Molecular and Structural Alterations of Skeletal Muscle Tissue Nuclei during Aging
Abstract
1. Introduction
2. Structural Organization of the Cell Nucleus
3. The Myonucleus in Aging
3.1. Number, Size, and Shape of Myonuclei
3.2. Myonuclei as Mechanosensors
3.3. Myonuclear Activity
3.4. Epigenetic Changes in Myonuclei
3.5. Telomeres in Myonuclei
4. The Satellite Cell Nucleus in Aging
4.1. Number of Satellite Cells
4.2. Myogenic Potential and Nuclear Activity of Satellite Cells
4.3. Telomeres in Satellite Cell Nuclei
4.4. Epigenetic Changes in Satellite Cell Nuclei
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hughes, V.A.; Frontera, W.R.; Roubenoff, R.; Evans, W.J.; Singh, M.A. Longitudinal changes in body composition in older men and women: Role of body weight change and physical activity. Am. J. Clin. Nutr. 2002, 76, 473–481. [Google Scholar] [CrossRef]
- Paddon-Jones, D.; Rasmussen, B.B. Dietary protein recommendations and the prevention of sarcopenia. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 86–90. [Google Scholar] [CrossRef]
- Mitchell, W.K.; Williams, J.; Atherton, P.; Larvin, M.; Lund, J.; Narici, M. Sarcopenia, dynapenia, and the impact of advancing age on human skeletal muscle size and strength; a quantitative review. Front. Physiol. 2012, 3, 260. [Google Scholar] [CrossRef]
- Thompson, L.D. Age-related muscle dysfunction. Exp. Gerontol. 2009, 44, 106–111. [Google Scholar] [CrossRef]
- Cruz-Jentoft, A.J.; Sayer, A.A. Sarcopenia. Lancet 2019, 393, 2636–2646. [Google Scholar] [CrossRef]
- Karakelides, H.; Sreekumaran Nair, K. Sarcopenia of aging and its metabolic impact. Curr. Top. Dev. Biol. 2005, 68, 123–148. [Google Scholar]
- Qiao, Y.S.; Chai, Y.H.; Gong, H.J.; Zhuldyz, Z.; Stehouwer, C.D.A.; Zhou, J.B.; Simó, R. The association between diabetes mellitus and risk of sarcopenia: Accumulated evidences from observational studies. Front. Endocrinol. 2021, 12, 782391. [Google Scholar] [CrossRef]
- Al Saedi, A.; Debruin, D.A.; Hayes, A.; Hamrick, M. Lipid metabolism in sarcopenia. Bone 2022, 164, 116539. [Google Scholar] [CrossRef]
- Ladang, A.; Beaudart, C.; Reginster, J.Y.; Al-Daghri, N.; Bruyère, O.; Burlet, N.; Cesari, M.; Cherubini, A.; da Silva, M.C.; Cooper, C.; et al. Biochemical Markers of Musculoskeletal Health and Aging to be Assessed in Clinical Trials of Drugs Aiming at the Treatment of Sarcopenia: Consensus Paper from an Expert Group Meeting Organized by the European Society for Clinical and Economic Aspects of Osteoporosis, Osteoarthritis and Musculoskeletal Diseases (ESCEO) and the Centre Académique de Recherche et d’Expérimentation en Santé (CARES SPRL), Under the Auspices of the World Health Organization Collaborating Center for the Epidemiology of Musculoskeletal Conditions and Aging. Calcif. Tissue Int. 2023, 112, 197–217. [Google Scholar]
- Saltin, B.; Gollnick, P.D. Handbook of Physiology: Skeletal Muscle; Peachy, L.D., Adnan, R., Geiger, S.R., Eds.; Springer: Berlin/Heidelberg, Germany, 1983; pp. 555–631. [Google Scholar]
- Bottinelli, R.; Pellegrino, M.A.; Canepari, M.; Rossi, R.; Reggiani, C. Specific contributions of various muscle fibre types to human muscle performance: An in vitro study. J. Electromyogr. Kinesiol. 1999, 9, 87–95. [Google Scholar] [CrossRef]
- Trappe, S.; Luden, N.; Minchev, K.; Raue, U.; Jemiolo, B.; Trappe, T.A. Skeletal muscle signature of a champion sprint runner. J. Appl. Physiol. 2015, 118, 1460–1466. [Google Scholar] [CrossRef]
- Giordani, L.; He, G.J.; Negroni, E.; Sakai, H.; Law, J.Y.C.; Siu, M.M.; Wan, R.; Corneau, A.; Tajbakhsh, S.; Cheung, T.H.; et al. High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol. Cell. 2019, 74, 609–621. [Google Scholar] [CrossRef]
- Rubenstein, A.B.; Smith, G.R.; Raue, U.; Begue, G.; Minchev, K.; Ruf-Zamojski, F.; Nair, V.D.; Wang, X.; Zhou, L.; Zaslavsky, E.; et al. Single-cell transcriptional profiles in human skeletal muscle. Sci Rep. 2020, 10, 229. [Google Scholar] [CrossRef]
- Combaret, L.; Dardevet, D.; Béchet, D.; Taillandier, D.; Mosoni, L.; Attaix, D. Skeletal muscle proteolysis in aging. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 37–41. [Google Scholar] [CrossRef]
- Sandri, M. Autophagy in skeletal muscle. FEBS Lett. 2010, 584, 1411–1416. [Google Scholar] [CrossRef]
- Brook, M.S.; Wilkinson, D.J.; Phillips, B.E.; Perez-Schindler, J.; Philp, A.; Smith, K.; Atherton, P.J. Skeletal muscle homeostasis and plasticity in youth and ageing: Impact of nutrition and exercise. Acta Physiol. 2016, 216, 15–41. [Google Scholar] [CrossRef]
- Jiao, J.; Demontis, F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr. Opin. Pharmacol. 2017, 34, 1–6. [Google Scholar] [CrossRef]
- Short, K.R.; Bigelow, M.L.; Kahl, J.; Singh, R.; Coenen-Schimke, J.; Raghavakaimal, S.; Sreekumaran Nair, K. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. USA 2005, 102, 5618–5623. [Google Scholar] [CrossRef]
- Always, S.E.; Mohamed, J.S.; Myers, M.J. Mitochondria initiate and regulate sarcopenia. Exerc. Sport Sci. Rev. 2017, 45, 58–69. [Google Scholar] [CrossRef]
- Dirks, A.J.; Leeuwenburgh, C. The role of apoptosis in age-related skeletal muscle atrophy. Sports Med. 2005, 35, 473–483. [Google Scholar] [CrossRef]
- Always, S.E.; Siu, P.M. Nuclear Apoptosis Contributes to Sarcopenia. Exerc. Sport Sci. Rev. 2008, 36, 51–57. [Google Scholar] [CrossRef]
