Overexpression of a ‘Beta’ MYB Factor Gene, VhMYB15, Increases Salinity and Drought Tolerance in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
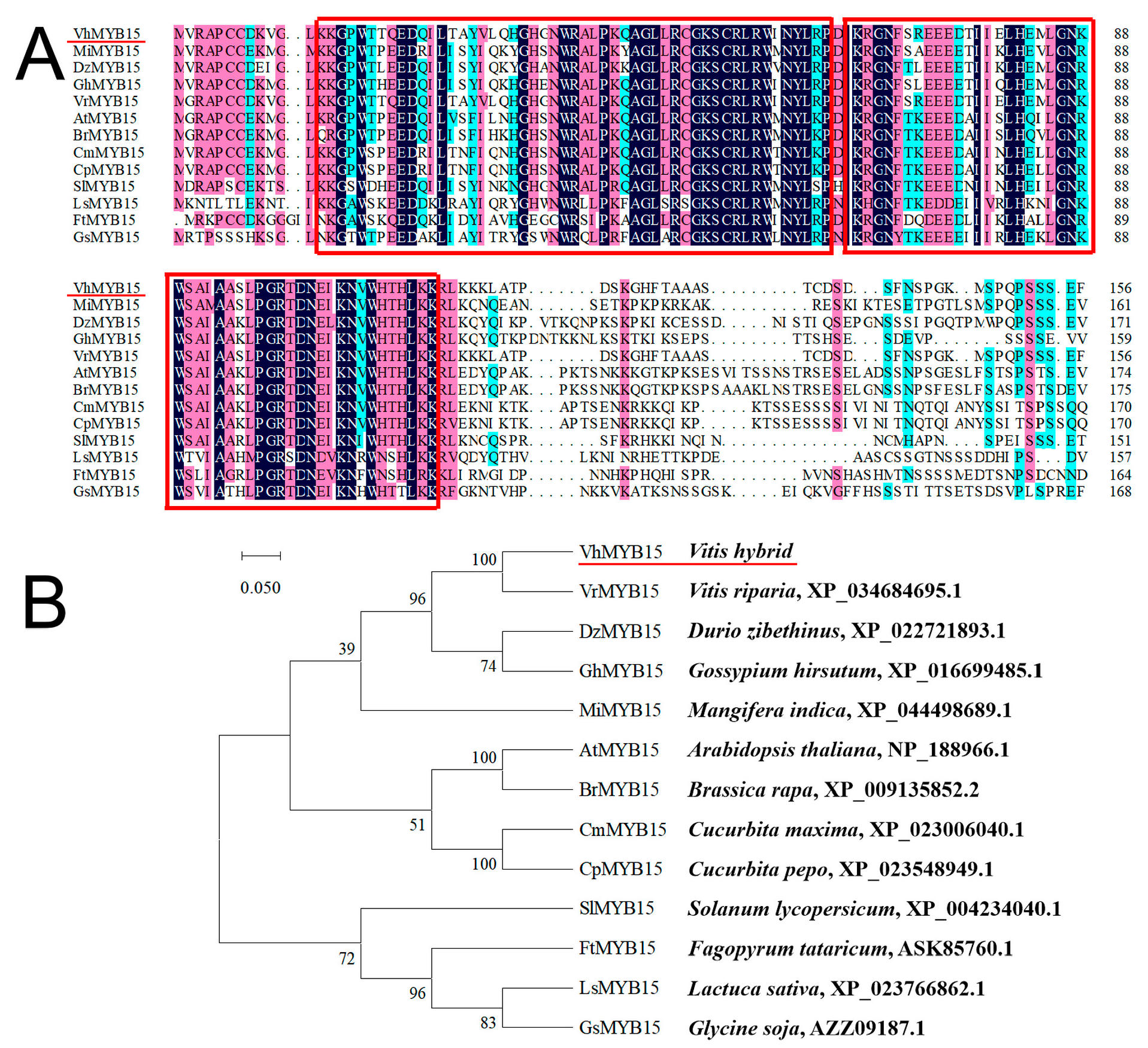
2.1. Isolation and Bioinformatics Analysis of VhMYB15
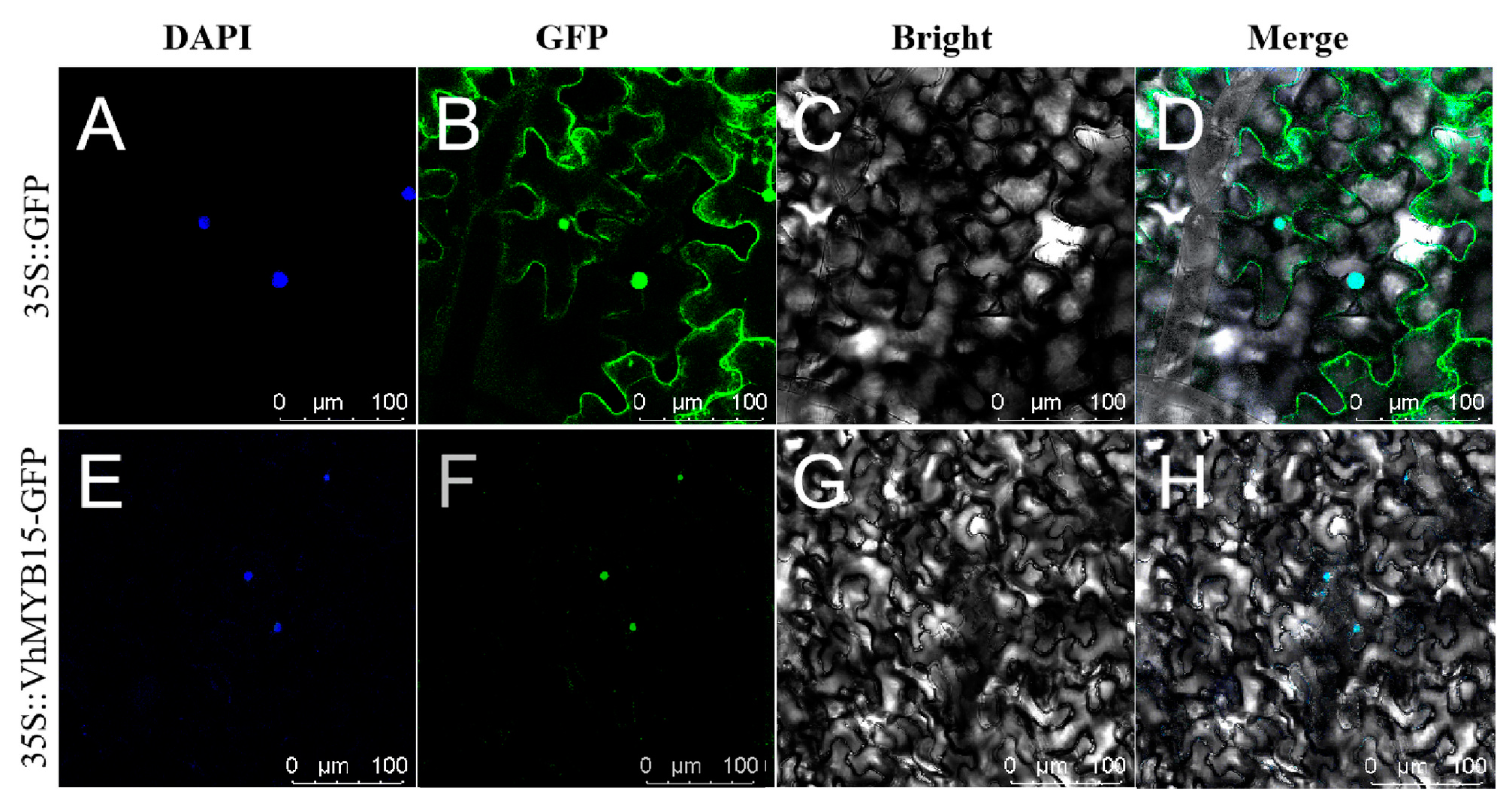
2.2. Subcellular Localization of VhMYB15 Protein
2.3. Analysis of the Expression Characteristics of VhMYB15 in ‘Beta’
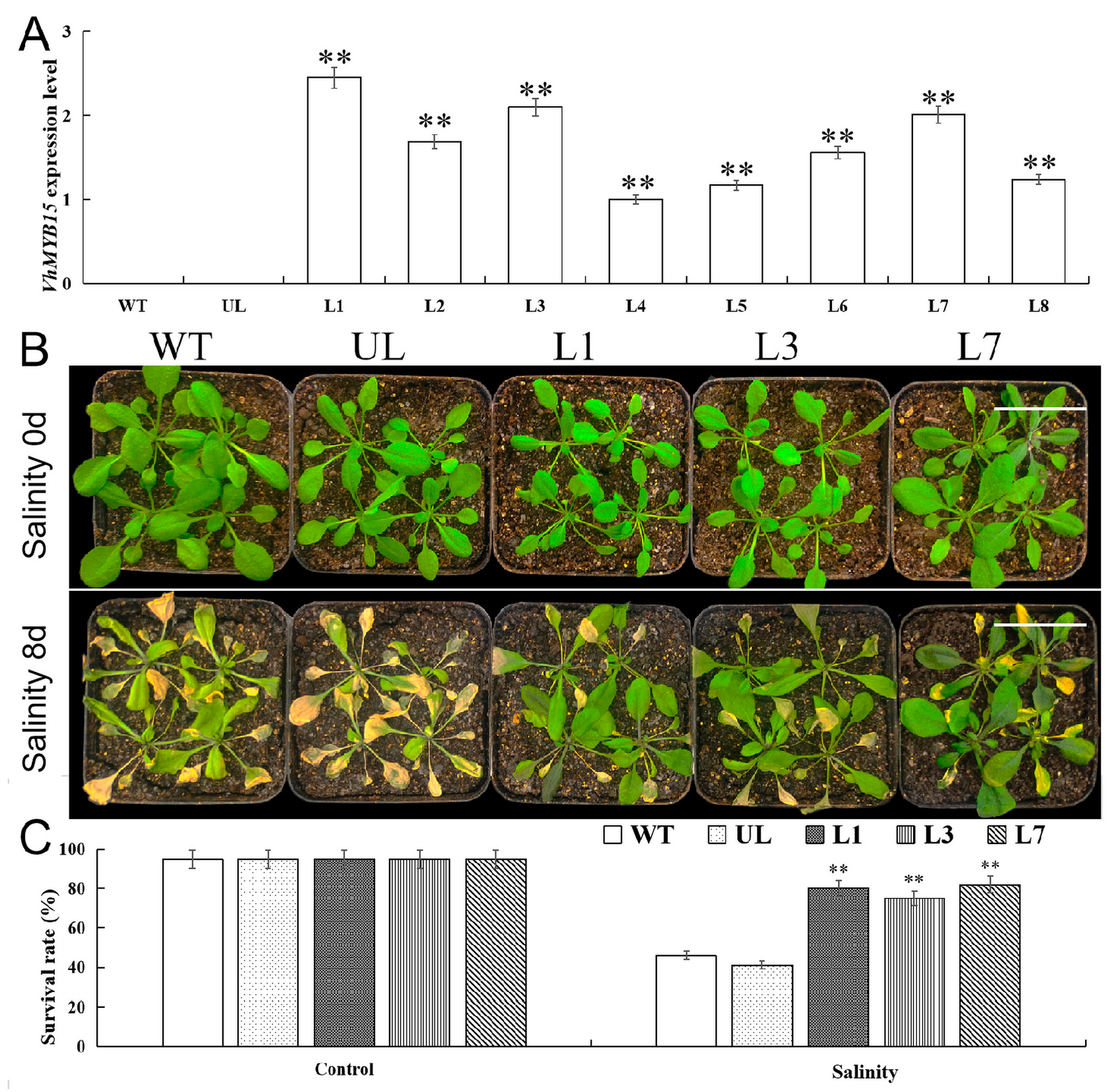
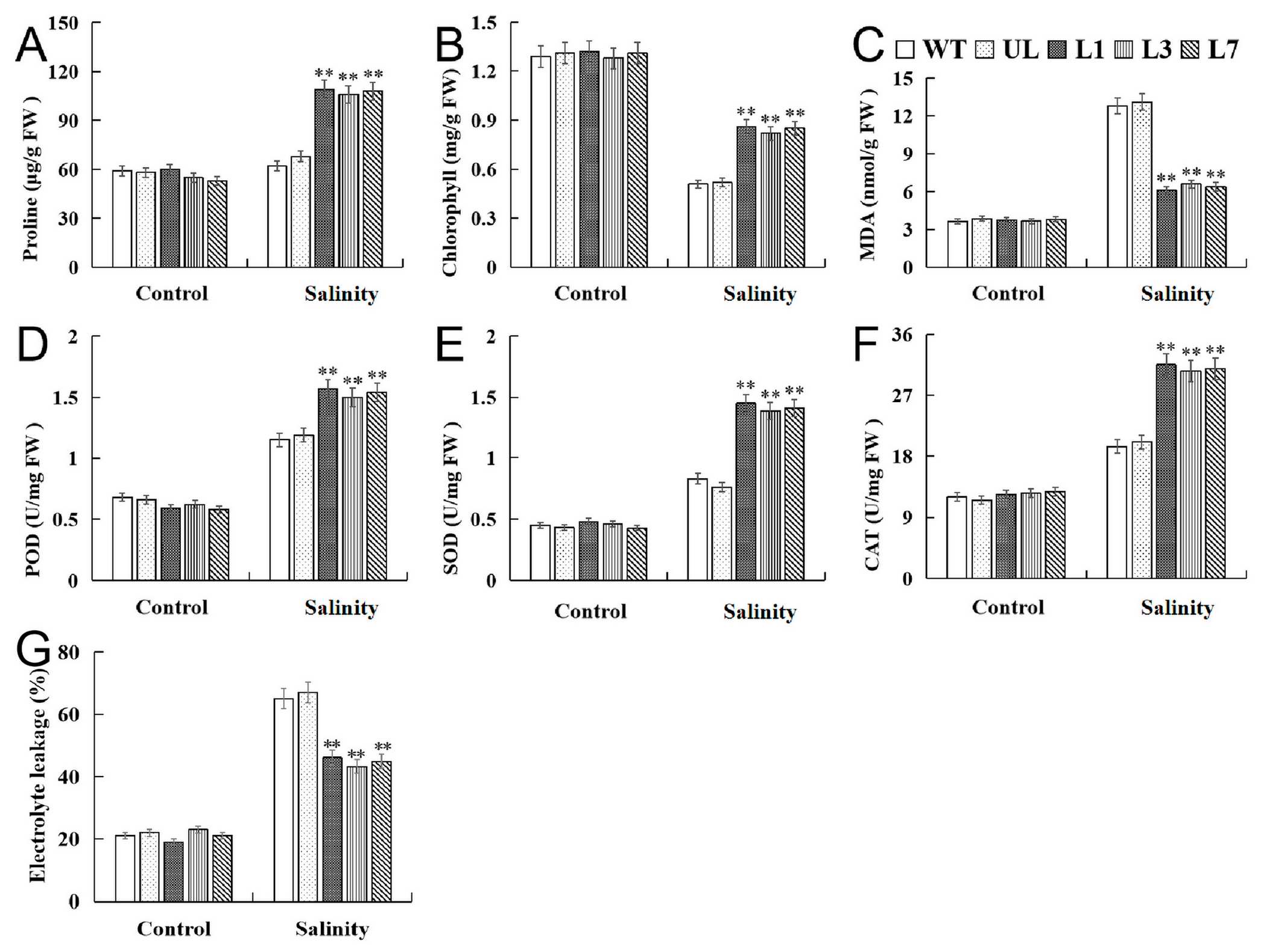
2.4. Heterologous Expression of VhMYB15 in Arabidopsis Improved Salinity Tolerance
2.5. VhMYB15 in Transgenic Arabidopsis Activated Salinity Tolerance-Related Genes
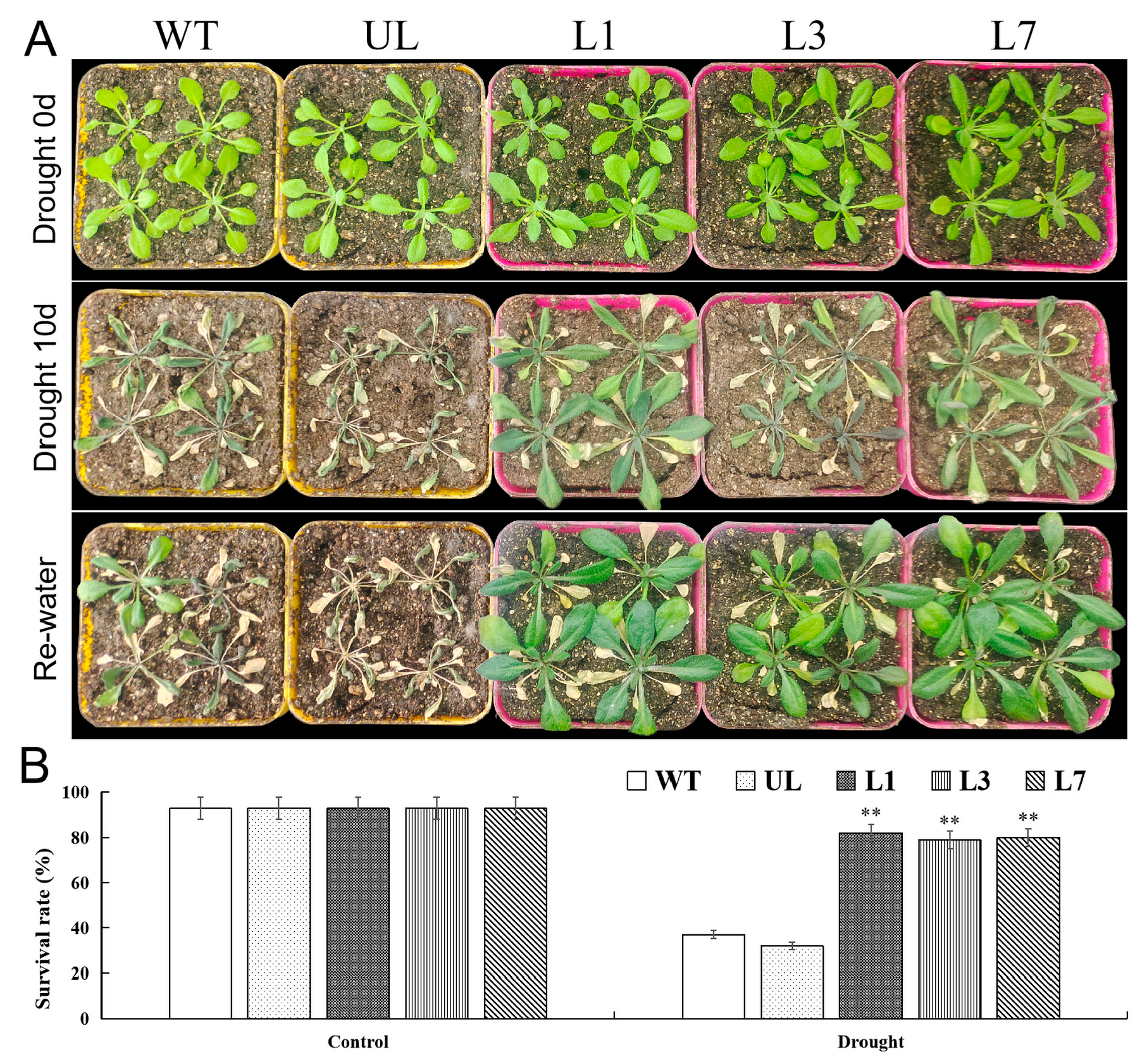
2.6. Heterologous Expression of VhMYB15 in Arabidopsis Improved Drought Tolerance
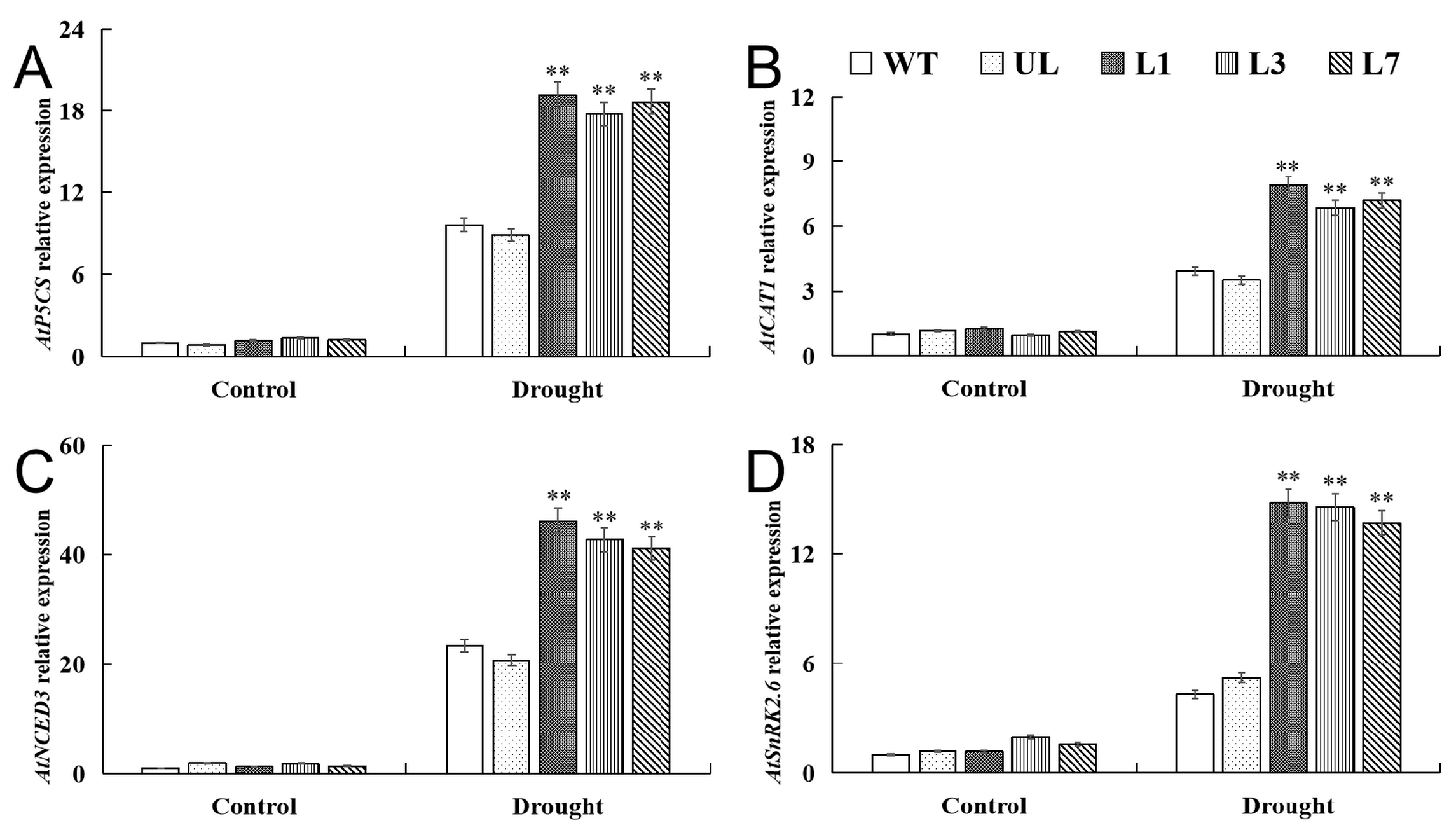
2.7. VhMYB15 in Transgenic Arabidopsis Activated Drought Tolerance-Related Genes
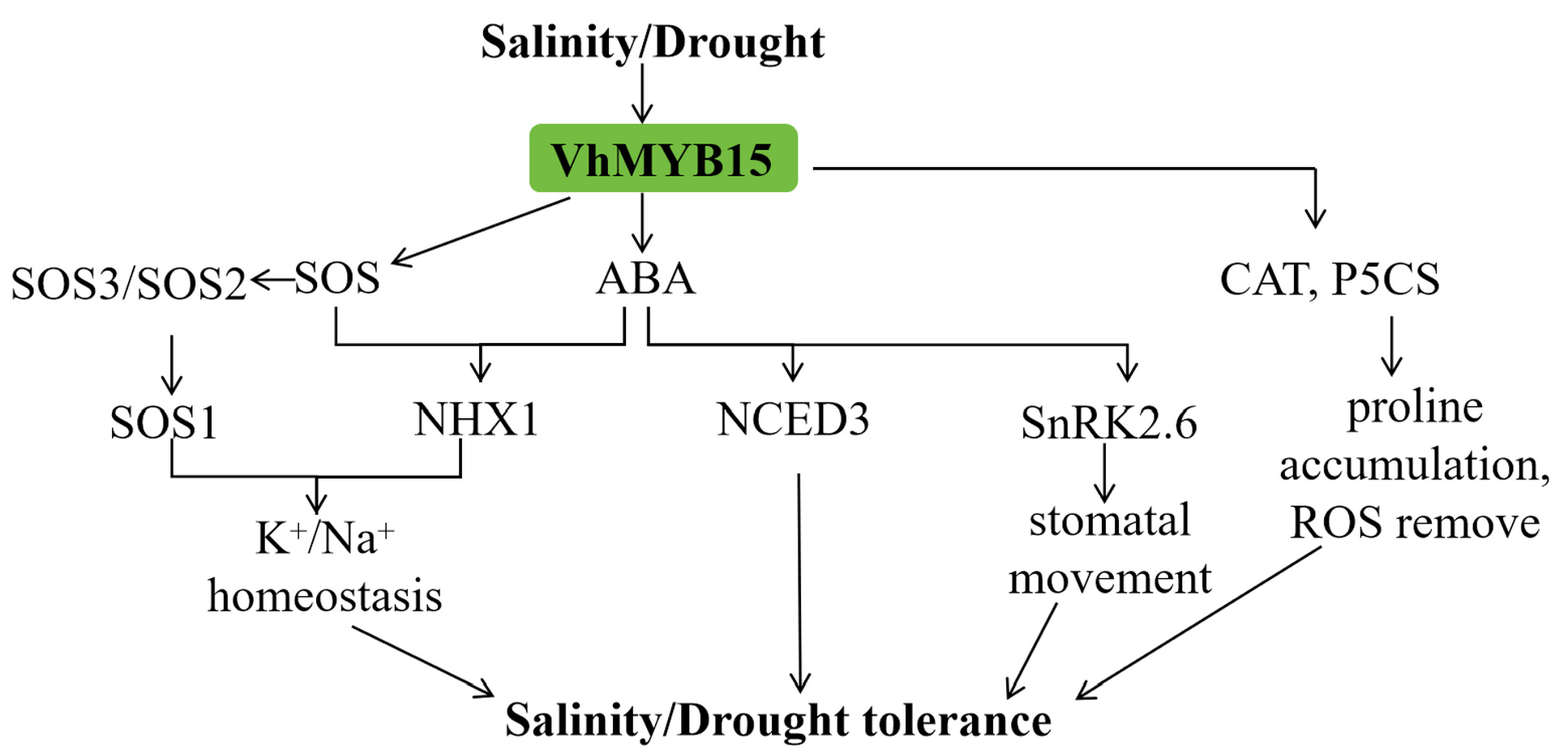
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Cloning and Bioinformatic Analysis of VhMYB15
4.3. Vector Construction and Subcellular Localization of VhMYB15
4.4. Expression Analysis of VhMYB15
4.5. Generation of Transgenic Lines
4.6. Analysis of Related Physiological Indexes in VhMYB15-Overexpressing Arabidopsis
4.7. Expression Analysis of Stress-Related Genes in VhMYB15-Overexpressing Arabidopsis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, D.; Wang, L.; Wang, Y.; Yang, G.; Gao, C.; Yu, Z.; Li, T.; Zhang, X.; Ma, L.; Xu, X.; et al. Overexpression of Malus xiaojinensis CS1 gene in tobacco affects plant development and increases iron stress tolerance. Sci. Hortic. 2013, 150, 65–72. [Google Scholar] [CrossRef]
- Zepeda, B.; Marcelis, L.F.M.; Kaiser, E.; Verdonk, J.C. Petunia as a model for MYB transcription factor action under salt stress. Front. Plant Sci. 2024, 14, 1286547. [Google Scholar] [CrossRef]
