Prognostic Markers in Tyrosine Kinases Specific to Basal-like 2 Subtype of Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Results
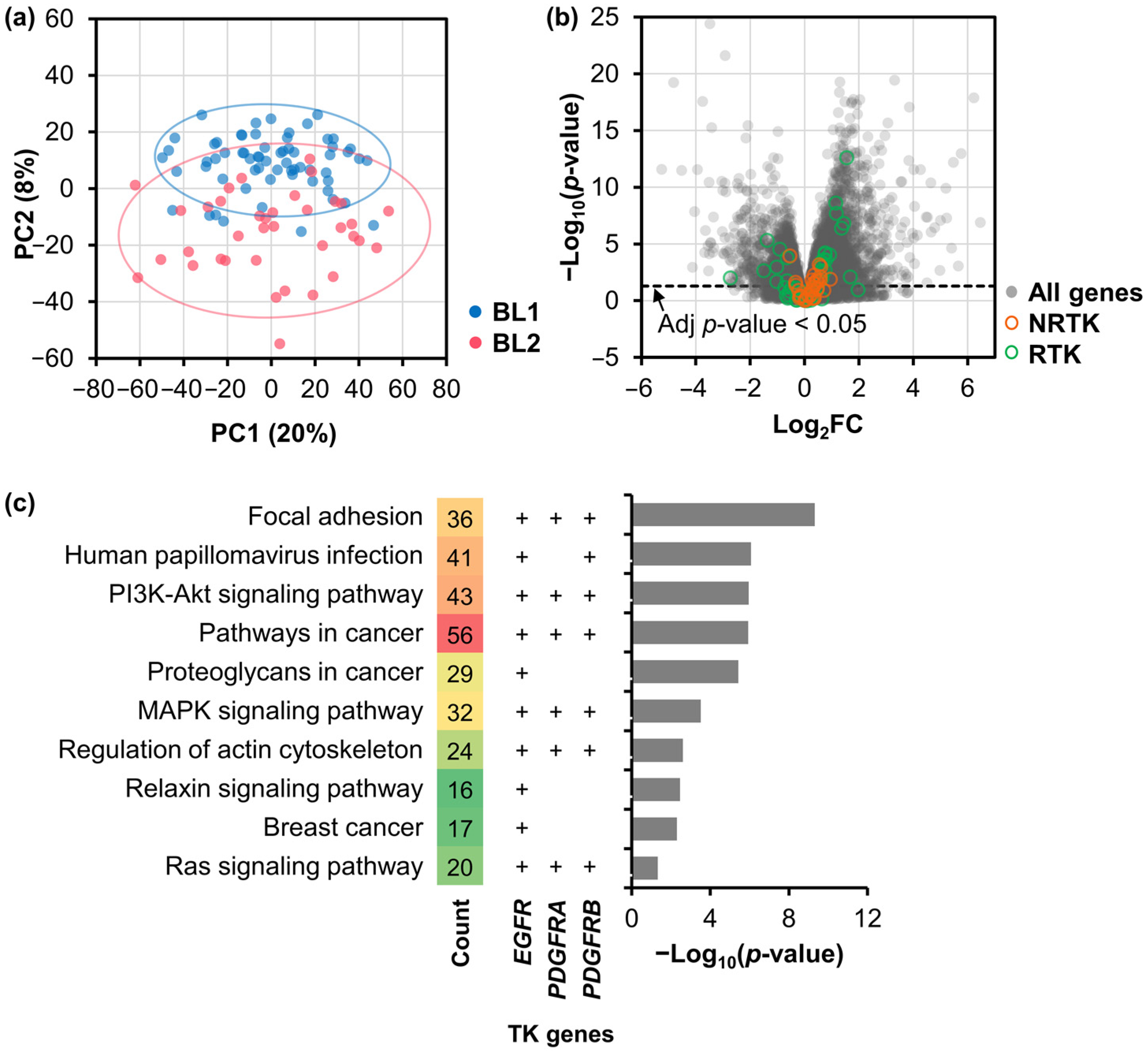
2.1. Differentially Expressed Genes and Their Functional Analysis
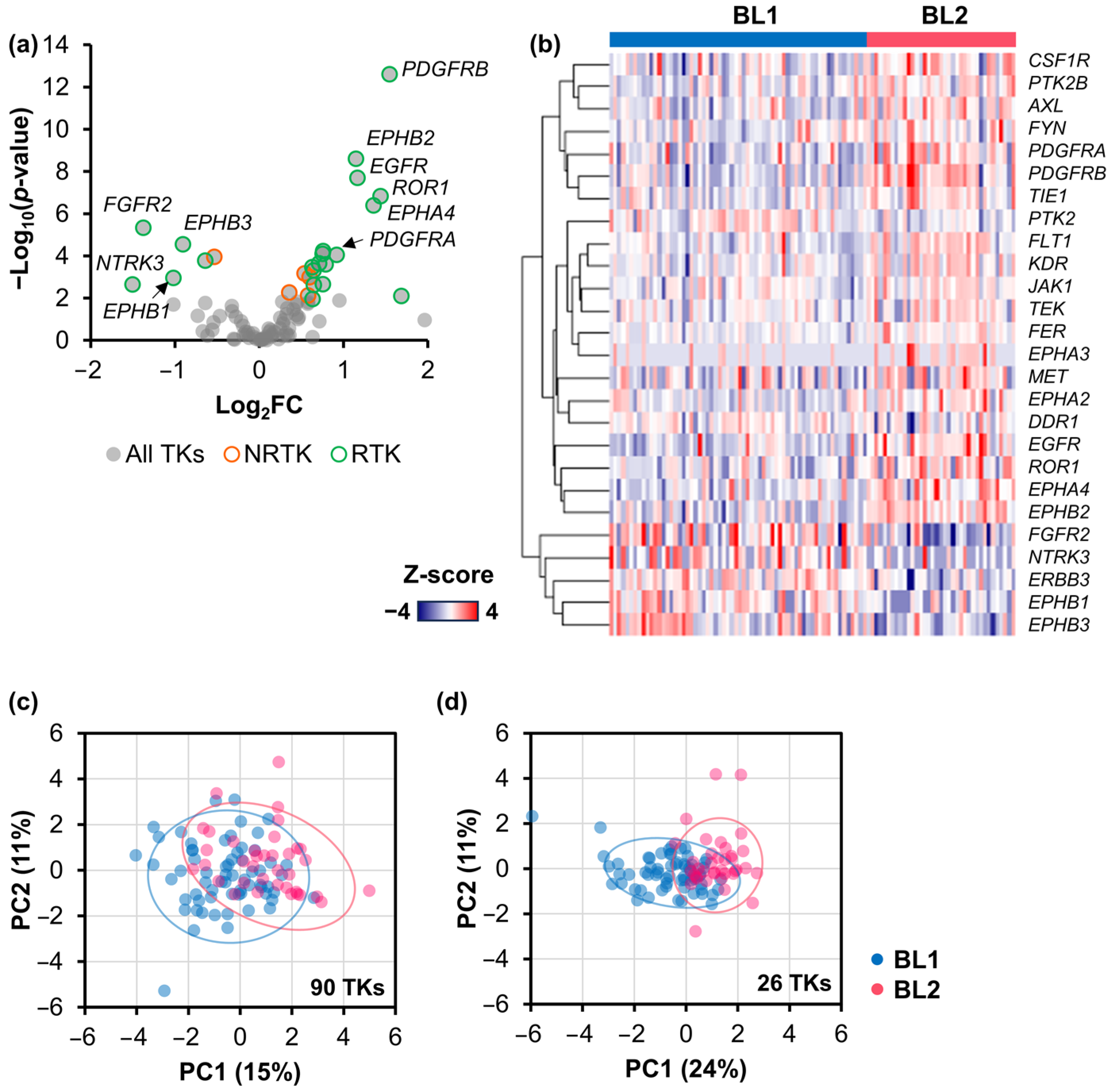
2.2. Signature TK Genes of the BL2 TNBC Subtype
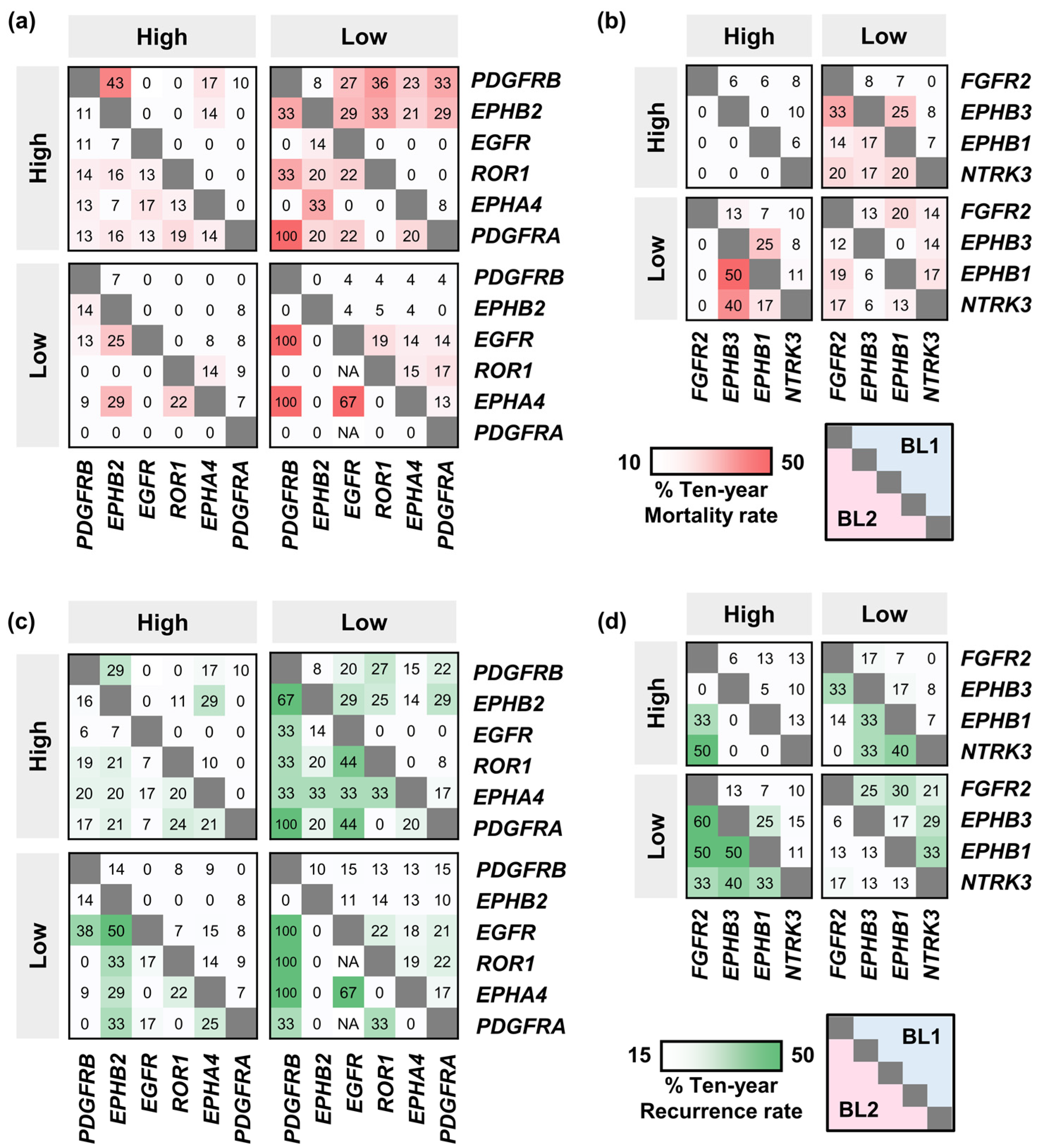
2.3. Mortality and Recurrence Rates of BL2 TNBC Patients Expressing Signature TK Genes
2.4. The Effect of the Co-Occurence of TK Gene Expression on Mortality and Recurrence Rates
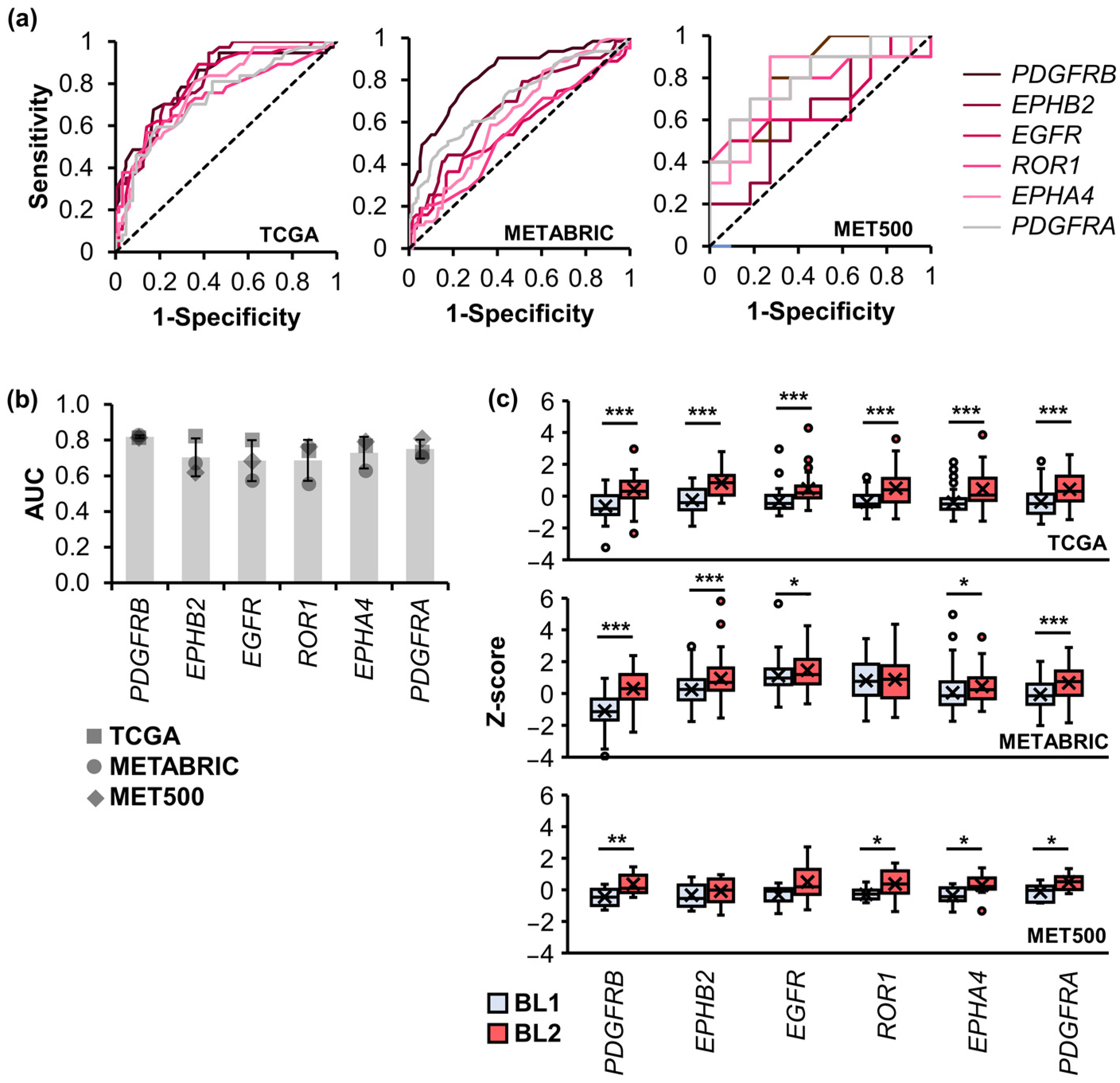
2.5. Validation of Signature TK Genes with TNBC Cohorts
2.6. RT-qPCR Validation of Signature Upregulated TK Genes in TNBC Cell Lines
3. Discussion
4. Materials and Methods
4.1. RNA-Seq Datasets
4.2. Gene Expression Analysis
4.3. Gene Ontology and KEGG Pathway Enrichment Analysis
4.4. Gene Expression Validation
4.5. Mortality and Recurrence Rates
4.6. Classification of Patients with High or Low TK Gene Expression
4.7. RT-qPCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Anders, C.K.; Carey, L.A. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer 2009, 9, S73–S81. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Colaprico, A.; Silva, T.C.; Chen, J.; An, H.; Ban, Y.; Huang, H.; Wang, L.; James, J.L.; Balko, J.M. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun. 2021, 12, 6276. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.I.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Hubalek, M.; Czech, T.; Müller, H. Biological subtypes of triple-negative breast cancer. Breast Care 2017, 12, 8–14. [Google Scholar] [CrossRef]
- Masuda, H.; Baggerly, K.A.; Wang, Y.; Zhang, Y.; Gonzalez-Angulo, A.M.; Meric-Bernstam, F.; Valero, V.; Lehmann, B.D.; Pietenpol, J.A.; Hortobagyi, G.N. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 2013, 19, 5533–5540. [Google Scholar] [CrossRef] [PubMed]
