Precise and Accurate DNA-3′/5-Ends Polishing with Thermus thermophilus Phage vb_Tt72 DNA Polymerase
Abstract
:1. Introduction
2. Results
2.1. The 3′→5′ Exonuclease Activity of the Tt72 DNA Polymerase
2.2. Fidelity of the Tt72 DNA Polymerase
2.3. DNA Ends Polishing with Tt72 DNA Polymerase
2.4. End Polishing of PCR Products with Single-Nucleotide 3′-Overhang
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmid, and Materials
4.2. Protein Purification
4.3. 3′→5′ Exonuclease Assay
4.4. DNA Polymerase Fidelity Assay
4.5. Preparation of pUC18 Linear Form
4.6. Filling in the 5′-Overhangs
4.7. Removal of the 3′ Overhangs
4.8. 3′-Ends Polishing of PCR Products
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kornberg, A.; Baker, T.A. DNA Replication, 2nd ed.; Freeman: New York, NY, USA, 1992. [Google Scholar]
- Prasad, R.; Longley, M.J.; Sharief, F.S.; Hou, E.W.; Copeland, W.C.; Wilson, S.H. Human DNA Polymerase θ Possesses 5′-dRP Lyase Activity and Functions in Single-Nucleotide Base Excision Repair in Vitro. Nucleic Acids Res. 2009, 37, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Garg, P.; Burgers, P.M.J. DNA Polymerases That Propagate the Eukaryotic DNA Replication Fork. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, Y.I.; Shcherbakova, P.V.; Rogozin, I.B. Roles of DNA Polymerases in Replication, Repair, and Recombination in Eukaryotes. Int. Rev. Cytol. 2006, 255, 41–132. [Google Scholar] [CrossRef] [PubMed]
- Ishino, S.; Ishino, Y. DNA Polymerases as Useful Reagents for Biotechnology—The History of Developmental Research in the Field. Front. Microbiol. 2014, 5, 465. [Google Scholar] [CrossRef]
- Chien, A.; Edgar, D.B.; Trela, J.M. Deoxyribonucleic Acid Polymerase from the Extreme Thermophile Thermus Aquaticus. J. Bacteriol. 1976, 127, 1550. [Google Scholar] [CrossRef]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-Directed Enzymatic Amplification of DNA with a Thermostable DNA Polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA in Vitro: The Polymerase Chain Reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51(Pt. 1), 263–273. [Google Scholar] [CrossRef]
- Myers, T.W.; Gelfand, D.H. Reverse Transcription and DNA Amplification by a Thermus thermophilus DNA Polymerase. Biochemistry 1991, 30, 7661–7666. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989. [Google Scholar]