- Zörb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and crop yield. Plant Biol. 2019, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a myb family gene, Osmyb6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Q.; Yan, C.; Sun, Q.; Wang, J.; Li, C.; Yuan, C.; Mou, Y.; Shan, S. The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut. Int. J. Biol. Macromol. 2024, 256, 128492. [Google Scholar] [CrossRef]
- Wang, Z.; Mao, Y.; Guo, Y.; Gao, J.; Liu, X.; Li, S.; Lin, Y.C.; Chen, H.; Wang, J.; Chiang, V.; et al. MYB transcription factor161 mediates feedback regulation of secondary wallassociated NAC-Domain1 family genes for wood formation. Plant Physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB transcription factors as regulators of secondary metabolism in plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Ai, P.; Xue, J.; Shi, Z.; Liu, Y.; Li, Z.; Li, T.; Zhao, W.; Khan, M.A.; Kang, D.; Wang, K.; et al. Genome-wide characterization and expression analysis of MYB transcription factors in Chrysanthemum nankingense. BMC Plant Biol. 2023, 23, 140. [Google Scholar] [CrossRef]
- Chen, N.; Yang, Q.; Pan, L.; Chi, X.; Chen, M.; Hu, D.; Yang, Z.; Wang, T.; Wang, M.; Yu, S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Gene 2014, 533, 332–345. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, R.; Yang, X.; Ju, Q.; Li, W.; Lu, S.; Tran, L.S.P.; Xu, J. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 2020, 43, 1925–1943. [Google Scholar] [CrossRef]
- Chen, T.Z.; Li, W.J.; Hu, X.H.; Guo, J.R.; Liu, A.M.; Zhang, B.L. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef]
- Cheng, L.; Li, X.; Huang, X.; Ma, T.; Liang, Y.; Ma, X.; Peng, X.; Jia, J.; Chen, S.; Chen, Y. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 70, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.C.J.; Schlechter, R.; Vannozzi, A.; Holl, J.; Hmmam, I.; Bogs, J.; Tornielli, G.B.; Castellarin, S.D.; Matus, J.T. A systemsoriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 2016, 23, 451–466. [Google Scholar] [CrossRef]