- Limsakul, P.; Choochuen, P.; Charupanit, G.; Charupanit, K. Transcriptomic Analysis of Subtype-Specific Tyrosine Kinases as Triple Negative Breast Cancer Biomarkers. Cancers 2023, 15, 403. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.S.; Van Etten, R.A. Tyrosine kinases as targets for cancer therapy. N. Engl. J. Med. 2005, 353, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Unni, N.; Peng, Y. The changing paradigm for the treatment of HER2-positive breast cancer. Cancers 2020, 12, 2081. [Google Scholar] [CrossRef] [PubMed]
- Iancu, G.; Serban, D.; Badiu, C.D.; Tanasescu, C.; Tudosie, M.S.; Tudor, C.; Costea, D.O.; Zgura, A.; Iancu, R.; Vasile, D. Tyrosine kinase inhibitors in breast cancer. Exp. Ther. Med. 2022, 23, 114. [Google Scholar] [CrossRef] [PubMed]
- Reis-Filho, J.S.; Tutt, A.N.J. Triple negative tumours: A critical review. Histopathology 2008, 52, 108–118. [Google Scholar] [CrossRef]
- Rakha, E.A.; El-Sayed, M.E.; Green, A.R.; Lee, A.H.S.; Robertson, J.F.; Ellis, I.O. Prognostic markers in triple-negative breast cancer. Cancer 2007, 109, 25–32. [Google Scholar] [CrossRef]
- de Nonneville, A.; Finetti, P.; Adelaide, J.; Lambaudie, É.; Viens, P.; Gonçalves, A.; Birnbaum, D.; Mamessier, E.; Bertucci, F. A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers. Cancers 2019, 11, 1158. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S.; Bendahl, P.-O.; Grabau, D.A.; Falck, A.-K.; Fernö, M.; Aaltonen, K.; Ryden, L. The three receptor tyrosine kinases c-KIT, VEGFR2 and PDGFRα, closely spaced at 4q12, show increased protein expression in triple-negative breast cancer. PLoS ONE 2014, 9, e102176. [Google Scholar] [CrossRef]
