Molecular Dynamics Simulations Suggest That Side-Chain Motions of Charged Amino Acids Determine Long-Range Effects in Proteins: An Egg of Coulomb
Abstract
1. Introduction
2. Results
2.1. Charged Side-Chain Dynamics in Human Ubiquitin
2.2. Charged Side-Chain Dynamics in the Ostrinia Furnacalis Chitinolytic Enzyme
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- McGuffee, S.R.; Elcock, A.H. Atomically detailed simulations of concentrated protein solutions: The effects of salt, pH, point mutations, and protein concentration in simulations of 1000-molecule systems. J. Am. Chem. Soc. 2006, 128, 12098–12110. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Skolnick, J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc. Natl. Acad. Sci. USA 2010, 107, 18457–18462. [Google Scholar] [CrossRef] [PubMed]
- Blundell, T.L.; Fernández-Recio, J. Cell biology: Brief encounters bolster contacts. Nature 2006, 444, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, H. Long-range coherence and energy storage in biological systems. Int. J. Quantum Chem. 1968, 2, 641–649. [Google Scholar] [CrossRef]
- Fröhlich, H. Long-range coherence in biological systems. Riv. Nuovo Cim. 1977, 7, 399–418. [Google Scholar] [CrossRef]
- Preto, J.; Pettini, M. Resonant long-range interactions between polar macromolecules. Phys. Lett. A 2013, 377, 587–591. [Google Scholar] [CrossRef]
- Preto, J.; Pettini, M.; Tuszynski, J.A. Possible role of electrodynamic interactions in long-distance biomolecular recognition. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2015, 91, 052710. [Google Scholar] [CrossRef] [PubMed]
- Nardecchia, I.; Lechelon, M.; Gori, M.; Donato, I.; Preto, J.; Floriani, E.; Jaeger, S.; Mailfert, S.; Marguet, D.; Ferrier, P.; et al. Detection of long-range electrostatic interactions between charged molecules by means of fluorescence correlation spectroscopy. Phys. Rev. E 2017, 96, 022403. [Google Scholar] [CrossRef] [PubMed]
- Lechelon, M.; Meriguet, Y.; Gori, M.; Ruffenach, S.; Nardecchia, I.; Floriani, E.; Coquillat, D.; Teppe, F.; Mailfert, S.; Marguet, D.; et al. Experimental evidence for long-distance electrodynamic intermolecular forces. Sci. Adv. 2022, 8, eabl5855. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alocci, D.; Bernini, A.; Niccolai, N. Atom depth analysis delineates mechanisms of protein intermolecular interactions. Biochem. Biophys. Res. Commun. 2013, 436, 725–729. [Google Scholar] [CrossRef]
- Varrazzo, D.; Bernini, A.; Spiga, O.; Ciutti, A.; Chiellini, S.; Venditti, V.; Bracci, L.; Niccolai, N. Three-dimensional computation of atom depth in complex molecular structures. Bioinformatics 2005, 21, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Perica, T.; Chothia, C. Ubiquitin—Molecular mechanisms for recognition of different structures. Curr. Opin. Struct. Biol. 2010, 20, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Kinoshita, K.; Nakamura, H. Hub promiscuity in protein-protein interaction networks. Int. J. Mol. Sci. 2010, 11, 1930–1943. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, H.; Liu, F.; Wu, Q.; Shen, X.; Yang, Q. Structural determinants of an insect beta-N-Acetyl-D-hexosaminidase specialized as a chitinolytic enzyme. J. Biol. Chem. 2011, 286, 4049–4058. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Sloper-Mould, K.E.; Jemc, J.C.; Pickart, C.M.; Hicke, L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001, 276, 30483–30489. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Lee, S.; Prag, G. Ubiquitin-binding domains. Biochem. J. 2006, 399, 361–372. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Esadze, A.; Li, D.W.; Wang, T.; Brüschweiler, R.; Iwahara, J. Dynamics of lysine side-chain amino groups in a protein studied by heteronuclear 1H–15N NMR spectroscopy. J. Am. Chem. Soc. 2011, 133, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://web.expasy.org/docs/relnotes/relstat.html (accessed on 1 July 2024).