- Jessica, A.R.; Richard, V.E.; Andrew, C.A. Genomic analysis uncovers functional variation in the C-terminus of anthocyaninactivating MYB transcription factors. Hortic. Res. 2021, 8, 77. [Google Scholar]
- Zhang, X.; Ma, W.; Guan, X.; Wang, F.; Fan, Z.; Gao, S.; Yao, Y. VvMYB14 participates in melatonin-induced proanthocyanidin biosynthesis by upregulating expression of VvMYBPA1 and VvMYBPA2 in grape seeds. Hortic. Res. 2023, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Vialet, S.; Verriès, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar]
- Amato, A.; Cavallini, E.; Walker, A.R.; Pezzotti, M.; Bliek, M.; Quattrocchio, F.; Koes, R.; Ruperti, B.; Bertini, E.; Zenoni, S.; et al. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 2019, 99, 1220–1241. [Google Scholar] [CrossRef] [PubMed]
- Poudel, P.R.; Koyama, K.; Goto-Yamamoto, N. Evaluating the influence of temperature on proanthocyanidin biosynthesis in developing grape berries (Vitis vinifera L.). Mol. Biol. Rep. 2020, 47, 3501–3510. [Google Scholar] [CrossRef]
- Yu, Y.; Guo, D.; Li, G.; Yang, Y.; Zhang, G.; Li, S.; Liang, Z. The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (Vd STS2). BMC Plant Biol. 2019, 19, 478. [Google Scholar] [CrossRef]
- Gao, Z.; Gao, L.; You, J.; Zhou, L.; Du, Z. Comparison of drought resistance of nine wine grape varieties grafted ‘Beida’ rootstocks. Mol. Plant Breed. 2021, 19, 3473–3480. [Google Scholar]
- Ma, X.; Ran, J.; Mei, G.; Hou, X.; You, X. Cloning and functional analysis of NoMYB60 gene involved in flavonoid biosynthesis in watercress (Nasturtium officinale R. Br.). Genes 2022, 13, 2109. [Google Scholar] [CrossRef] [PubMed]