- Millis, S.Z.; Gatalica, Z.; Winkler, J.; Vranic, S.; Kimbrough, J.; Reddy, S.; O’Shaughnessy, J.A. Predictive biomarker profiling of >6000 breast cancer patients shows heterogeneity in TNBC, with treatment implications. Clin. Breast Cancer 2015, 15, 473–481. [Google Scholar] [CrossRef]
- Gromova, I.; Espinoza, J.A.; Grauslund, M.; Santoni-Rugiu, E.; Møller Talman, M.-L.; van Oostrum, J.; Moreira, J.M.A. Functional Proteomic Profiling of Triple-Negative Breast Cancer. Cells 2021, 10, 2768. [Google Scholar] [CrossRef]
- Cui, N.-P.; Qiao, S.; Jiang, S.; Hu, J.-L.; Wang, T.-T.; Liu, W.-W.; Qin, Y.; Wang, Y.-N.; Zheng, L.-S.; Zhang, J.-C. Protein tyrosine kinase 7 regulates EGFR/Akt signaling pathway and correlates with malignant progression in triple-negative breast cancer. Front. Oncol. 2021, 11, 699889. [Google Scholar] [CrossRef]
- Tryfonopoulos, D.; Walsh, S.; Collins, D.M.; Flanagan, L.; Quinn, C.; Corkery, B.; McDermott, E.W.; Evoy, D.; Pierce, A.; O’Donovan, N. Src: A potential target for the treatment of triple-negative breast cancer. Ann. Oncol. 2011, 22, 2234–2240. [Google Scholar] [CrossRef]
- Costa, R.; Shah, A.N.; Santa-Maria, C.A.; Cruz, M.R.; Mahalingam, D.; Carneiro, B.A.; Chae, Y.K.; Cristofanilli, M.; Gradishar, W.J.; Giles, F.J. Targeting Epidermal Growth Factor Receptor in triple negative breast cancer: New discoveries and practical insights for drug development. Cancer Treat. Rev. 2017, 53, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Hung, M.-C.; Yamaguchi, H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am. J. Cancer Res. 2016, 6, 1609. [Google Scholar] [PubMed]
- Liu, C.-Y.; Huang, T.-T.; Chu, P.-Y.; Huang, C.-T.; Lee, C.-H.; Wang, W.-L.; Lau, K.-Y.; Tsai, W.-C.; Chao, T.-I.; Su, J.-C. The tyrosine kinase inhibitor nintedanib activates SHP-1 and induces apoptosis in triple-negative breast cancer cells. Exp. Mol. Med. 2017, 49, e366. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, T.; Kusumoto, S.; Ando, K.; Ohba, M.; Ohmori, T. Receptor tyrosine kinase-targeted cancer therapy. Int. J. Mol. Sci. 2018, 19, 3491. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Turner, N.C. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann. Oncol. 2019, 30, 1051–1060. [Google Scholar] [CrossRef]
- De Luca, A.; Maiello, M.R.; D’Alessio, A.; Pergameno, M.; Normanno, N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther. Targets 2012, 16, S17–S27. [Google Scholar] [CrossRef]
- Giltnane, J.M.; Balko, J.M. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov. Med. 2014, 17, 275–283. [Google Scholar]
- Linklater, E.S.; Tovar, E.A.; Essenburg, C.J.; Turner, L.; Madaj, Z.; Winn, M.E.; Melnik, M.K.; Korkaya, H.; Maroun, C.R.; Christensen, J.G. Targeting MET and EGFR crosstalk signaling in triple-negative breast cancers. Oncotarget 2016, 7, 69903. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.-M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell 2020, 183, 1436–1456. [Google Scholar] [CrossRef] [PubMed]
- Espinosa Fernandez, J.R.; Eckhardt, B.L.; Lee, J.; Lim, B.; Pearson, T.; Seitz, R.S.; Hout, D.R.; Schweitzer, B.L.; Nielsen, T.J.; Lawrence, O.R. Identification of triple-negative breast cancer cell lines classified under the same molecular subtype using different molecular characterization techniques: Implications for translational research. PLoS ONE 2020, 15, e0231953. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Pietenpol, J.A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 2014, 232, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Trapani, D.; Viale, G.; Criscitiello, C.; Curigliano, G. Practical classification of triple-negative breast cancer: Intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020, 6, 54. [Google Scholar] [CrossRef]