- Rahmati, M.; McCarthy, J.J.; Malakoutinia, F. Myonuclear permanence in skeletal muscle memory: A systematic review and meta-analysis of human and animal studies. J. Cachexia Sarcopenia Muscle 2022, 13, 2276–2297. [Google Scholar] [CrossRef]
- Musarò, A.; Giacinti, C.; Pelosi, L.; Dobrowolny, G.; Barberi, L.; Nardis, C.; Coletti, D.; Scicchitano, B.M.; Adamo, S.; Molinaro, M. Stem cell mediated muscle regeneration and repair in aging and neuromuscular diseases. Eur. J. Histochem. 2007, 51 (Suppl. S1), 35–44. [Google Scholar]
- Garcıa-Prat, L.; Sousa-Victor, P.; Muñoz-Canoves, P. Functional dysregulation of stem cells during aging: A focus on skeletal muscle stem cells. FEBS J. 2013, 280, 4051–4062. [Google Scholar] [CrossRef]
- Always, S.E.; Myers, M.J.; Mohamed, J.S. Regulation of satellite cell function in sarcopenia. Front. Aging Neurosci. 2014, 6, 246. [Google Scholar] [CrossRef]
- Ansved, T.; Larsson, L. Effects of ageing on enzyme-histochemical, morphometrical and contractile properties of the soleus muscle in the rat. J. Neurol. Sci. 1989, 93, 105–124. [Google Scholar] [CrossRef]
- Barns, M.; Gondro, C.; Tellam, R.L.; Radley-Crabb, H.G.; Grounds, M.D.; Shavlakadze, T. Molecular analyses provide insight into mechanisms underlying sarcopenia and myofibre denervation in old skeletal muscles of mice. Int. J. Biochem. Cell Biol. 2014, 53, 174–185. [Google Scholar] [CrossRef]
- Payne, G.W.; Bearden, S.E. The microcirculation of skeletal muscle in aging. Microcirculation 2006, 13, 275–277. [Google Scholar] [CrossRef]
- Lopes, K.G.; Farinatti, P.; Bottino, D.A.; de Souza, M.D.G.C.; Maranhão, P.A.; Bouskela, E.; Lourenço, R.A.; de Oliveira, R.B. Sarcopenia in the elderly versus microcirculation, inflammation status, and oxidative stress: A cross-sectional study. Clin. Hemorheol. Microcirc. 2022, 80, 185–195. [Google Scholar] [CrossRef]
- Tenover, J.L. Testosterone and the aging male. J. Androl. 1997, 18, 103–106. [Google Scholar] [CrossRef]
- Léger, B.; Derave, W.; De Bock, K.; Hespel, P.; Russell, A.P. Human sarcopenia reveals an increase in SOCS-3 and myostatin and a reduced efficiency of Akt phosphorylation. Rejuvenation Res. 2008, 11, 163B–175B. [Google Scholar] [CrossRef]
- Bruunsgaard, H.; Pedersen, M.; Pedersen, B.K. Aging and proinflammatory cytokines. Curr. Opin. Hematol. 2001, 8, 131–136. [Google Scholar] [CrossRef]
- Schaap, L.A.; Pluijm, S.M.; Deeg, D.J.; Visser, M. Inflammatory markers and loss of muscle mass (sarcopenia) and strength. Am. J. Med. 2006, 119, e517. [Google Scholar] [CrossRef]
- Pan, L.; Xie, W.; Fu, X.; Lu, W.; Jin, H.; Lai, J.; Zhang, A.; Yu, Y.; Li, Y.; Xiao, W. Inflammation and sarcopenia: A focus on circulating inflammatory cytokines. Exp. Gerontol. 2021, 154, 111544. [Google Scholar] [CrossRef]
- Misteli, T.; Spector, D.L. Applications of the green fluorescent protein in cell biology and biotechnology. Nature Biotechnol. 1997, 15, 961–964. [Google Scholar] [CrossRef]
- Masiello, I.; Siciliani, S.; Biggiogera, M. Perichromatin region: A moveable feast. Histochem. Cell Biol. 2018, 150, 227–233. [Google Scholar] [CrossRef]
- Pegoraro, G.; Misteli, T. The central role of chromatin maintenance in aging. Aging 2009, 1, 1017–1022. [Google Scholar] [CrossRef]
- Tiku, V.; Antebi, A. Nucleolar function in lifespan regulation. Trends. Cell Biol. 2018, 28, 662–672. [Google Scholar] [CrossRef]
- Malatesta, M.; Cardani, R.; Pellicciari, C.; Meola, G. RNA transcription and maturation in skeletal muscle cells are similarly impaired in myotonic dystrophy and sarcopenia: The ultrastructural evidence. Front. Aging Neurosci. 2014, 6, 196. [Google Scholar] [CrossRef]
- Gerace, L.; Burke, B. Functional organization of the nuclear envelope. Annu. Rev. Cell Biol. 1988, 4, 335–374. [Google Scholar] [CrossRef]
- D’Angelo, M.A.; Raices, M.; Panowski, S.H.; Hetzer, M.W. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009, 136, 284–295. [Google Scholar] [CrossRef]
- Dahl, K.N.; Kahn, S.M.; Wilson, K.L.; Discher, D.E. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004, 117, 4779–4786. [Google Scholar] [CrossRef]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.-W.; Tewari, M.; et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef]
- Stephens, A.D.; Banigan, E.J.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 2017, 28, 1984–1996. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Goldberg, M.; Harel, A.; Gruenbaum, Y. The nuclear lamina: Molecular organization and interaction with chromatin. Crit. Rev. Eukaryot. Gene Expr. 1999, 9, 285–293. [Google Scholar] [CrossRef]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef]
- Ho, C.Y.; Jaalouk, D.E.; Vartiainen, M.K.; Lammerding, J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 2013, 497, 507–511. [Google Scholar] [CrossRef]
- Houben, F.; Ramaekers, F.C.S.; Snoeckx, L.H.E.H.; Broers, J.L.V. Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim. Biophys Acta Mol. Cell Res. 2007, 1773, 675–686. [Google Scholar] [CrossRef]
- Ramage, L.; Nuki, G.; Salter, D.M. Signalling cascades in mechanotransduction: Cell-matrix interactions and mechanical loading. Scand J. Med. Sci. Sport. 2009, 19, 457–469. [Google Scholar] [CrossRef]
- Poh, Y.C.; Shevtsov, S.P.; Chowdhury, F.; Wu, D.C.; Na, S.; Dundr, M.; Wang, N. Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nat. Commun. 2012, 3, 810–866. [Google Scholar] [CrossRef]
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017, 216, 305–315. [Google Scholar] [CrossRef]
