Efficacy of doublecortin as a marker to analyse the absolute number and dendritic growth of newly generated neurons in the adult dentate gyrus. Eur. J. Neurosci. 2004, 19, 234–246. [Google Scholar] [CrossRef]
- Kempfle, J.S.; Luu, N.C.; Petrillo, M.; Al-Asad, R.; Zhang, A.; Edge, A.S.B. Lin28 reprograms inner ear glia to a neuronal fate. Stem Cells 2020, 38, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.; Yang, W.H.; Wu, J.; Roback, J.D.; Gregory, S.G.; Chi, J.T. Single Cell RNA-Seq Analysis of Human Red Cells. Front. Physiol. 2022, 13, 828700. [Google Scholar] [CrossRef]
- Biben, C.; Weber, T.S.; Potts, K.S.; Choi, J.; Miles, D.C.; Carmagnac, A.; Sargeant, T.; de Graaf, C.A.; Fennell, K.A.; Farley, A.; et al. In vivo clonal tracking reveals evidence of haemangioblast and haematomesoblast contribution to yolk sac haematopoiesis. Nat. Commun. 2023, 14, 41. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e821. [Google Scholar] [CrossRef]
- Yamamoto, R.; Ohnishi, H.; Omori, K.; Yamamoto, N. In silico analysis of inner ear development using public whole embryonic body single-cell RNA-sequencing data. Dev. Biol. 2021, 469, 160–171. [Google Scholar] [CrossRef]
- Silbereis, J.C.; Pochareddy, S.; Zhu, Y.; Li, M.; Sestan, N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 2016, 89, 248–268. [Google Scholar] [CrossRef] [PubMed]
- Morabito, S.; Reese, F.; Rahimzadeh, N.; Miyoshi, E.; Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 2023, 3, 100498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, J.; Maire, P.; Xu, P.X. Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium. PLoS Genet. 2017, 13, e1006967. [Google Scholar] [CrossRef] [PubMed]
- Legan, P.K.; Lukashkina, V.A.; Goodyear, R.J.; Kössi, M.; Russell, I.J.; Richardson, G.P. A targeted deletion in alpha-tectorin reveals that the tectorial membrane is required for the gain and timing of cochlear feedback. Neuron 2000, 28, 273–285. [Google Scholar] [CrossRef]
- Ishii, N.; Wanaka, A.; Ohno, K.; Matsumoto, K.; Eguchi, Y.; Mori, T.; Tsujimoto, Y.; Tohyama, M. Localization of bcl-2, bax, and bcl-x mRNAs in the developing inner ear of the mouse. Brain Res. 1996, 726, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Zelarayan, L.C.; Vendrell, V.; Alvarez, Y.; Domínguez-Frutos, E.; Theil, T.; Alonso, M.T.; Maconochie, M.; Schimmang, T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev. Biol. 2007, 308, 379–391. [Google Scholar] [CrossRef]
- Filova, I.; Pysanenko, K.; Tavakoli, M.; Vochyanova, S.; Dvorakova, M.; Bohuslavova, R.; Smolik, O.; Fabriciova, V.; Hrabalova, P.; Benesova, S.; et al. ISL1 is necessary for auditory neuron development and contributes toward tonotopic organization. Proc. Natl. Acad. Sci. USA 2022, 119, e2207433119. [Google Scholar] [CrossRef]
- Geng, R.; Noda, T.; Mulvaney, J.F.; Lin, V.Y.; Edge, A.S.; Dabdoub, A. Comprehensive Expression of Wnt Signaling Pathway Genes during Development and Maturation of the Mouse Cochlea. PLoS ONE 2016, 11, e0148339. [Google Scholar] [CrossRef]
- Tornari, C.; Towers, E.R.; Gale, J.E.; Dawson, S.J. Regulation of the orphan nuclear receptor Nr2f2 by the DFNA15 deafness gene Pou4f3. PLoS ONE 2014, 9, e112247. [Google Scholar] [CrossRef]
- Raft, S.; Andrade, L.R.; Shao, D.; Akiyama, H.; Henkemeyer, M.; Wu, D.K. Ephrin-B2 governs morphogenesis of endolymphatic sac and duct epithelia in the mouse inner ear. Dev. Biol. 2014, 390, 51–67. [Google Scholar] [CrossRef][Green Version]
- Dvorakova, M.; Macova, I.; Bohuslavova, R.; Anderova, M.; Fritzsch, B.; Pavlinkova, G. Early ear neuronal development, but not olfactory or lens development, can proceed without SOX2. Dev. Biol. 2020, 457, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, M.; Fujioka, M.; Okano, H.; Ozawa, H. Mapping of Notch signaling in the developing organ of Corti in common marmosets. Front. Neuroanat. 2023, 17, 1188886. [Google Scholar] [CrossRef] [PubMed]
- Szeto, I.Y.Y.; Chu, D.K.H.; Chen, P.; Chu, K.C.; Au, T.Y.K.; Leung, K.K.H.; Huang, Y.H.; Wynn, S.L.; Mak, A.C.Y.; Chan, Y.S.; et al. SOX9 and SOX10 control fluid homeostasis in the inner ear for hearing through independent and cooperative mechanisms. Proc. Natl. Acad. Sci. USA 2022, 119, e2122121119. [Google Scholar] [CrossRef]
- Cox, B.C.; Liu, Z.; Lagarde, M.M.; Zuo, J. Conditional gene expression in the mouse inner ear using Cre-loxP. J. Assoc. Res. Otolaryngol. 2012, 13, 295–322. [Google Scholar] [CrossRef]
- Tateya, T.; Sakamoto, S.; Ishidate, F.; Hirashima, T.; Imayoshi, I.; Kageyama, R. Three-dimensional live imaging of Atoh1 reveals the dynamics of hair cell induction and organization in the developing cochlea. Development 2019, 146, dev177881. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Owen, T.; Zhang, L.; Zuo, J. Dynamic expression pattern of Sonic hedgehog in developing cochlear spiral ganglion neurons. Dev. Dyn. 2010, 239, 1674–1683. [Google Scholar] [CrossRef]