- Kumar, P.; Aggarwal, R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 2016, 293, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Merkher, Y.; Chen, L.; Liu, N.; Leonov, S.; Chen, Y. Recent advances in therapeutic strategies for triple-negative breast cancer. J. Hematol. Oncol. 2022, 15, 121. [Google Scholar] [CrossRef]
- Tam, W.L.; Lu, H.; Buikhuisen, J.; Soh, B.S.; Lim, E.; Reinhardt, F.; Wu, Z.J.; Krall, J.A.; Bierie, B.; Guo, W. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 2013, 24, 347–364. [Google Scholar] [CrossRef]
- Camorani, S.; Hill, B.S.; Collina, F.; Gargiulo, S.; Napolitano, M.; Cantile, M.; Di Bonito, M.; Botti, G.; Fedele, M.; Zannetti, A. Targeted imaging and inhibition of triple-negative breast cancer metastases by a PDGFRβ aptamer. Theranostics 2018, 8, 5178. [Google Scholar] [CrossRef]
- Sun, T.; Aceto, N.; Meerbrey, K.L.; Kessler, J.D.; Zhou, C.; Migliaccio, I.; Nguyen, D.X.; Pavlova, N.N.; Botero, M.; Huang, J. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 2011, 144, 703–718. [Google Scholar] [CrossRef] [PubMed]
- You, K.S.; Yi, Y.W.; Cho, J.; Park, J.-S.; Seong, Y.-S. Potentiating therapeutic effects of epidermal growth factor receptor inhibition in triple-negative breast cancer. Pharmaceuticals 2021, 14, 589. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, S.; Zhao, W.; Qin, S.; Chu, Q.; Wu, K. EGFR-TKIs resistance via EGFR-independent signaling pathways. Mol. Cancer 2018, 17, 53. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Basho, R.K.; Gilcrease, M.; Murthy, R.K.; Helgason, T.; Karp, D.D.; Meric-Bernstam, F.; Hess, K.R.; Herbrich, S.M.; Valero, V.; Albarracin, C. Targeting the PI3K/AKT/mTOR pathway for the treatment of mesenchymal triple-negative breast cancer: Evidence from a phase 1 trial of mTOR inhibition in combination with liposomal doxorubicin and bevacizumab. JAMA Oncol. 2017, 3, 509–515. [Google Scholar] [CrossRef]