- Piao, W.; Sakuraba, Y.; Paek, N.C. Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thalian. BMB Rep. 2019, 52, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Sun, L.; Yu, P.; Yang, Z.; Zhang, P.; Zhang, Y.; Wu, W.; Chen, D.; Zhan, X.; Khan, R. The MYB transcription factor Baymax1 plays a critical role in rice male fertility. Theor. Appl. Genet. 2021, 134, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Wang, Y.; Kuzma, M.; Chalifoux, M.; Tremblay, L.; Yang, S.; Ying, J.; Sample, A.; Wang, H.M.; Griffiths, R.; et al. Activation tagging identifies Arabidopsis transcription factor AtMYB68 for heat and drought tolerance at yield determining reproductive stages. Plant J. 2020, 104, 1535–1550. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Han, J.; Xu, T.; Li, X.; Yao, C.; Li, T.; Sun, X.; Wang, X.; Yang, G. Overexpression of MbERF12, an ERF gene from Malus baccata (L.) Borkh increases cold and salt tolerance in Arabidopsis thaliana associated with the ROS scavenging through ethylene signal transduction. In Vitr. Cell. Dev. Biol.-Plant 2021, 57, 760–770. [Google Scholar] [CrossRef]
- Leng, B.; Wang, X.; Yuan, F.; Zhang, H.; Lu, C.; Chen, M.; Wang, B. Heterologous expression of the Limonium bicolor MYB transcription factor LbTRY in Arabidopsis thaliana increases salt sensitivity by modifying root hair development and osmotic homeostasis. Plant Sci. 2021, 302, 110704. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, R.; Imran, M.; Amanullah, S.; Rothenberg, D.O.; Wang, Q.; Wang, L.; Fan, Y. Functional characterization of Hedychium coronarium J. Koenig MYB132 confers the potential role in floral aroma synthesis. Plants 2021, 10, 2014. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, M.; Zhang, X.; Liu, Y.; Xue, F.; Sun, J.; Li, Y. Cloning and expression analysis of GhMYB113 gene in Gossypium hirsutum. Acta Bot. Boreali-Occident. Sin. 2013, 33, 878–884. [Google Scholar]
- Dai, Y.; Lu, Y.; Chen, Y.; Gao, Y. Cloning and characterization of ZmMYB002, a MYB transcription factor gene responsive to abiotic stresses in maize (Zea mays L.). J. Yangzhou Univ. 2013, 34, 60–63. [Google Scholar]