- Kunkel, T.A. Rapid and Efficient Site-Specific Mutagenesis without Phenotypic Selection. Proc. Natl. Acad. Sci. USA 1985, 82, 488–492. [Google Scholar] [CrossRef]
- Bebenek, K.; Kunkel, T.A. The Use of Native T7 DNA Polymerase for Site-Directed Mutagenesis. Nucleic Acids Res. 1989, 17, 5408. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Koop, B.F.; Hood, L. A Simple Method Using T4 DNA Polymerase to Clone Polymerase Chain Reaction Products. Biotechniques 1994, 17, 236–238. [Google Scholar] [PubMed]
- Challberg, M.D.; Englund, P.T. Specific Labeling of 3’ Termini with T4 DNA Polymerase. Methods Enzym. 1980, 65, 39–43. [Google Scholar] [CrossRef]
- Aslanidis, C.; de Jong, P.J. Ligation-Independent Cloning of PCR Products (LIC-PCR). Nucleic Acids Res. 1990, 18, 6069–6074. [Google Scholar] [CrossRef] [PubMed]
- Bodescot, M.; Brison, O. Efficient Second-Strand cDNA Synthesis Using T7 DNA Polymerase. DNA Cell Biol. 1994, 13, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.A.; Dipasquale, B.; Youle, R.J. In Situ Labeling of Granule Cells for Apoptosis-Associated DNA Fragmentation Reveals Different Mechanisms of Cell Loss in Developing Cerebellum. Neuron 1993, 11, 621–632. [Google Scholar] [CrossRef]
- Zhu, B. Bacteriophage T7 DNA Polymerase—Sequenase. Front. Microbiol. 2014, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Blanco, L.; Bernad, A.; Lázaro, J.M.; Martín, G.; Garmendia, C.; Salas, M. Highly Efficient DNA Synthesis by the Phage Phi 29 DNA Polymerase. Symmetrical Mode of DNA Replication. J. Biol. Chem. 1989, 264, 8935–8940. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.A.; Salas, M.; Blanco, L. Fidelity of Phi 29 DNA Polymerase. Comparison between Protein-Primed Initiation and DNA Polymerization. J. Biol. Chem. 1993, 268, 2719–2726. [Google Scholar] [CrossRef]
- Berman, A.J.; Kamtekar, S.; Goodman, J.L.; Lázaro, J.M.; de Vega, M.; Blanco, L.; Salas, M.; Steitz, T.A. Structures of Phi29 DNA Polymerase Complexed with Substrate: The Mechanism of Translocation in B-Family Polymerases. EMBO J. 2007, 26, 3494–3505. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Nelson, J.R.; Giesler, T.L.; Lasken, R.S. Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Res. 2001, 11, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Krzywkowski, T.; Kühnemund, M.; Wu, D.; Nilsson, M. Limited Reverse Transcriptase Activity of Phi29 DNA Polymerase. Nucleic Acids Res. 2018, 46, 3625–3632. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Hosono, S.; Fang, L.; Wu, X.; Faruqi, A.F.; Bray-Ward, P.; Sun, Z.; Zong, Q.; Du, Y.; Du, J.; et al. Comprehensive Human Genome Amplification Using Multiple Displacement Amplification. Proc. Natl. Acad. Sci. USA 2002, 99, 5261–5266. [Google Scholar] [CrossRef] [PubMed]