- Li, H.; Prever, L.; Hirsch, E.; Gulluni, F. Targeting PI3K/AKT/mTOR signaling pathway in breast cancer. Cancers 2021, 13, 3517. [Google Scholar] [CrossRef]
- Ali, R.; Wendt, M.K. The paradoxical functions of EGFR during breast cancer progression. Signal Transduct. Target. Ther. 2017, 2, 16042. [Google Scholar] [CrossRef]
- Finn, R.S.; Bengala, C.; Ibrahim, N.; Roché, H.; Sparano, J.; Strauss, L.C.; Fairchild, J.; Sy, O.; Goldstein, L.J. Dasatinib as a single agent in triple-negative breast cancer: Results of an open-label phase 2 study. Clin. Cancer Res. 2011, 17, 6905–6913. [Google Scholar] [CrossRef]
- Curigliano, G.; Pivot, X.; Cortés, J.; Elias, A.; Cesari, R.; Khosravan, R.; Collier, M.; Huang, X.; Cataruozolo, P.E.; Kern, K.A. Randomized phase II study of sunitinib versus standard of care for patients with previously treated advanced triple-negative breast cancer. Breast 2013, 22, 650–656. [Google Scholar] [CrossRef]
- Sun, S.; Jiang, Y.; Zhang, G.; Song, H.; Zhang, X.; Zhang, Y.; Liang, X.; Sun, Q.; Pang, D. Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J. Surg. Oncol. 2012, 105, 773–779. [Google Scholar] [CrossRef]
- Lee, H.J.; Seo, A.N.; Park, S.Y.; Kim, J.Y.; Park, J.Y.; Yu, J.H.; Ahn, J.-H.; Gong, G. Low prognostic implication of fibroblast growth factor family activation in triple-negative breast cancer subsets. Ann. Surg. Oncol. 2014, 21, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Routh, E.D.; Pullikuth, A.; Jin, G.; Su, J.; Chou, J.W.; Hoadley, K.A.; Print, C.; Knowlton, N.; Black, M.A. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology 2018, 7, e1490854. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chang, I.-Y.; You, H.J. Interactions between EGFR and EphA2 promote tumorigenesis through the action of Ephexin1. Cell Death Dis. 2022, 13, 528. [Google Scholar] [CrossRef] [PubMed]