- Kaatze, G.A.; Behrends, R.; Pottel, R. Hydrogen network fluctuations and dielectric spectrometry of liquids. J. Non-Cryst. Solids 2002, 305, 19–29. [Google Scholar] [CrossRef]
- Ferguson, B.; Zhang, X.C. Materials for terahertz science and technology. Nat. Mater. 2002, 1, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Falconer, R.J. Terahertz spectroscopy’s application to protein chemistry. Spectrosc. Eur. 2012, 24, 12–14. [Google Scholar]
- Berggård, T.; Linse, S.; James, P. Methods for the detection and analysis of protein-protein interactions. Proteomics 2007, 7, 2833–2842. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Grigerra, J.R.; Straatsma, T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Kortemme, T. De novo protein design—From new structures to programmable functions. Cell 2024, 187, 526–544. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

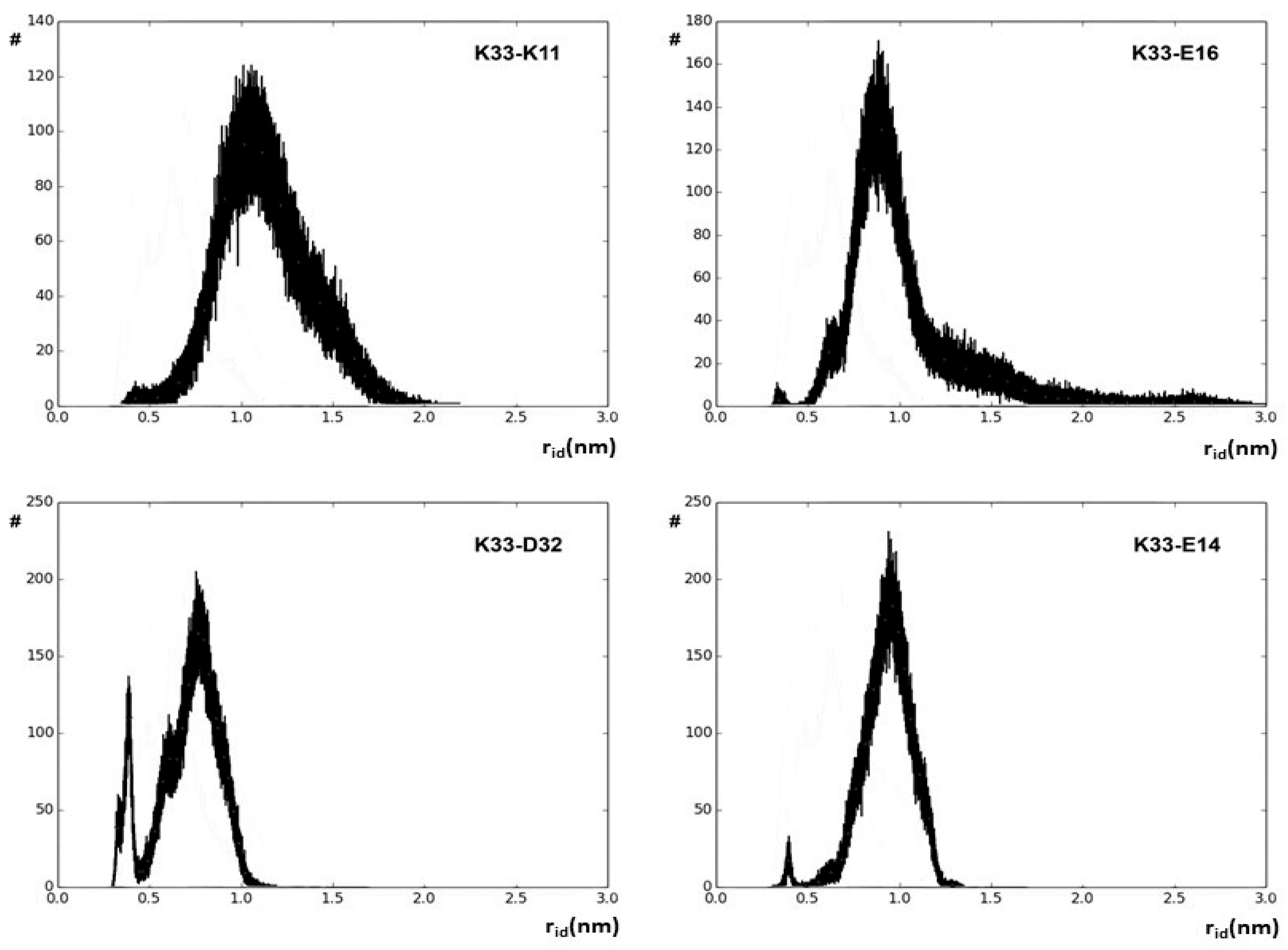
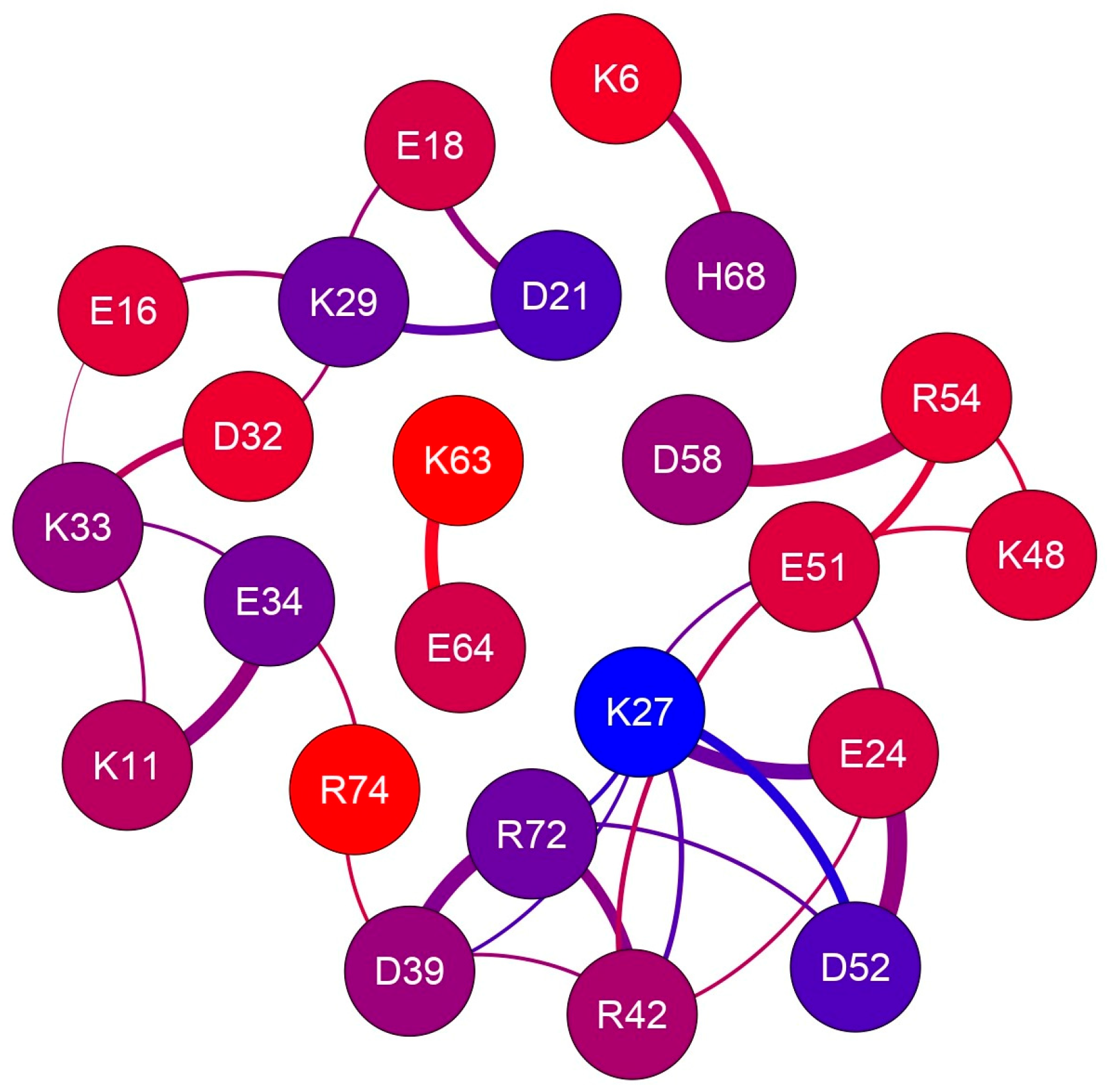


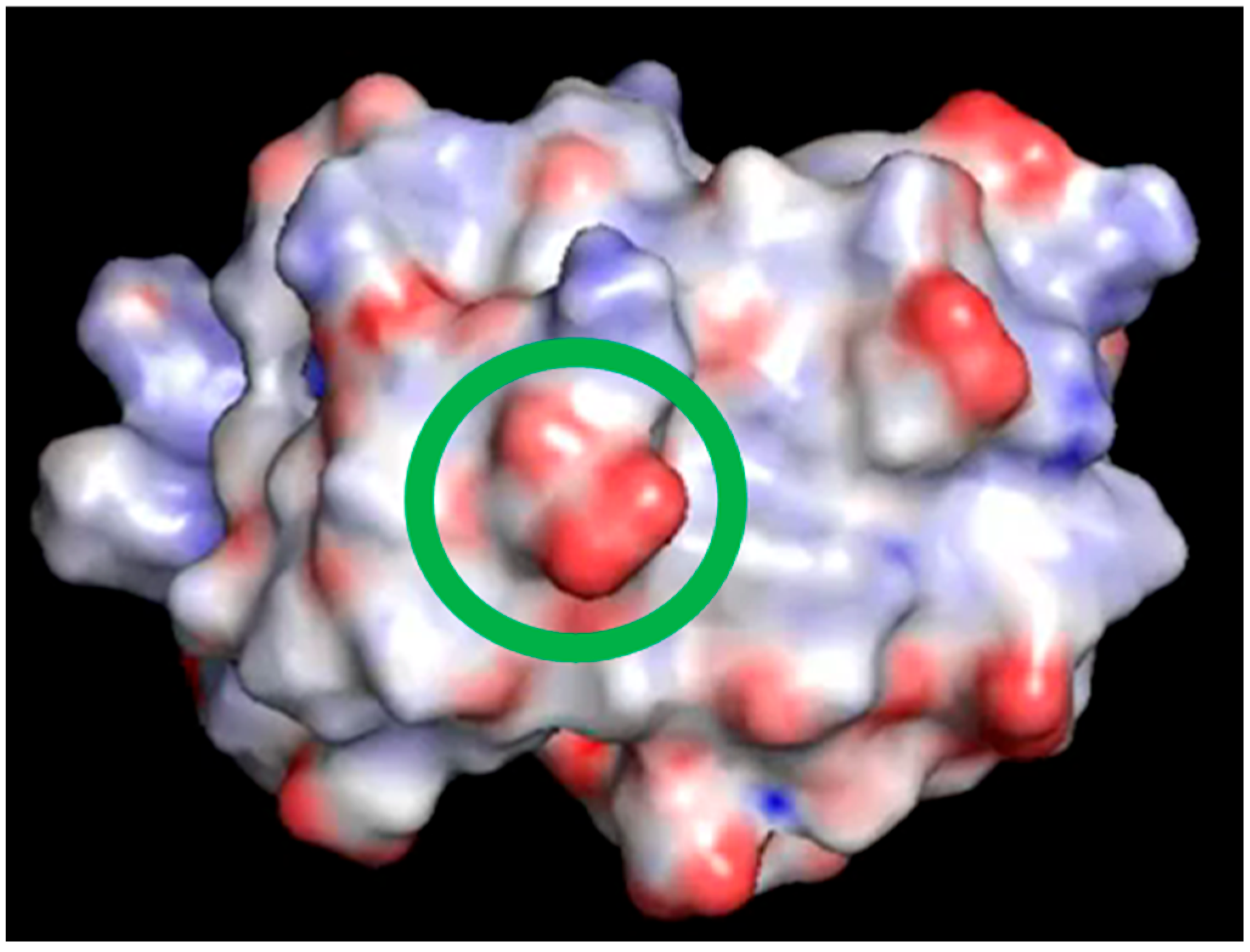
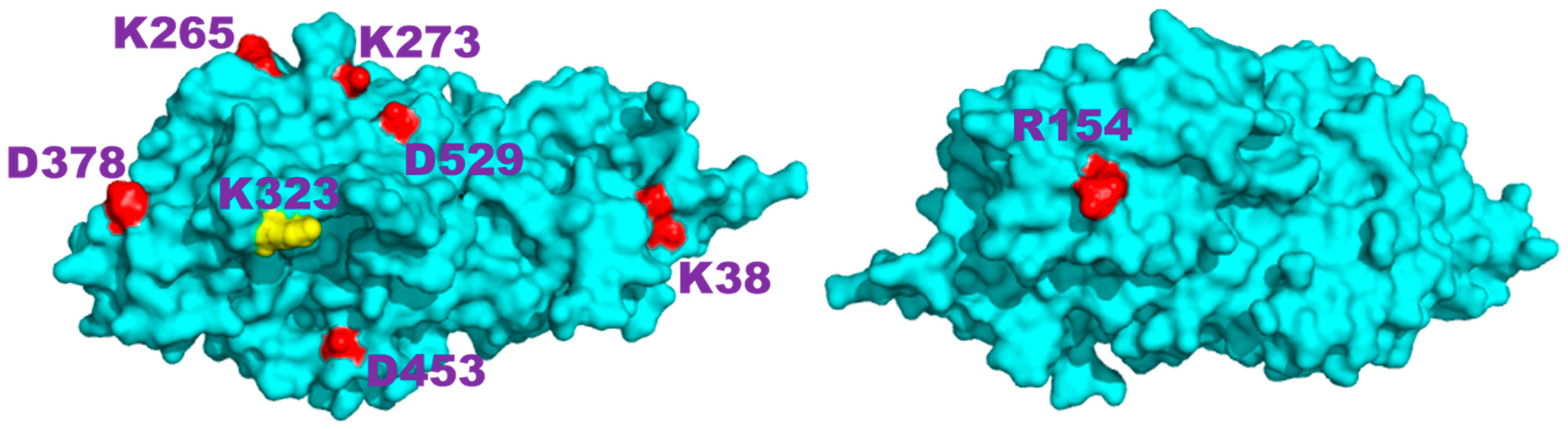
| Atom | No. of Interactions | Di,9 a | DSSP b |
|---|---|---|---|
| NZ K6 | 1 | 1.29 | E |
| NZ K11 | 2 | 1.06 | N/A |
| CD E16 | 2 | 1.22 | E |
| CD E18 | 2 | 1.16 | N/A |
| CG D21 | 2 | 0.60 | S |
| CD E24 | 3 | 1.17 | H |
| NZ K27 | 6 | 0.35 | H |
| NZ K29 | 4 | 0.77 | H |