- Machida, S.; Takizawa, Y.; Ishimaru, M.; Sugita, Y.; Sekine, S.; Nakayama, J.I.; Wolf, M.; Kurumizaka, H. Structural basis of heterochromatin formation by human HP1. Mol. Cell 2018, 69, 385–397. [Google Scholar] [CrossRef]
- Kornberg, K.D.; Lorch, Y. Irresistible force meets immovable object: Transcription and the nucleosome. Cell. 1991, 67, 833–836. [Google Scholar] [CrossRef]
- Beato, M.; Eisfeld, K. Transcription factor access to chromatin. Nucleic Acids Res. 1997, 25, 3559–3563. [Google Scholar] [CrossRef]
- Adams, C.C.; Workman, J.L. Nucleosome displacement in transcription. Cell 1993, 72, 305–308. [Google Scholar] [CrossRef]
- Lemon, B.; Tjian, R. Orchestrated response: A symphony of transcription factors for gene control. Genes Dev. 2000, 14, 2551–2569. [Google Scholar] [CrossRef]
- Fakan, S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994, 4, 86–90. [Google Scholar] [CrossRef]
- Olson, M.O.; Hingorani, K.; Szebeni, A. Conventional and nonconventional roles of the nucleolus. Int. Rev. Cytol. 2002, 219, 199–266. [Google Scholar]
- Ahmad, Y.; Boisvert, F.M.; Gregor, P.; Cobley, A.; Lamond, A.I. NOPdb: Nucleolar proteome database. Nucleic Acids Res. 2009, 37, 181–184. [Google Scholar] [CrossRef]
- Borowik, A.K.; Davidyan, A.; Peelor, F.F.; Voloviceva, E.; Doidge, S.M.; Bubak, M.P.; Mobley, C.B.; McCarthy, J.J.; Dupont-Versteegden, E.E.; Miller, B.F. Skeletal Muscle Nuclei in Mice are not Post-mitotic. Function 2022, 4, zqac059. [Google Scholar] [CrossRef]
- Sanes, J.R.; Lichtman, J.W. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2001, 2, 791–805. [Google Scholar] [CrossRef]
- Hippenmeyer, S.; Huber, R.M.; Ladle, D.R.; Murphy, K.; Arber, S. ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron 2007, 55, 726–740. [Google Scholar] [CrossRef]
- Burden, S.J.; Huijbers, M.G.; Remedio, L. Fundamental molecules and mechanisms for forming and maintaining neuromuscular synapses. Int. J. Mol. Sci. 2018, 19, 490. [Google Scholar] [CrossRef]
- Li, L.; Xiong, W.C.; Mei, L. Neuromuscular junction formation, aging, and disorders. Annu. Rev. Physiol. 2018, 80, 159–188. [Google Scholar] [CrossRef]
- Charvet, B.; Ruggiero, F.; Le Guellec, D. The development of the myotendinous junction. A review. Musc. Lig. Tend. J. 2012, 2, 53–63. [Google Scholar]
- Kim, M.; Franke, V.; Brandt, B.; Lowenstein, E.D.; Schöwel, V.; Spuler, S.; Akalin, A.; Birchmeier, C. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat. Commun. 2020, 11, 6375. [Google Scholar] [CrossRef]
- Petrany, M.J.; Swoboda, C.O.; Sun, C.; Chetal, K.; Chen, X.; Weirauch, M.T.; Salomonis, N.; Millay, D.P. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 2020, 11, 6374. [Google Scholar] [CrossRef]
- Bruusgaard, J.C.; Liestol, K.; Gundersen, K. Distribution of myonuclei and microtubules in live muscle fibers of young, middle-aged, and old mice. J. Appl. Physiol. 2006, 100, 2024–2030. [Google Scholar] [CrossRef]
- Cramer, A.A.W.; Prasad, V.; Eftestøl, E.; Song, T.; Hansson, K.-A.; Dugdale, H.F.; Sadayappan, S.; Ochala, J.; Gundersen, K.; Millay, D.P. Nuclear numbers in syncytial muscle fibers promote size but limit the development of larger myonuclear domains. Nat. Commun. 2020, 11, 6287. [Google Scholar] [CrossRef]
- Blau, H.M.; Pavlath, G.K.; Rich, K.; Webster, S.G. Localization of muscle gene products in nuclear domains: Does this constitute a problem for myoblast therapy? Adv. Exp. Med. Biol. 1990, 280, 167–172. [Google Scholar]
- Hall, Z.W.; Ralston, E. Nuclear domains in muscle cells. Cell 1989, 59, 771–772. [Google Scholar] [CrossRef] [PubMed]
- Pavlath, G.K.; Rich, K.; Webster, S.G.; Blau, H.M. Localization of muscle gene products in nuclear domains. Nature 1989, 337, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Qaisar, R.; Larsson, L. What determines myonuclear domain size? Indian J. Physiol. Pharmacol. 2014, 58, 1–12. [Google Scholar] [PubMed]
- Vassilopoulos, D.; Lumb, E.M.; Emery, A.E. Karyometric changes in human muscle with age. Eur. Neurol. 1977, 16, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Kadi, F.; Schjerling, P.; Andersen, L.L.; Charifi, N.; Madsen, J.L.; Christensen, L.R.; Andersen, J.L. The effects of heavy resistance training and detraining on satellite cells in human skeletal muscles. Physiol. J. 2004, 558, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Cristea, A.; Qaisar, R.; Edlund, P.K.; Lindblad, J.; Bengtsson, E.; Larsson, L. Effects of aging and gender on the spatial organization of nuclei in single human skeletal muscle cells. Aging Cell 2010, 9, 685–697. [Google Scholar] [CrossRef]
- Malatesta, M.; Perdoni, F.; Muller, S.; Zancanaro, C.; Pellicciari, C. Nuclei of aged myofibres undergo structural and functional changes suggesting impairment in RNA processing. Eur. J. Histochem. 2009, 53, 97–106. [Google Scholar] [CrossRef]
- Manta, P.; Vassilopoulos, D.; Spengos, M. Nucleo-cytoplasmic ratio in ageing skeletal muscle. Eur. Arch. Psychiatry Clin. Neurosci. 1987, 236, 235–236. [Google Scholar] [CrossRef]
- Brook, M.S.; Wilkinson, D.J.; Tarum, J.; Mitchell, K.W.; Lund, J.L.; Phillips, B.E.; Szewczyk, N.J.; Kadi, F.; Greenhaff, P.L.; Smith, K.; et al. Neither myonuclear accretion nor a myonuclear domain size ceiling is a feature of the attenuated hypertrophic potential of aged human skeletal muscle. GeroScience 2023, 45, 451–462. [Google Scholar] [CrossRef]