- El Zawily, A.; Vizeacoumar, F.S.; Dahiya, R.; Banerjee, S.L.; Bhanumathy, K.K.; Elhasasna, H.; Hanover, G.; Sharpe, J.C.; Sanchez, M.G.; Greidanus, P. A Multipronged Unbiased Strategy Guides the Development of an Anti-EGFR/EPHA2–Bispecific Antibody for Combination Cancer Therapy. Clin. Cancer Res. 2023, 29, 2686–2701. [Google Scholar] [CrossRef]
- Broekman, F.; Giovannetti, E.; Peters, G.J. Tyrosine kinase inhibitors: Multi-targeted or single-targeted? World J. Clin. Oncol. 2011, 2, 80. [Google Scholar] [CrossRef]
- Shyam Sunder, S.; Sharma, U.C.; Pokharel, S. Adverse effects of tyrosine kinase inhibitors in cancer therapy: Pathophysiology, mechanisms and clinical management. Signal Transduct. Target. Ther. 2023, 8, 262. [Google Scholar] [CrossRef]
- Hantschel, O. Unexpected off-targets and paradoxical pathway activation by kinase inhibitors. ACS Chem. Biol. 2015, 10, 234–245. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer cell lines are useful model systems for medical research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, M.; Wu, L.; Yang, H.; Yao, Y.; Yang, Q.; Du, J.; Liu, L.; Li, Y.; Bai, Y. Stromal cells in the tumor microenvironment: Accomplices of tumor progression? Cell Death Dis. 2023, 14, 587. [Google Scholar] [CrossRef]
- Alkasalias, T.; Moyano-Galceran, L.; Arsenian-Henriksson, M.; Lehti, K. Fibroblasts in the tumor microenvironment: Shield or spear? Int. J. Mol. Sci. 2018, 19, 1532. [Google Scholar] [CrossRef]
- Bai, F.; Liu, S.; Liu, X.; Hollern, D.P.; Scott, A.; Wang, C.; Zhang, L.; Fan, C.; Fu, L.; Perou, C.M. PDGFRβ is an essential therapeutic target for BRCA1-deficient mammary tumors. Breast Cancer Res. 2021, 23, 10. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef]