- Kroneis, T.; El-Heliebi, A. Whole Genome Amplification by Isothermal Multiple Strand Displacement Using Phi29 DNA Polymerase. Methods Mol. Biol. 2015, 1347, 111–117. [Google Scholar] [CrossRef]
- Korlach, J.; Marks, P.J.; Cicero, R.L.; Gray, J.J.; Murphy, D.L.; Roitman, D.B.; Pham, T.T.; Otto, G.A.; Foquet, M.; Turner, S.W. Selective Aluminum Passivation for Targeted Immobilization of Single DNA Polymerase Molecules in Zero-Mode Waveguide Nanostructures. Proc. Natl. Acad. Sci. USA 2008, 105, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single Molecule Real-Time (SMRT) Sequencing Comes of Age: Applications and Utilities for Medical Diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Manrao, E.A.; Derrington, I.M.; Laszlo, A.H.; Langford, K.W.; Hopper, M.K.; Gillgren, N.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Reading DNA at Single-Nucleotide Resolution with a Mutant MspA Nanopore and Phi29 DNA Polymerase. Nat. Biotechnol. 2012, 30, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Dorawa, S.; Werbowy, O.; Plotka, M.; Kaczorowska, A.-K.; Makowska, J.; Kozlowski, L.P.; Fridjonsson, O.H.; Hreggvidsson, G.O.; Aevarsson, A.; Kaczorowski, T. Molecular Characterization of a DNA Polymerase from Thermus thermophilus MAT72 Phage vB_Tt72: A Novel Type-A Family Enzyme with Strong Proofreading Activity. Int. J. Mol. Sci. 2022, 23, 7945. [Google Scholar] [CrossRef]
- Keith, B.J.; Jozwiakowski, S.K.; Connolly, B.A. A Plasmid-Based lacZα Gene Assay for DNA Polymerase Fidelity Measurement. Anal. Biochem. 2013, 433, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Jozwiakowski, S.K.; Connolly, B.A. Plasmid-Based lacZα Assay for DNA Polymerase Fidelity: Application to Archaeal Family-B DNA Polymerase. Nucleic Acids Res. 2009, 37, e102. [Google Scholar] [CrossRef]
- Clark, J.M. Novel Non-Templated Nucleotide Addition Reactions Catalyzed by Procaryotic and Eucaryotic DNA Polymerases. Nucleic Acids Res. 1988, 16, 9677–9686. [Google Scholar] [CrossRef]