- Liang, X.; Luo, G.; Li, W.; Yao, A.; Liu, W.; Xie, L.; Han, M.; Li, X.; Han, D. Overexpression of a Malus baccata CBF transcription factor gene, MbCBF1, increases cold and salinity tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2022, 192, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, K.; Jia, X.; Fu, C.; Yu, H.; Wang, Y. Antioxidant peptides, the guardian of life from oxidative stress. Med. Res. Rev. 2024, 44, 275–364. [Google Scholar] [CrossRef]
- Hou, Q.; Shen, T.; Yu, R.; Deng, H.; Wen, X.; Qiao, G. Functional analysis of sweet cherry PavbHLH106 in the regulation of cold stress. Plant Cell Rep. 2024, 43, 7. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Zhang, H.; Shen, Y.; Cui, Y.; Luo, P. ERF54 regulates cold tolerance in Rosa multiflora through DREB/COR signalling pathways. Plant Cell Environ. 2024. [Google Scholar] [CrossRef]
- Zhou, X.; Lei, D.; Yao, W.; Li, S.; Wang, H.; Lu, J.; Zhang, Y.; Lin, Y.; Wang, Y.; He, W.; et al. A novel R2R3-MYB transcription factor PbMYB1L of Pyrus bretschneideri regulates cold tolerance and anthocyanin accumulation. Plant Cell Rep. 2024, 43, 34. [Google Scholar] [CrossRef]
- Jiao, C.; Sun, J.; Wei, Y. SlWRKY31 enhances chilling tolerance by interacting with SlSIZ1 in tomato fruit. Postharvest Biol. Technol. 2024, 207, 112631. [Google Scholar] [CrossRef]
- Dounavi, A.; Netzer, F.; Celepirovic, N.; Ivankovic, M.; Burger, J.; Figueroa, A.G.; Schön, S.; Simon, J.; Cremer, E.; Fussi, B.; et al. Genetic and physiological differences of European beech provenances (F-sylvatica L.) exposed to drought stress. For. Ecol. Manag. 2016, 361, 226–236. [Google Scholar] [CrossRef]
- Luo, T.; Pei, Y. Physiological responses of Vitellaria paradors seedlings to different water stress intensity. J. West China For. Sci. 2020, 49, 21–27. [Google Scholar]
- Jiang, X.; Mu, L.; Wang, X.; Xu, W. Physiological responses of three kinds of slope protection shrubs to drought stress. Pratacultural Sci. 2013, 30, 678–686. [Google Scholar]