- Gallegly, J.C.; Turesky, N.A.; Strotman, B.A.; Gurley, C.M.; Peterson, C.A.; Dupont-Versteegden, E.E. Satellite cell regulation of muscle mass is altered at old age. J. Appl. Physiol. 2004, 97, 1082–1090. [Google Scholar] [CrossRef]
- Brack, A.S.; Bildsoe, H.; Hughes, S.M. Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibres during murine age-related muscle atrophy. J. Cell Sci. 2005, 118, 4813–4821. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Hepple, R.T. Elevated caspase and AIF gene expression correlate with progression of sarcopenia during aging in male F344BN rats. Exp. Gerontol. 2006, 41, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Marzetti, E.; Privitera, G.; Simili, V.; Wohlgemuth, S.E.; Aulisa, L.; Pahor, M.; Leeuwenburgh, C. Multiple pathways to the same end: Mechanisms of myonuclear apoptosis in sarcopenia of aging. Sci. World J. 2010, 10, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Zancanaro, C.; Mariotti, R.; Perdoni, F.; Nicolato, E.; Malatesta, M. Physical training is associated with changes in NMR and morphometrical parameters of the skeletal muscle in senescent mice. Eur. J. Histochem. 2007, 51, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.R.; Hsia, R.-C.; Folker, E.S.; Lovering, R.M. Age-dependent changes in nuclear-cytoplasmic signaling in skeletal muscle. Exp. Gerontol. 2021, 150, 111338. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.L.; Roy, R.R.; Edgerton, V.R. Myonuclear domains in muscle adaptation and disease. Muscle Nerve 1999, 22, 1350–1360. [Google Scholar] [CrossRef]
- Levy, Y.; Ross, J.A.; Niglas, M.; Snetkov, V.A.; Lynham, S.; Liao, C.-Y.; Puckelwartz, M.J.; Hsu, Y.-M.; McNally, E.M.; Alsheimer, M.; et al. Prelamin A causes aberrant myonuclear arrangement and results in muscle fiber weakness. JCI Insight 2018, 3, e120920. [Google Scholar] [CrossRef]
- Larsson, L.; Li, X.; Frontera, W.R. Effects of aging on shortening velocity and myosin isoform composition in single human skeletal muscle cells. Am. J. Physiol. 1997, 272, C638–C649. [Google Scholar] [CrossRef]
- D’Antona, G.; Pellegrino, M.A.; Adami, R.; Rossi, R.; Carlizzi, C.N.; Canepari, M.; Saltin, B.; Bottinelli, R. The effect of ageing and immobiliza tion on structure and function of human skeletal muscle fibres. J. Physiol. 2003, 552, 499–511. [Google Scholar] [CrossRef]
- Kirby, T.J.; Lammerding, J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 2018, 20, 373–381. [Google Scholar] [CrossRef]
- Maniotis, A.J.; Chen, C.S.; Ingber, D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA 1997, 94, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.; Jaalouk, D.E.; Shanahan, C.M.; Burke, B.; Roux, K.J.; Lammerding, J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011, 286, 26743–26753. [Google Scholar] [CrossRef] [PubMed]
- Guilluy, C.; Osborne, L.D.; Van, L.L.; Sharek, L.; Superfine, R.; Garcia-Mata, R.; Burridge, K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 2014, 16, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Harr, J.C.; Luperchio, T.R.; Wong, X.; Cohen, E.; Wheelan, S.J.; Reddy, K.L. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 2015, 208, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Dahl, K.N.; Ribeiro, A.J.; Lammerding, J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008, 102, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Llorian, M.; Smith, C.W.J. Decoding muscle alternative splicing. Curr. Opin. Genet. Develop. 2011, 21, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Malatesta, M.; Giagnacovo, M.; Costanzo, M.; Cisterna, B.; Cardani, R.; Meola, G. Muscleblind-like1 undergoes ectopic relocation in the nuclei of skeletal muscles in myotonic dystrophy and sarcopenia. Eur. J Histochem. 2013, 57, e15. [Google Scholar] [CrossRef][Green Version]
- Malatesta, M.; Fattoretti, P.; Giagnacovo, M.; Pellicciari, C.; Zancanaro, C. Physical training modulates structural and functional features of cell nuclei in type II myofibers of old mice. Rejuvenation Res. 2011, 14, 543–552. [Google Scholar] [CrossRef]
- Day, K.; Waite, L.L.; Thalacker-Mercer, A.; West, A.; Bamman, M.M.; Brooks, J.D.; Myers, R.M.; Absher, D. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013, 14, R102. [Google Scholar] [CrossRef]
- Zykovich, A.; Hubbard, A.; Flynn, J.M.; Tarnopolsky, M.; Fraga, M.F.; Kerksick, C.; Ogborn, D.; MacNeil, L.; Mooney, S.D.; Melov, S. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell. 2014, 13, 360–366. [Google Scholar] [CrossRef]
- Sharples, A.P.; Seaborne, R.A.; Stewart, C.E. Epigenetics of skeletal muscle aging. In Epigenetics of Aging and Longevity; Vaiserman, A.M., Ed.; Academic Press: London, UK, 2018. [Google Scholar]
- Sati, S.; Tanwar, V.S.; Kumar, K.A.; Patowary, A.; Jain, V.; Ghosh, S.; Ahmad, S.; Singh, M.; Reddy, S.U.; Chandak, G.R.; et al. High resolution methylome map of rat indicates role of intragenic DNA methylation in identification of coding region. PLoS ONE 2012, 7, e31621. [Google Scholar] [CrossRef]
- Maunakea, A.K.; Nagarajan, R.P.; Bilenky, M.; Ballinger, T.J.; D’Souza, C.; Fouse, S.D.; Johnson, B.E.; Hong, C.; Nielsen, C.; Zhao, Y.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Murach, K.A.; Dimet-Wiley, A.L.; Wen, Y.; Brightwell, C.R.; Latham, C.M.; Dungan, C.M.; Fry, C.S.; Watowich, S.J. Late-life exercise mitigates skeletal muscle epigenetic aging. Aging Cell 2022, 21, e13527. [Google Scholar] [CrossRef] [PubMed]