- Lopatina, A.; Medvedeva, S.; Artamonova, D.; Kolesnik, M.; Sitnik, V.; Ispolatov, Y.; Severinov, K. Natural Diversity of CRISPR Spacers of Thermus: Evidence of Local Spacer Acquisition and Global Spacer Exchange. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180092. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.X.; Slater, M.R.; Ackermann, H.-W. Isolation and Characterization of Thermus Bacteriophages. Arch. Virol. 2006, 151, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Tamakoshi, M.; Murakami, A.; Sugisawa, M.; Tsuneizumi, K.; Takeda, S.; Saheki, T.; Izumi, T.; Akiba, T.; Mitsuoka, K.; Toh, H.; et al. Genomic and Proteomic Characterization of the Large Myoviridae Bacteriophage ϕTMA of the Extreme Thermophile Thermus thermophilus. Bacteriophage 2011, 1, 152–164. [Google Scholar] [CrossRef]
- Naryshkina, T.; Liu, J.; Florens, L.; Swanson, S.K.; Pavlov, A.R.; Pavlova, N.V.; Inman, R.; Minakhin, L.; Kozyavkin, S.A.; Washburn, M.; et al. Thermus thermophilus Bacteriophage ϕYS40 Genome and Proteomic Characterization of Virions. J. Mol. Biol. 2006, 364, 667–677. [Google Scholar] [CrossRef]
- Nagayoshi, Y.; Kumagae, K.; Mori, K.; Tashiro, K.; Nakamura, A.; Fujino, Y.; Hiromasa, Y.; Iwamoto, T.; Kuhara, S.; Ohshima, T.; et al. Physiological Properties and Genome Structure of the Hyperthermophilic Filamentous Phage φOH3 Which Infects Thermus thermophilus HB8. Front. Microbiol. 2016, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, I.; Yanase, H. The Genomic Structure of Thermus Bacteriophage ϕIN93. J. Biochem. 2009, 146, 775–785. [Google Scholar] [CrossRef]
- Lin, L.; Hong, W.; Ji, X.; Han, J.; Huang, L.; Wei, Y. Isolation and Characterization of an Extremely Long Tail Thermus Bacteriophage from Tengchong Hot Springs in China. J. Basic. Microbiol. 2010, 50, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.-G.; Jenkins, H.T.; Chechik, M.; Blagova, E.V.; Lopatina, A.; Klimuk, E.; Minakhin, L.; Severinov, K.; Greive, S.J.; Antson, A.A. Viral Genome Packaging Terminase Cleaves DNA Using the Canonical RuvC-like Two-Metal Catalysis Mechanism. Nucleic Acids Res. 2017, 45, 3580–3590. [Google Scholar] [CrossRef] [PubMed]
- Aevarsson, A.; Kaczorowska, A.-K.; Adalsteinsson, B.T.; Ahlqvist, J.; Al-Karadaghi, S.; Altenbuchner, J.; Arsin, H.; Átlasson, Ú.Á.; Brandt, D.; Cichowicz-Cieślak, M.; et al. Going to Extremes—A Metagenomic Journey into the Dark Matter of Life. FEMS Microbiol. Lett. 2021, 368, fnab067. [Google Scholar] [CrossRef]