- Gong, Z.; Koiwa, H.; Cushman, M.A.; Ray, A.; Bufford, D.; Koreeda, S.; Matsumoto, T.K.; Zhu, J.; Cushman, J.C.; Bressan, R.A.; et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001, 126, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.K.; Song, R.F.; Guo, J.X.; Zhang, Y.; Zuo, J.X.; Chen, H.H.; Liao, C.Y.; Hu, X.Y.; Ren, F.; Lu, Y.T.; et al. CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell 2023, 35, 2570–2591. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fu, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na+/H+ antiporter salt-overly-sensitive 1 (AtSOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef]
- Madadi, K.; Ahmadabadi, M.; Pazhouhandeh, M. Heterologous expression of Arabidopsis SOS3 increases salinity tolerance in Petunia. Mol. Biol. Rep. 2022, 49, 6553–6562. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis AtSOS2 protein kinase physically interacts with and is activated by the calcium-binding protein AtSOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of AtSOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by AtSOS2 and AtSOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wan, S.; Jiang, Y.; Xia, Y.Q.; Chen, X.H.; Gao, M.Z.; Cao, Y.X.; Luo, Y.H.; Zhou, Y.; Jiang, X.Y. Over-expression of a plasma membrane H+-ATPase SpAHA1 conferred salt tolerance to transgenic Arabidopsis. Protoplasma 2018, 255, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Yarra, R.; Kirti, P.B. Expressing class I wheat NHX (TaNHX2) gene in eggplant (Solanum melongena L.) improves plant performance under saline condition. Funct. Integr. Genom. 2019, 19, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Tian, X.X.; Mao, P.C.; Meng, L. Overexpression of Iris lactea tonoplast Na+/H+ antiporter gene IlNHX confers improved salt tolerance in tobacco. Biol. Plant. 2020, 64, 50–57. [Google Scholar] [CrossRef]