- Ubaida-Mohien, C.; Lyashkov, A.; Gonzalez-Freire, M.; Tharakan, R.; Shardell, M.; Moaddel, R.; Semba, R.D.; Chia, C.W.; Gorospe, M.; Sen, R. Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. Elife 2019, 8, e49874. [Google Scholar] [CrossRef] [PubMed]
- Murach, K.A.; Mobley, C.B.; Zdunek, C.J.; Frick, K.K.; Jones, S.R.; McCarthy, J.J.; Peterson, C.A.; Dungan, C.M. Muscle memory: Myonuclear accretion, maintenance, morphology, and miRNA levels with training and detraining in adult mice. J. Cachexia Sarcopenia Muscle 2020, 11, 1705–1722. [Google Scholar] [CrossRef] [PubMed]
- Loiselle, J.J.; Sutherland, L.C. RBM10: Harmful or helpful-many factors to consider. J. Cell. Biochem. 2018, 119, 3809–3818. [Google Scholar] [CrossRef] [PubMed]
- Mohan, N.; Kumar, V.; Kandala, D.T.; Kartha, C.C.; Laishram, R.S. A splicing-independent function of RBM10 controls specific 3′ UTR processing to regulate cardiac hypertrophy. Cell Rep. 2018, 24, 3539–3553. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020, 10, 15360. [Google Scholar] [CrossRef]
- Voisin, S.; Jacques, M.; Landen, S.; Harvey, N.R.; Haupt, L.M.; Griffiths, L.R.; Gancheva, S.; Ouni, M.; Jähnert, M.; Ashton, K.J. Meta-analysis of genome-wide DNA methylation and integrative omics of age in human skeletal muscle. J. Cachexia Sarcopenia Muscle 2021, 12, 1064–1078. [Google Scholar] [CrossRef]
- Figueiredo, V.C.; Wen, Y.; Alkner, B.; Fernandez-Gonzalo, R.; Norrbom, J.; Vechetti, I.J., Jr.; Valentino, T.; Mobley, C.B.; Zentner, G.E.; Peterson, C.A.; et al. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. Physiol. J. 2021, 599, 3363–3384. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef] [PubMed]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef] [PubMed]
- Hahn, U.; Desai-Hahn, R.; Rüterjans, H. 1H and 15N NMR investigation of the interaction of pyrimidine nucleotides with ribonuclease A. Eur. J. Biochem. 1985, 146, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hu, G.F. Angiogenin-mediated rRNA transcription in cancer and neurodegeneration. Int. J. Biochem. Mol. Biol. 2010, 1, 26–35. [Google Scholar] [PubMed]
- Kelpsch, D.J.; Tootle, T.L. Nuclear actin: From discovery to function. Anat. Rec. 2018, 301, 1999–2013. [Google Scholar] [CrossRef] [PubMed]
- Lacavalla, M.A.; Cisterna, B.; Zancanaro, C.; Malatesta, M. Ultrastructural immunocytochemistry shows impairment of RNA pathways in skeletal muscle nuclei of old mice: A link to sarcopenia? Eur. J. Histochem. 2021, 65, 3229. [Google Scholar] [CrossRef] [PubMed]
- Cutler, A.A.; Dammer, E.B.; Doung, D.M.; Seyfried, N.T.; Corbett, A.H.; Pavlath, G.K. Biochemical isolation of myonuclei employed to define changes to the myonuclear proteome that occur with aging. Aging Cell 2017, 16, 738–749. [Google Scholar] [CrossRef]
- Kirby, T.J.; Lee, J.D.; England, J.H.; Chaillou, T.; Esser, K.A.; McCarthy, J.J. Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J. Appl. Physiol. (1985) 2015, 119, 321–327. [Google Scholar] [CrossRef]
- Wilkie, G.S.; Schirmer, E.C. Purification of nuclei and preparation of nuclear envelopes from skeletal muscle. Methods Mol. Biol. 2008, 463, 23–41. [Google Scholar]
- Dimauro, I.; Pearson, T.; Caporossi, D.; Jackson, M.J. A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res. Notes 2012, 5, 513. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, Y.; Mallappa, C.; Vallaster, C.S.; Imbalzano, A.N. Isolation of nuclei from skeletal muscle satellite cells and myofibers for use in chromatin immunoprecipitation assays. Methods Mol. Biol. 2012, 798, 517–530. [Google Scholar]
- Zhang, X.; Habiballa, L.; Aversa, Z.; Ng, Y.E.; Sakamoto, A.E.; Englund, D.A.; Pearsall, V.M.; White, T.A.; Robinson, M.M.; Rivas, D.A.; et al. Characterization of cellular senescence in aging skeletal muscle. Nat. Aging 2022, 2, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Perez, K.; Ciotlos, S.; McGirr, J.; Limbad, C.; Doi, R.; Nederveen, J.P.; Nilsson, M.I.; Winer, D.A.; Evans, W.; Tarnopolsky, M.; et al. Single nuclei profiling identifies cell specific markers of skeletal muscle aging, frailty, and senescence. Aging 2022, 14, 9393–9422. [Google Scholar] [CrossRef]
- Kayo, T.; Allison, D.B.; Weindruch, R.; Prolla, T.A. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc. Natl. Acad. Sci. USA 2001, 98, 5093–5098. [Google Scholar] [CrossRef] [PubMed]
- Welle, S.; Brooks, A.I.; Delehanty, J.M.; Needler, N.; Bhatt, K.; Shah, B.; Thornton, C.A. Skeletal muscle gene expression profiles in 20-29 year old and 65-71 year old women. Exp. Gerontol. 2004, 39, 369–377. [Google Scholar] [CrossRef]
- Edwards, M.G.; Anderson, R.M.; Yuan, M.; Kendziorski, C.M.; Weindruch, R.; Prolla, T.A. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genom. 2007, 8, 80. [Google Scholar] [CrossRef]
- Goodman, C.A.; McNally, R.M.; Hoffmann, F.M.; Hornberger, T.A. Smad3 induces atrogin-1, inhibits mTOR and protein synthesis, and promotes muscle atrophy in vivo. Mol. Endocrinol. 2013, 27, 1946–1957. [Google Scholar] [CrossRef]
- Sartori, R.; Gregorevic, P.; Sandri, M. TGFbeta and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends Endocrinol. Metab. 2014, 25, 464–471. [Google Scholar] [CrossRef]
- Goode, J.M.; Pearen, M.A.; Tuong, Z.K.; Wang, S.-C.M.; Oh, T.G.; Shao, M.X.; Muscat, G.E.O. The nuclear receptor, Nor-1, induces the physiological responses associated with exercise. Mol. Endocrinol. 2016, 30, 660–676. [Google Scholar] [CrossRef]