- Hanashima, C.; Li, S.C.; Shen, L.; Lai, E.; Fishell, G. Foxg1 suppresses early cortical cell fate. Science 2004, 303, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Xuan, S.; Baptista, C.A.; Balas, G.; Tao, W.; Soares, V.C.; Lai, E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 1995, 14, 1141–1152. [Google Scholar] [CrossRef]
- Cargnin, F.; Kwon, J.S.; Katzman, S.; Chen, B.; Lee, J.W.; Lee, S.K. FOXG1 Orchestrates Neocortical Organization and Cortico-Cortical Connections. Neuron 2018, 100, 1083–1096.e1085. [Google Scholar] [CrossRef]
- Liu, J.; Yang, M.; Su, M.; Liu, B.; Zhou, K.; Sun, C.; Ba, R.; Yu, B.; Zhang, B.; Zhang, Z.; et al. FOXG1 sequentially orchestrates subtype specification of postmitotic cortical projection neurons. Sci. Adv. 2022, 8, eabh3568. [Google Scholar] [CrossRef]
- Chen, J.; Gao, D.; Sun, L.; Yang, J. Kölliker’s organ-supporting cells and cochlear auditory development. Front. Mol. Neurosci. 2022, 15, 1031989. [Google Scholar] [CrossRef] [PubMed]
- Kagoshima, H.; Ohnishi, H.; Yamamoto, R.; Yasumoto, A.; Tona, Y.; Nakagawa, T.; Omori, K.; Yamamoto, N. EBF1 Limits the Numbers of Cochlear Hair and Supporting Cells and Forms the Scala Tympani and Spiral Limbus during Inner Ear Development. J. Neurosci. 2024, 44, e1060232023. [Google Scholar] [CrossRef] [PubMed]
- Powers, K.G.; Wilkerson, B.A.; Beach, K.E.; Seo, S.S.; Rodriguez, J.S.; Baxter, A.N.; Hunter, S.E.; Bermingham-McDonogh, O. Deletion of the Ebf1, a mouse deafness gene, causes a dramatic increase in hair cells and support cells of the organ of Corti. Development 2024, 151, dev202816. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R Package Version 0.7.0. 2021. Available online: https://cran.r-project.org/package=rstatix (accessed on 23 September 2024).
- Kassambara, A. ggpubr:‘Ggplot2’based Publication Ready Plots. 2020. Available online: https://cran.r-project.org/package=ggpubr (accessed on 23 September 2024).
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Micali, N.; Ma, S.; Li, M.; Kim, S.K.; Mato-Blanco, X.; Sindhu, S.K.; Arellano, J.I.; Gao, T.; Shibata, M.; Gobeske, K.T.; et al. Molecular programs of regional specification and neural stem cell fate progression in macaque telencephalon. Science 2023, 382, eadf3786. [Google Scholar] [CrossRef]
- Carlson, M.; Falcon, S. org.Mm.eg.db: Genome Wide Annotation for Mouse. R Package Version 3.8.2. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Mm.eg.db.html (accessed on 23 September 2024).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- DiStefano, M.T.; Hughes, M.Y.; Patel, M.J.; Wilcox, E.H.; Oza, A.M. Expert interpretation of genes and variants in hereditary hearing loss. Med. Genet. 2020, 32, 109–115. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics; Version 2. 2016, pp. 1–189. Available online: https://cran.r-project.org/package=ggplot2 (accessed on 23 September 2024).
- Nakazawa, M.; Nakazawa, M.M. Package ‘fmsb’. 2019. Available online: https://cran.r-project.org/web/packages/fmsb/fmsb.pdf (accessed on 23 September 2024).
- Csardi, G.; Nepusz, T. The igraph software. Complex Syst. 2006, 1695, 862049. [Google Scholar]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. R Package Version 2019, 1. Available online: https://bioconductor.org/packages/release/bioc/html/enrichplot.html (accessed on 23 September 2024).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, Y.; Ryu, J.; Yang, H.; Ma, Y.; Nam, K.-H.; Jang, S.-W.; Shim, S. Molecular Characterization of Subdomain Specification of Cochlear Duct Based on Foxg1 and Gata3. Int. J. Mol. Sci. 2024, 25, 12700. https://doi.org/10.3390/ijms252312700
Gil Y, Ryu J, Yang H, Ma Y, Nam K-H, Jang S-W, Shim S. Molecular Characterization of Subdomain Specification of Cochlear Duct Based on Foxg1 and Gata3. International Journal of Molecular Sciences. 2024; 25(23):12700. https://doi.org/10.3390/ijms252312700
Chicago/Turabian StyleGil, Yongjin, Jiho Ryu, Hayoung Yang, Yechan Ma, Ki-Hoan Nam, Sung-Wuk Jang, and Sungbo Shim. 2024. "Molecular Characterization of Subdomain Specification of Cochlear Duct Based on Foxg1 and Gata3" International Journal of Molecular Sciences 25, no. 23: 12700. https://doi.org/10.3390/ijms252312700
APA StyleGil, Y., Ryu, J., Yang, H., Ma, Y., Nam, K.-H., Jang, S.-W., & Shim, S. (2024). Molecular Characterization of Subdomain Specification of Cochlear Duct Based on Foxg1 and Gata3. International Journal of Molecular Sciences, 25(23), 12700. https://doi.org/10.3390/ijms252312700