- Su, M.; Li, X.F.; Ma, X.Y.; Peng, X.J.; Zhao, A.G.; Cheng, L.Q.; Chen, S.Y.; Liu, G.S. Cloning two P5CS genes from bioenergy sorghum and their expression profiles under abiotic stresses and MeJA treatment. Plant Sci. 2011, 181, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.F.; Ji, J.; Guan, W.Z.; Feng, Y.H.; Li, X.Z.; Jin, C.; Li, J.; Wang, Y.R.; Wang, G. A Lycium chinense-derived P5CS-like gene is regulated by water deficit-induced endogenous abscisic acid and overexpression of this gene enhances tolerance to water deficit stress in Arabidopsis. Mol. Breed. 2014, 34, 1109–1124. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Challabathula, D.; Analin, B.; Mohanan, A.; Bakka, K. Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and-tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J. Plant Physiol. 2022, 268, 153583. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Q.; Guo, Y.; Yang, J.; Wang, M.; Duan, X.; Niu, J.; Liu, S.; Zhang, J.; Lu, Y. Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OST1. J. Exp. Bot. 2018, 69, 4403–4417. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Li, X.; Li, W.; Yang, Q.; Li, Z.; Cheng, Z.; Lv, L.; Zhang, L.; Han, D. Isolation and preliminary functional analysis of FvICE1, involved in cold and drought tolerance in Fragaria vesca through overexpression and CRISPR/Cas9 technologies. Plant Physiol. Biochem. 2023, 196, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhou, Z.; Du, M.; Li, T.; Wu, X.; Yu, J.; Zhang, P.; Yang, G. Overexpression of a Malus xiaojinensis WRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitr. Cell. Dev. Biol.-Plant 2020, 56, 600–609. [Google Scholar] [CrossRef]
- Han, D.; Wang, Y.; Zhang, L.; Ma, L.; Zhang, X.; Xu, X.; Han, Z. Isolation and functional characterization of MxCS1: A gene encoding a citrate synthase in Malus xiaojinensis. Biol. Plantarum 2012, 56, 50–56. [Google Scholar] [CrossRef]
- Han, D.; Zhang, Z.; Ni, B.; Ding, H.; Liu, W.; Li, W.; Chai, L.; Yang, G. Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 2018, 13, 433–441. [Google Scholar] [CrossRef]