- Pillon, N.J.; Gabriel, B.M.; Dollet, L.; Smith, J.A.B.; Puig, L.S.; Botella, J.; Bishop, D.J.; Krook, A.; Zierath, J.Z. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 2020, 11, 470. [Google Scholar] [CrossRef]
- Jing, Y.; Zuo, Y.; Yu, Y.; Sun, L.; Yu, Z.; Ma, S.; Zhao, Q.; Sun, G.; Hu, H.; Li, J.; et al. Single-nucleus profiling unveils a geroprotective role of the FOXO3 in primate skeletal muscle aging. Protein Cell 2023, 14, 499–514. [Google Scholar] [CrossRef]
- Morris, B.J.; Willcox, D.C.; Donlon, T.A.; Willcox, B.J. FOXO3: A major gene for human longevity—A mini-review. Gerontology 2015, 61, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Li, Q.; Wang, L.; Lu, P.; Suzuki, K.; Liu, Z.; Lei, J.; Li, W.; He, X.; Wang, S.; et al. FOXO3-engineered human ESC-derived vascular cells promote vascular protection and regeneration. Cell Stem Cell 2019, 24, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, S.; Yan, P.; Ren, J.; Song, M.; Li, J.; Lei, J.; Pan, H.; Wang, S.; Ma, X.; et al. A single-cell transcriptomic landscape of primate arterial aging. Nat. Commun. 2020, 11, 2202. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Soerensen, M.; Nygaard, M.; Dato, S.; Stevnsner, T.; Bohr, V.A.; Christensen, K.; Christiansen, L. Association study of FOXO3A SNPs and aging phenotypes in Danish oldest-old individuals. Aging Cell 2015, 14, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Burbano, M.S.J.; Robin, J.D.; Bauwens, S.; Martin, M.; Donati, E.; Martínez, L.; Lin, P.; Sacconi, S.; Magdinier, F.; Gilson, E. Non-canonical telomere protection role of FOXO3a of human skeletal muscle cells regulated by the TRF2-redox axis. Commun. Biol. 2023, 6, 561. [Google Scholar]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Marzetti, E.; Calvani, R.; Tosato, M.; Cesari, M.; Di Bari, M.; Cherubini, A.; Broccatelli, M.; Savera, G.; D’Elia, M.; Pahor, M.; et al. Physical activity and exercise as countermeasures to physical frailty and sarcopenia. Aging Clin. Exp. Res. 2017, 29, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Eckstrom, E.; Neukam, S.; Kalin, L.; Wright, J. Physical Activity and Healthy Aging. Clin. Geriatr. Med. 2020, 36, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Kadi, F.; Ponsot, E.; Piehl-Aulin, K.; Mackey, A.; Kjaer, M.; Oskarsson, E.; Holm, L. The effects of regular strength training on telomere length in human skeletal muscle. Med. Sci. Sports Exerc. 2008, 40, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Kadi, F.; Ponsot, E. The biology of satellite cells and telomeres in human skeletal muscle: Effects of aging and physical activity. Scand. J. Med. Sci. Sports 2010, 20, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Arsenis, N.C.; You, T.; Ogawa, E.F.; Tinsley, G.M.; Zuo, L. Physical activity and telomere length: Impact of aging and potential mechanisms of action. Oncotarget 2017, 8, 45008–45019. [Google Scholar] [CrossRef]
- Ponsot, E.; Lexell, J.; Kadi, F. Skeletal muscle telomere length is not impaired in healthy physically active old women and men. Muscle Nerve 2008, 37, 467–472. [Google Scholar] [CrossRef]
- Renault, V.; Thorne, L.-E.; Eriksson, P.-O.; Butler-Browne, G.; Mouly, V. Regenerative potential of human skeletal muscle during aging. Aging Cell 2002, 1, 132–139. [Google Scholar] [CrossRef]
- Semeraro, M.D.; Almer, G.; Renner, W.; Gruber, H.-J.; Herrmann, M. Telomere length in leucocytes and solid tissues of young and aged rats. Aging 2022, 14, 1713–1728. [Google Scholar] [CrossRef]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.C.; Rudnicki, M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.R.; Kanji, A.H.; Allbrook, D. The structure of the satellite cells in skeletal muscle. J. Anat. 1965, 99, 435–444. [Google Scholar] [PubMed]
- Schultz, E.; Gibson, M.C.; Champion, T. Satellite cells are mitotically quiescent in mature mouse muscle: An EM and radioautographic study. J. Exp. Zool. 1978, 206, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Parise, G.; McKinnell, I.W.; Rudnicki, M.A. Muscle satellite cell and atypical myogenic progenitor response following exercise. Muscle Nerve 2008, 37, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Lepper, C.; Partridge, T.A.; Fan, C.M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 2011, 138, 3639–3646. [Google Scholar] [CrossRef] [PubMed]
- Relaix, F.; Zammit, P.S. Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 2012, 139, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Snijders, T.; Nederveen, J.P.; McKay, B.R.; Joanisse, S.; Verdijk, L.B.; van Loon, L.J.; Parise, G. Satellite cells in human skeletal muscle plasticity. Front. Physiol. 2015, 6, 283. [Google Scholar] [CrossRef]
- Forcina, L.; Miano, C.; Pelosi, L.; Musarò, A. An Overview about the biology of skeletal muscle satellite cells. Cur. Genomics 2019, 20, 24–37. [Google Scholar] [CrossRef]
- Brancaccio, A.; Palacios, D. Chromatin signaling in muscle stem cells: Interpreting the regenerative microenvironment. Front. Aging Neurosci. 2015, 7, 36. [Google Scholar] [CrossRef]
- Hikida, R.S. Aging changes in satellite cells and their functions. Curr. Aging Sci. 2011, 4, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Hikida, R.S.; Walsh, S.; Barylski, N.; Campos, G.; Hagerman, F.C.; Staron, R.S. Is hypertrophy limited in elderly muscle fibers? A comparison of elderly and young strength-trained men. Basic. Appl. Myol. 1998, 8, 419–427. [Google Scholar]