- Ahlqvist, J.; Linares-Pastén, J.A.; Håkansson, M.; Jasilionis, A.; Kwiatkowska-Semrau, K.; Friðjónsson, Ó.H.; Kaczorowska, A.K.; Dabrowski, S.; Ævarsson, A.; Hreggviðsson, G.Ó.; et al. Crystal Structure and Initial Characterization of a Novel Archaeal-like Holliday Junction-Resolving Enzyme from Thermus thermophilus Phage Tth15-6. Acta Crystallogr. D Struct. Biol. 2022, 78(Pt. 2), 212–227. [Google Scholar] [CrossRef]
- Beese, L.S.; Steitz, T.A. Structural Basis for the 3′-5′ Exonuclease Activity of Escherichia coli DNA Polymerase I: A Two Metal Ion Mechanism. EMBO J. 1991, 10, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, V.; Freemont, P.S.; Sanderson, M.R.; Beese, L.; Friedman, J.M.; Joyce, C.M.; Steitz, T.A. Genetic and Crystallographic Studies of the 3′,5′-Exonucleolytic Site of DNA Polymerase I. Science 1988, 240, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A. DNA Replication Fidelity. J. Biol. Chem. 1992, 267, 18251–18254. [Google Scholar] [CrossRef] [PubMed]
- Bebenek, K.; Joyce, C.M.; Fitzgerald, M.P.; Kunkel, T.A. The Fidelity of DNA Synthesis Catalyzed by Derivatives of Escherichia coli DNA Polymerase I. J. Biol. Chem. 1990, 265, 13878–13887. [Google Scholar] [CrossRef] [PubMed]
- Tindall, K.R.; Kunkel, T.A. Fidelity of DNA Synthesis by the Thermus aquaticus DNA Polymerase. Biochemistry 1988, 27, 6008–6013. [Google Scholar] [CrossRef]
- Gao, Y.; He, Y.; Ivanov, I.; Yang, X.; Tian, H.; Liu, X. Expression and Functional Study of VpV262 Pol, a Moderately Halophilic DNA Polymerase from the Vibrio Parahaemolyticus Phage VpV262. Enzym. Microb. Technol. 2020, 139, 109588. [Google Scholar] [CrossRef]
- Yang, S.; Li, X.; Ding, D.; Hou, J.; Jin, Z.; Yu, X.; Bo, T.; Li, W.; Li, M. A Method for Filling in the Cohesive Ends of Double-Stranded DNA Using Pfu DNA Polymerase. Biotechnol. Appl. Biochem. 2005, 42(Pt. 3), 223–226. [Google Scholar] [CrossRef]
- Hu, G. DNA Polymerase-Catalyzed Addition of Nontemplated Extra Nucleotides to the 3′ of a DNA Fragment. DNA Cell Biol. 1993, 12, 763–770. [Google Scholar] [CrossRef]
- Costa, G.L.; Weiner, M.P. Polishing with T4 or Pfu Polymerase Increases the Efficiency of Cloning of PCR Fragments. Nucleic Acids Res. 1994, 22, 2423. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nojima, H.; Okayama, H. High Efficiency Transformation of Escherichia coli with Plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Mruk, I.; Kaczorowski, T.; Witczak, A. Natural Tuning of Restriction Endonuclease Synthesis by Cluster of Rare Arginine Codons. Sci. Rep. 2019, 9, 5808. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