- Han, D.; Shi, Y.; Wang, B.; Liu, W.; Yu, Z.; Lv, B.; Yang, G. Isolation and preliminary functional analysis of MxCS2: A gene encoding a citrate synthase in Malus xiaojinensis. Plant Mol. Biol. Rep. 2015, 33, 133–142. [Google Scholar] [CrossRef]
- Han, D.; Shi, Y.; Yu, Z.; Liu, W.; Lv, B.; Wang, B.; Yang, G. Isolation and functional analysis of MdCS1: A gene encoding a citrate synthase in Malus domestica (L.) Borkh. Plant Growth Regul. 2015, 75, 209–218. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, H.; Su, C.; Qi, Y.; Liu, X.; Pu, J. Genome-wide identification and expression profile analysis of the NAC transcription factor family during abiotic and biotic stress in woodland strawberry. PLoS ONE 2018, 13, e0197892. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Dai, J.; Chen, Z.; Li, W.; Li, X.; Zhang, L.; Yao, A.; Zhang, B.; Han, D. Overexpression of a ‘Beta’ MYB Factor Gene, VhMYB15, Increases Salinity and Drought Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 1534. https://doi.org/10.3390/ijms25031534
Han J, Dai J, Chen Z, Li W, Li X, Zhang L, Yao A, Zhang B, Han D. Overexpression of a ‘Beta’ MYB Factor Gene, VhMYB15, Increases Salinity and Drought Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 2024; 25(3):1534. https://doi.org/10.3390/ijms25031534
Chicago/Turabian StyleHan, Jiaxin, Jing Dai, Zhe Chen, Wenhui Li, Xingguo Li, Lihua Zhang, Anqi Yao, Bingxiu Zhang, and Deguo Han. 2024. "Overexpression of a ‘Beta’ MYB Factor Gene, VhMYB15, Increases Salinity and Drought Tolerance in Arabidopsis thaliana" International Journal of Molecular Sciences 25, no. 3: 1534. https://doi.org/10.3390/ijms25031534
APA StyleHan, J., Dai, J., Chen, Z., Li, W., Li, X., Zhang, L., Yao, A., Zhang, B., & Han, D. (2024). Overexpression of a ‘Beta’ MYB Factor Gene, VhMYB15, Increases Salinity and Drought Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences, 25(3), 1534. https://doi.org/10.3390/ijms25031534