- Roth, S.M.; Martel, G.F.; Ivey, F.M.; Lemmer, J.T.; Metter, E.J.; Hurley, B.F.; Rogers, M.A. Skeletal muscle satellite cell populations in healthy young and older men and women. Anat. Rec. 2000, 260, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Verdijk, L.B.; Koopman, R.; Schaart, G.; Meijer, K.; Savelberg, H.H.; van Loon, L.J. Satellite cell content is specifically reduced in type II skeletal muscle fibers in the elderly. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E151–E157. [Google Scholar] [CrossRef] [PubMed]
- Brooks, N.E.; Schuenke, M.D.; Hikida, R.S. No change in skeletal muscle satellite cells in young and aging rat soleus muscle. J. Physiol. Sci. 2009, 59, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Schmalbruch, H.; Hellhammer, U. The number of satellite cells in normal human muscle. Anat. Rec. 1976, 185, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.C.; Schultz, E. Age-related differences in absolute numbers of skeletal muscle satellite cells. Muscle Nerve 1983, 6, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Nnodim, J.O. Satellite cell numbers in senile rat levator ani muscle. Mech. Ageing Dev. 2000, 112, 99–111. [Google Scholar] [CrossRef]
- Kadi, F.; Charifi, N.; Denis, C.; Lexell, J. Satellite cells and myonuclei in young and elderly women and men. Muscle Nerve 2004, 29, 120–127. [Google Scholar] [CrossRef]
- Shefer, G.; Van de Mark, D.P.; Richardson, J.B.; Yablonka-Reuveni, Z. Satellite-cell pool size does matter: Defining the myogenic potency of aging skeletal muscle. Dev. Biol. 2006, 294, 50–66. [Google Scholar] [CrossRef]
- Collins, C.A.; Zammit, P.S.; Ruiz, A.P.; Morgan, J.E.; Partridge, T.A. A population of myogenic stem cells that survives skeletal muscle aging. Stem Cells 2007, 25, 885–894. [Google Scholar] [CrossRef]
- Fry, C.S.; Lee, J.D.; Mula, J.; Kirby, T.J.; Jackson, J.R.; Liu, F.; Yang, L.; Mendias, C.L.; Dupont-Versteegden, E.E.; McCarthy, J.J.; et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat. Med. 2015, 21, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, D.; Vasso, M.; De Palma, S.; Fania, C.; Torretta, E.; Cammarata, F.P.; Magnaghi, V.; Procacci, P.; Gelfi, C. Specific protein changes contribute to the differential muscle mass loss during ageing. Proteomics 2016, 16, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Yablonka-Reuveni, Z.; Day, K.; Vine, A.; Shefer, G. Defining the transcriptional signature of skeletal muscle stem cells. J. Anim. Sci. 2008, 86, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, E.; Sousa-Victor, P.; Ballestar, E.; Muñoz-Cánoves, P. Epigenetic regulation of myogenesis. Epigenetics 2009, 4, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Fujimaki, S.; Hidaka, R.; Asashima, M.; Takemasa, T.; Kuwabara, T. Wnt protein-mediated satellite cell conversion in adult and aged mice following voluntary wheel running. J. Biol. Chem. 2014, 289, 7399–7412. [Google Scholar] [CrossRef]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002, 3, 397–409. [Google Scholar] [CrossRef]
- Conboy, I.M.; Conboy, M.J.; Smythe, G.M.; Rando, T.A. Notch-mediated restoration of regenerative potential to aged muscle. Science 2003, 302, 1575–1577. [Google Scholar] [CrossRef]
- Bhasin, S.; Storer, T.W.; Berman, N.; Callegari, C.; Clevenger, B.; Phillips, J.; Bunnell, T.J.; Tricker, R.; Shirazi, A.; Casaburi, R. The effects of supraphysiologic doses of testosterone on muscle size and strength in normal men. N. Engl. J. Med. 1996, 335, 1–7. [Google Scholar] [CrossRef]
- Niel, L.; Willemsen, K.R.; Volante, S.N.; Monks, D.A. Sexual dimorphism and androgen regulation of satellite cell population in differentiating rat levator ani muscle. Dev. Neurobiol. 2008, 68, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Sinha-Hikim, I.; Taylor, W.E.; Gonzalez-Cadavid, N.F.; Zheng, W.; Bhasin, S. Androgen receptor in human skeletal muscle and cultured muscle satellite cells: Up-regulation by androgen treatment. J. Clin. Endocrinol. Metab. 2004, 89, 5245–5255. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, M.; Moretti, A.; Sorrentino, C.; Toro, G.; Gentile, G.; Iolascon, G.; Castoria, G.; Migliaccio, A. Filamin A cooperates with the androgen receptor in preventing skeletal muscle senescence. Cell Death Discov. 2023, 9, 437. [Google Scholar] [CrossRef] [PubMed]
- Malatesta, M.; Perdoni, F.; Muller, S.; Pellicciari, C.; Zancanaro, C. Pre-mRNA processing is partially impaired in satellite cell nuclei from aged muscles. J. Biomed. Biotechnol. 2010, 2010, 410405. [Google Scholar] [CrossRef] [PubMed]
- Cisterna, B.; Giagnacovo, M.; Costanzo, M.; Fattoretti, P.; Zancanaro, C.; Pellicciari, C.; Malatesta, M. Adapted physical exercise enhances activation and differentiation potential of satellite cells in the skeletal muscle of old mice. J. Anat. 2016, 228, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Tichy, E.D.; Sidibe, D.K.; Tierney, M.T.; Stec, M.J.; Sharifi-Sanjani, M.; Hosalkar, H.; Mubarak, S.; Johnson, F.B.; Sacco, A.; Mourkioti, F. Single stem cell imaging and analysis reveals telomere length differences in diseased human and mouse skeletal muscles. Stem Cell Reports 2017, 9, 1328–1341. [Google Scholar] [CrossRef] [PubMed]
- Decary, S.; Mouly, V.; Hamida, C.B.; Sautet, A.; Barbet, J.P.; Butler-Browne, G.S. Replicative potential and telomere length in human skeletal muscle: Implications for satellite cell-mediated gene therapy. Hum. Gene Ther. 1997, 8, 1429–1438. [Google Scholar] [CrossRef]