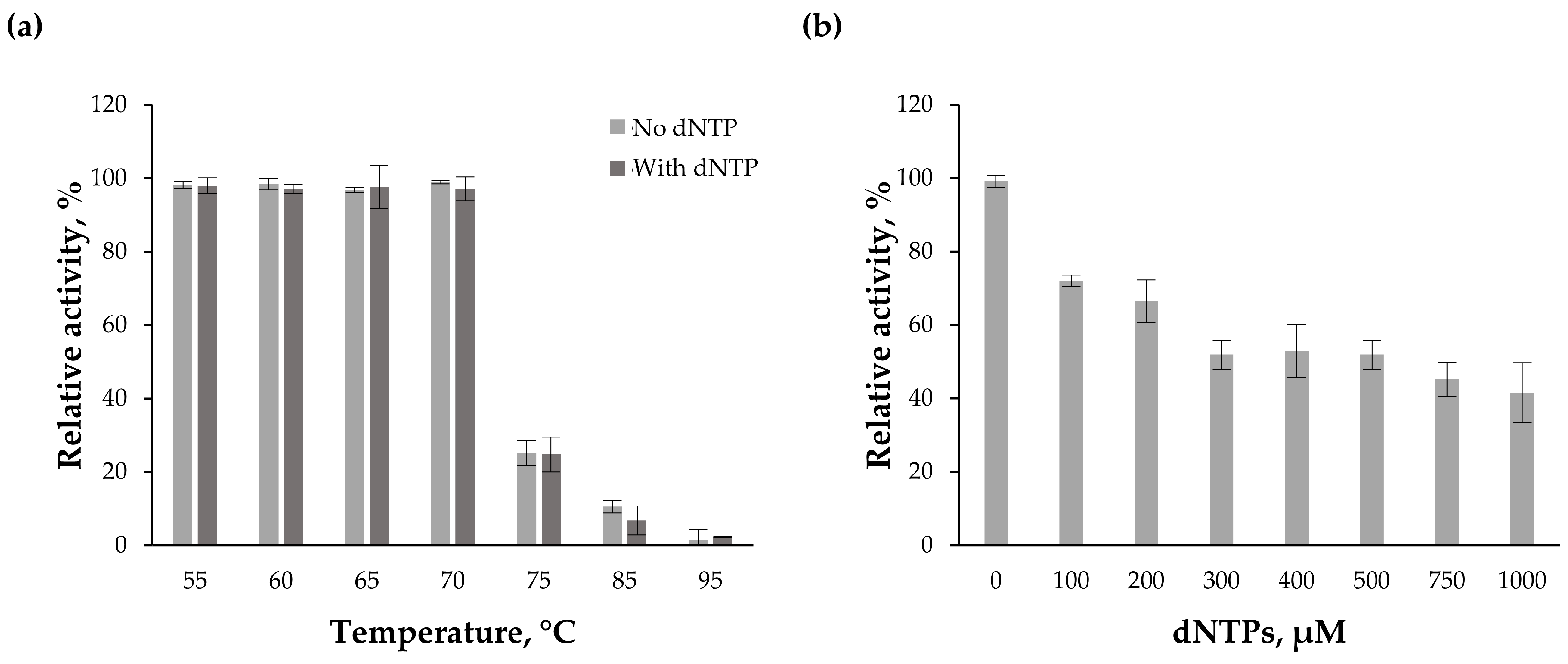

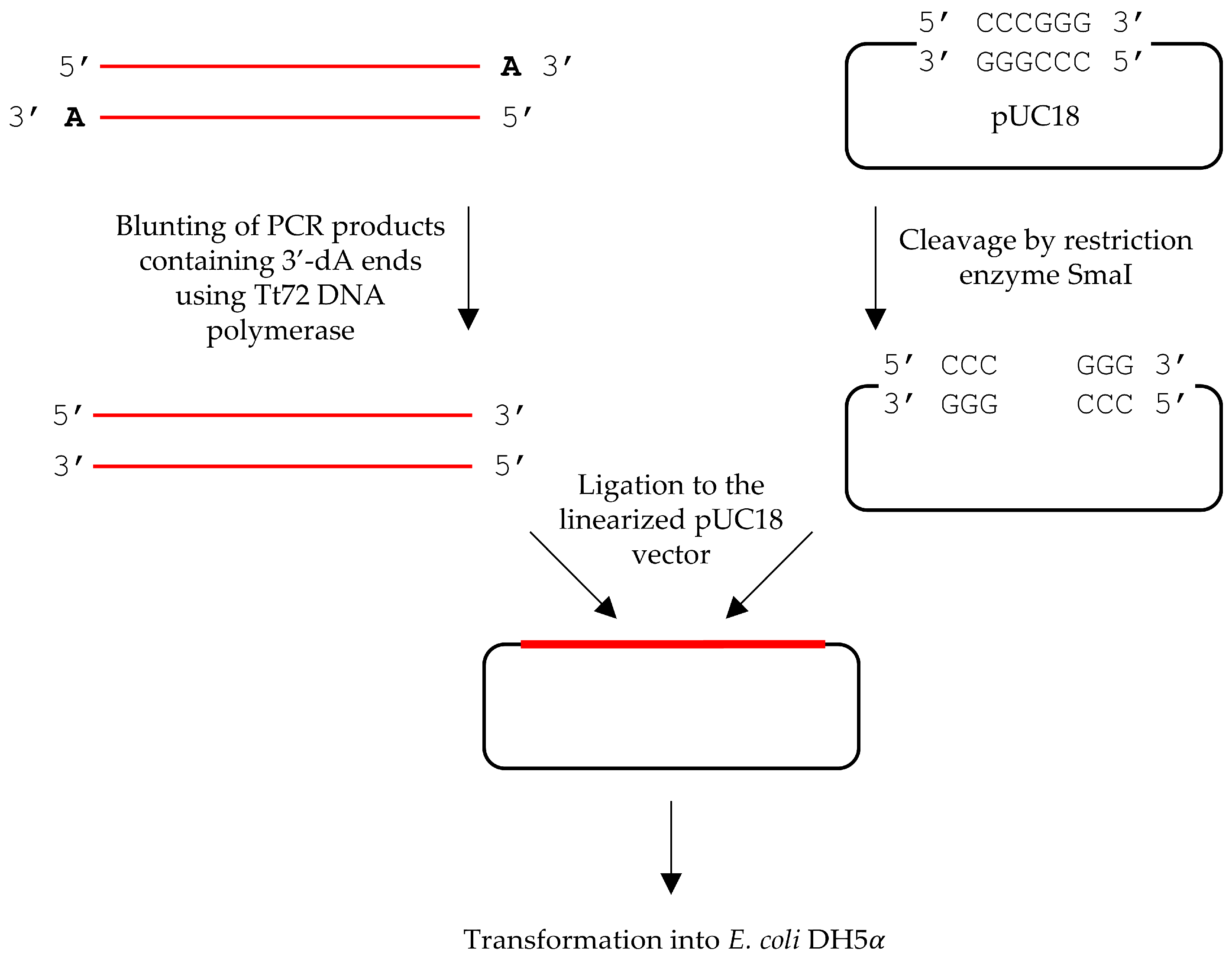
| Polymerase | Total Number of Colonies 1 | Number of White Colonies | Mutation Frequency 2 | Error Rate 3 |
|---|---|---|---|---|
| Tt72 DNA polymerase | 26,884 | 68 | 2.06 × 10−3 | 1.41 × 10−5 |
| Tt72 DNA polymerase exo− (D78A) | 17,682 | 119 | 6.23 × 10−3 | 4.29 × 10−5 |
| Taq DNA polymerase | 39,661 | 266 | 6.24 × 10−3 | 4.27 × 10−5 |
| Pfu DNA polymerase | 28,866 | 30 | 5.72 × 10−4 | 3.91 × 10−6 |
| Polymerase | Number of Colonies 1 | Number of White Colonies 1 | Efficiency (%) 2 |
|---|---|---|---|
| Tt72 DNA polymerase | 14,889 | 12,749 | 85.7 ± 0.90 |
| T4 DNA polymerase | 14,055 | 12,774 | 90.9 ± 0.23 |
| Klenow Fragment | 10,404 | 8237 | 79.8 ± 1.57 |
| Pfu DNA polymerase | 13,947 | 10,649 | 76.6 ± 1.69 |
| Polymerase | Number of Colonies 1 | Number of White Colonies 1 | Efficiency (%) 2 |
|---|---|---|---|
| Tt72 DNA polymerase | 15,468 | 14,944 | 96.6 ± 0.17 |
| T4 DNA polymerase | 12,606 | 12,294 | 97.5 ± 0.04 |
| Klenow Fragment | 10,132 | 7800 | 77.0 ± 1.35 |
| Polymerase | Number of Colonies 1 | Number of White Colonies 1 | Efficiency (%) 2 |
|---|---|---|---|
| Tt72 DNA polymerase | 19,589 | 1458 | 7.4 ± 0.62 |
| T4 DNA polymerase | 10,525 | 797 | 7.6 ± 0.25 |
| Klenow Fragment | 23,912 | 631 | 2.6 ± 0.40 |
| Pfu DNA polymerase | 20,876 | 1100 | 5.3 ± 0.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorawa, S.; Kaczorowski, T. Precise and Accurate DNA-3′/5-Ends Polishing with Thermus thermophilus Phage vb_Tt72 DNA Polymerase. Int. J. Mol. Sci. 2024, 25, 13544. https://doi.org/10.3390/ijms252413544
Dorawa S, Kaczorowski T. Precise and Accurate DNA-3′/5-Ends Polishing with Thermus thermophilus Phage vb_Tt72 DNA Polymerase. International Journal of Molecular Sciences. 2024; 25(24):13544. https://doi.org/10.3390/ijms252413544
Chicago/Turabian StyleDorawa, Sebastian, and Tadeusz Kaczorowski. 2024. "Precise and Accurate DNA-3′/5-Ends Polishing with Thermus thermophilus Phage vb_Tt72 DNA Polymerase" International Journal of Molecular Sciences 25, no. 24: 13544. https://doi.org/10.3390/ijms252413544
APA StyleDorawa, S., & Kaczorowski, T. (2024). Precise and Accurate DNA-3′/5-Ends Polishing with Thermus thermophilus Phage vb_Tt72 DNA Polymerase. International Journal of Molecular Sciences, 25(24), 13544. https://doi.org/10.3390/ijms252413544