- Abdolahi, S.; Ghazvinian, Z.; Muhammadnejad, S.; Saleh, M.; Asadzadeh Aghdaei, H.; Baghaei, K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022, 20, 206. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Rueda, O.M.; Sammut, S.-J.; Seoane, J.A.; Chin, S.-F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef]


| Condition 1 | Condition 2 | p-Value | |
|---|---|---|---|
| Mortality | Recurrence | ||
| FGFR2 H/EPHB3 L | FGFR2 L/EPHB3 L | ns | 0.024 |
| EGFR H/NTRK3 L | EGFR L/NTRK3 L | ns | 0.028 |
| EGFR H/EPHB1 L | EGFR L/EPHB1 L | ns | 0.032 |
| EGFR H/EPHB2 H | EGFR L/EPHB2 H | ns | 0.039 |
| EGFR H/EPHA4 L | EGFR L/EPHA4 L | 0.045 | 0.045 |
| EGFR H/FGFR2 L | EGFR L/FGFR2 H | ns | 0.046 |
| EGFR H/ROR1 H | EGFR L/ROR1 H | ns | 0.047 |
| EGFR H/PDGFRA H | EGFR L/PDGFRA H | ns | 0.047 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Limsakul, P.; Choochuen, P.; Jungrungrueang, T.; Charupanit, K. Prognostic Markers in Tyrosine Kinases Specific to Basal-like 2 Subtype of Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2024, 25, 1405. https://doi.org/10.3390/ijms25031405
Limsakul P, Choochuen P, Jungrungrueang T, Charupanit K. Prognostic Markers in Tyrosine Kinases Specific to Basal-like 2 Subtype of Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2024; 25(3):1405. https://doi.org/10.3390/ijms25031405
Chicago/Turabian StyleLimsakul, Praopim, Pongsakorn Choochuen, Thawirasm Jungrungrueang, and Krit Charupanit. 2024. "Prognostic Markers in Tyrosine Kinases Specific to Basal-like 2 Subtype of Triple-Negative Breast Cancer" International Journal of Molecular Sciences 25, no. 3: 1405. https://doi.org/10.3390/ijms25031405
APA StyleLimsakul, P., Choochuen, P., Jungrungrueang, T., & Charupanit, K. (2024). Prognostic Markers in Tyrosine Kinases Specific to Basal-like 2 Subtype of Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 25(3), 1405. https://doi.org/10.3390/ijms25031405