| CG D32 | 2 | 1.23 | H |
| NZ K33 | 4 | 0.93 | H |
| CD E34 | 3 | 0.76 | H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niccolai, N.; Morandi, E.; Bernini, A. Molecular Dynamics Simulations Suggest That Side-Chain Motions of Charged Amino Acids Determine Long-Range Effects in Proteins: An Egg of Coulomb. Int. J. Mol. Sci. 2024, 25, 13375. https://doi.org/10.3390/ijms252413375
Niccolai N, Morandi E, Bernini A. Molecular Dynamics Simulations Suggest That Side-Chain Motions of Charged Amino Acids Determine Long-Range Effects in Proteins: An Egg of Coulomb. International Journal of Molecular Sciences. 2024; 25(24):13375. https://doi.org/10.3390/ijms252413375
Chicago/Turabian StyleNiccolai, Neri, Edoardo Morandi, and Andrea Bernini. 2024. "Molecular Dynamics Simulations Suggest That Side-Chain Motions of Charged Amino Acids Determine Long-Range Effects in Proteins: An Egg of Coulomb" International Journal of Molecular Sciences 25, no. 24: 13375. https://doi.org/10.3390/ijms252413375
APA StyleNiccolai, N., Morandi, E., & Bernini, A. (2024). Molecular Dynamics Simulations Suggest That Side-Chain Motions of Charged Amino Acids Determine Long-Range Effects in Proteins: An Egg of Coulomb. International Journal of Molecular Sciences, 25(24), 13375. https://doi.org/10.3390/ijms252413375