- Barberi, L.; Scicchitano, B.M.; De Rossi, M.; Bigot, A.; Duguez, S.; Wielgosik, A.; Stewart, C.; McPhee, J.; Conte, M.; Narici, M.; et al. Age-dependent alteration in muscle regeneration: The critical role of tissue niche. Biogerontology 2013, 14, 273–292. [Google Scholar] [CrossRef]
- Daniali, L.; Benetos, A.; Susser, E.; Kark, J.D.; Labat, C.; Kimura, M.; Desai, K.; Granick, M.; Aviv, A. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun. 2013, 4, 1597. [Google Scholar] [CrossRef]
- Balan, E.; De Groote, E.; Bouillon, M.; Viceconte, N.; Mahieu, M.; Naslain, D.; Nielens, H.; Decottignies, A.; Deldicque, L. No effect of the endurance training status on senescence despite reduced inflammation in skeletal muscle of older individuals. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E447–E454. [Google Scholar] [CrossRef]
- Zvereva, M.I.; Shcherbakova, D.M.; Dontsova, O.A. Telomerase: Structure, functions, and activity regulation. Biochemistry 2010, 75, 1563–1583. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.S.; Carlson, M.E.; Conboy, I.M. Differentiation rather than aging of muscle stem cells abolishes their telomerase activity. Biotechnol. Prog. 2009, 25, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Prowse, K.R.; Greider, C.W. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. USA 1995, 92, 4818–4822. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.E.; Shay, J.W. Telomere dynamics in cancer progression and prevention: Fundamental differences in human and mouse telomere biology. Nat. Med. 2000, 6, 849–851. [Google Scholar] [CrossRef]
- Liu, L.; Rando, T.A. Manifestations and mechanisms of stem cell aging. J. Cell Biol. 2011, 193, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cheung, T.H.; Charville, G.W.; Hurgo, B.M.; Leavitt, T.; Shih, J.; Brunet, A.; Rando, T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013, 4, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Schwörer, S.; Becker, F.; Feller, C.; Baig, A.H.; Köber, U.; Henze, H.; Kraus, J.M.; Xin, B.; Lechel, A.; Lipka, D.B.; et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 2016, 540, 428–432. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Cañamero, M.; Maraver, A.; Gómez-López, G.; Contreras, J.; Murillo-Cuesta, S.; Rodríguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed cell senescence during mammalian embryonic development. Cell 2013, 155, 1104–1118. [Google Scholar] [CrossRef]
- Zhao, Y.; Ding, Y.; He, L.; Zhou, Q.; Chen, X.; Li, Y.; Alfonsi, M.V.; Wu, Z.; Sun, H.; Wang, H. Multiscale 3D genome reorganization during skeletal muscle stem cell lineage progression and aging. Sci. Adv. 2023, 9, eabo1360. [Google Scholar] [CrossRef]
- Lazure, F.; Farouni, R.; Sahinyan, K.; Blackburn, D.M.; Hernández-Corchado, A.; Perron, G.; Lu, T.; Osakwe, A.; Ragoussis, J.; Crist, C.; et al. Transcriptional reprogramming of skeletal muscle stem cells by the niche environment. Nat. Commun. 2023, 14, 535. [Google Scholar] [CrossRef]
- Soto-Palma, C.; Niedernhofer, L.J.; Faulk, C.D.; Dong, X. Epigenetics, DNA damage, and aging. J. Clin. Invest 2022, 132, e158446. [Google Scholar] [CrossRef] [PubMed]
- Seale, K.; Horvath, S.; Teschendorff, A.; Eynon, N.; Voisin, S. Making sense of the ageing methylome. Nat. Rev. Genet. 2022, 23, 585–605. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.-P. Aging and epigenetic drift: A vicious cycle. J. Clin. Investig. 2014, 124, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, H.; Jeong, H.S.; Keith, K.; Maegawa, S.; Calendo, G.; Madzo, J.; Jelinek, J.; Issa, J.J. DNA methylation entropy as a measure of stem cell replication and aging. Genome Biol. 2023, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Bigot, A.; Duddy, W.J.; Ouandaogo, Z.G.; Negroni, E.; Mariot, V.; Ghimbovschi, S.; Harmon, B.; Wielgosik, A.; Loiseau, C.; Devaney, J.; et al. Age-Associated Methylation Suppresses SPRY1, Leading to a Failure of Re-quiescence and Loss of the Reserve Stem Cell Pool in Elderly Muscle. Cell Rep. 2015, 13, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Burton, M.A.; Garratt, E.S.; Hewitt, M.O.; Sharkh, H.Y.; Antoun, E.; Westbury, L.D.; Dennison, E.M.; Harvey, N.C.; Cooper, C.; MacIsaac, J.L.; et al. DNA methylation of insulin signaling pathways is associated with HOMA2-IR in primary myoblasts from older adults. Skelet. Muscle 2023, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Zhu, C.F. Causal relationship between insulin resistance and sarcopenia. Diabetol. Metab. Syndr. 2023, 15, 46. [Google Scholar] [CrossRef] [PubMed]
- Loyola, A.; Bonaldi, T.; Roche, D.; Imhof, A.; Almouzni, G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 2006, 24, 309–316. [Google Scholar] [CrossRef]
- Liu, L.; Rodriguez-Mateo, C.; Huang, P.; Huang, A.; Lieu, A.; Mao, M.; Chung, M.; Yang, S.; Yu, K.; Charville, G.W.; et al. Hairless regulates heterochromatin maintenance and muscle stem cell function as a histone demethylase antagonist. Proc. Natl. Acad. Sci. USA 2021, 118, e2025281118. [Google Scholar] [CrossRef]
- Sahu, A.; Mamiya, H.; Shinde, S.N.; Cheikhi, A.; Winter, L.L.; Vo, N.V.; Stolz, D.; Roginskaya, V.; Tang, W.Y.; St Croix, C.; et al. Age-related declines in α-Klotho drive progenitor cell mitochondrial dysfunction and impaired muscle regeneration. Nat. Commun. 2018, 9, 4859. [Google Scholar] [CrossRef]
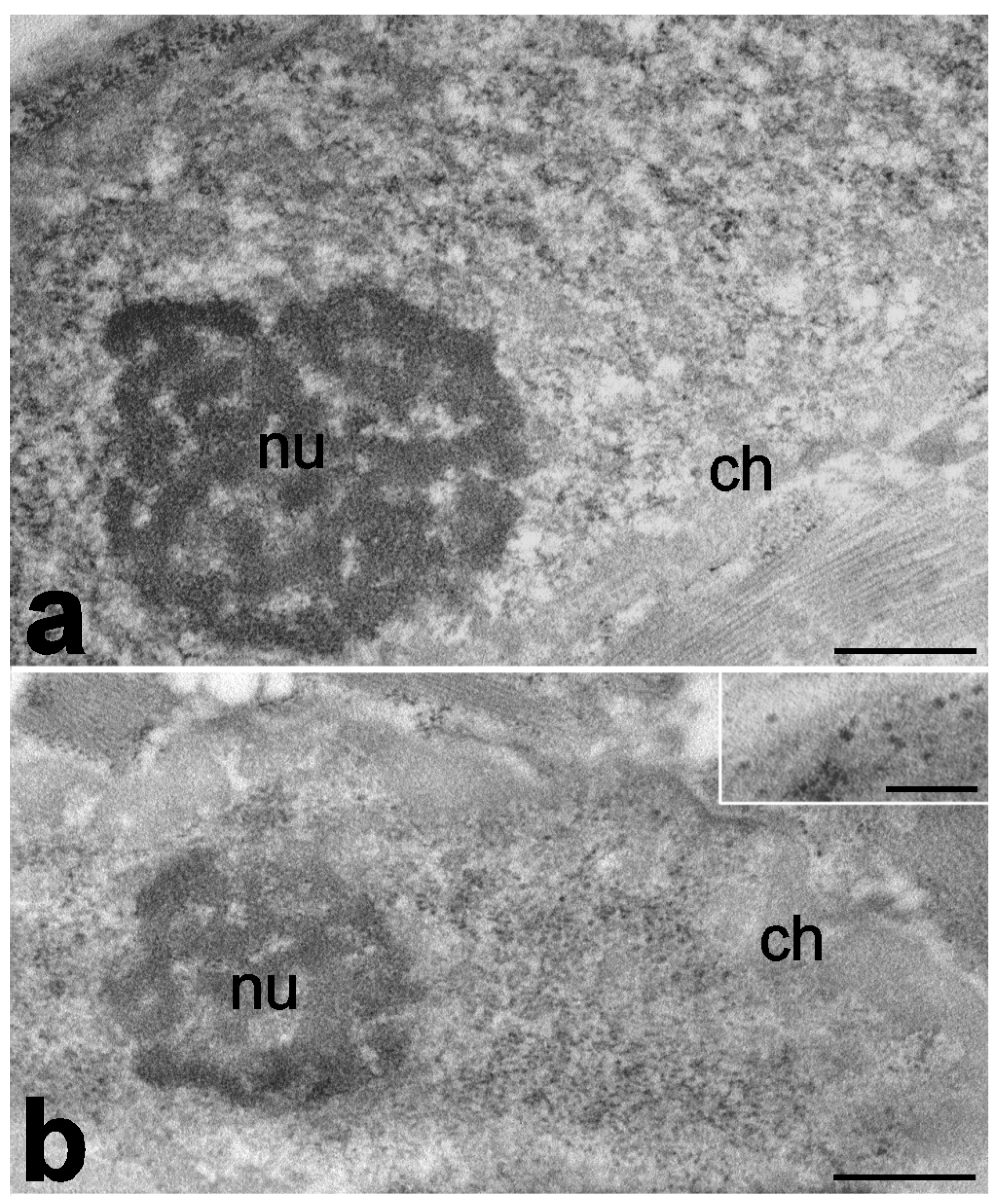

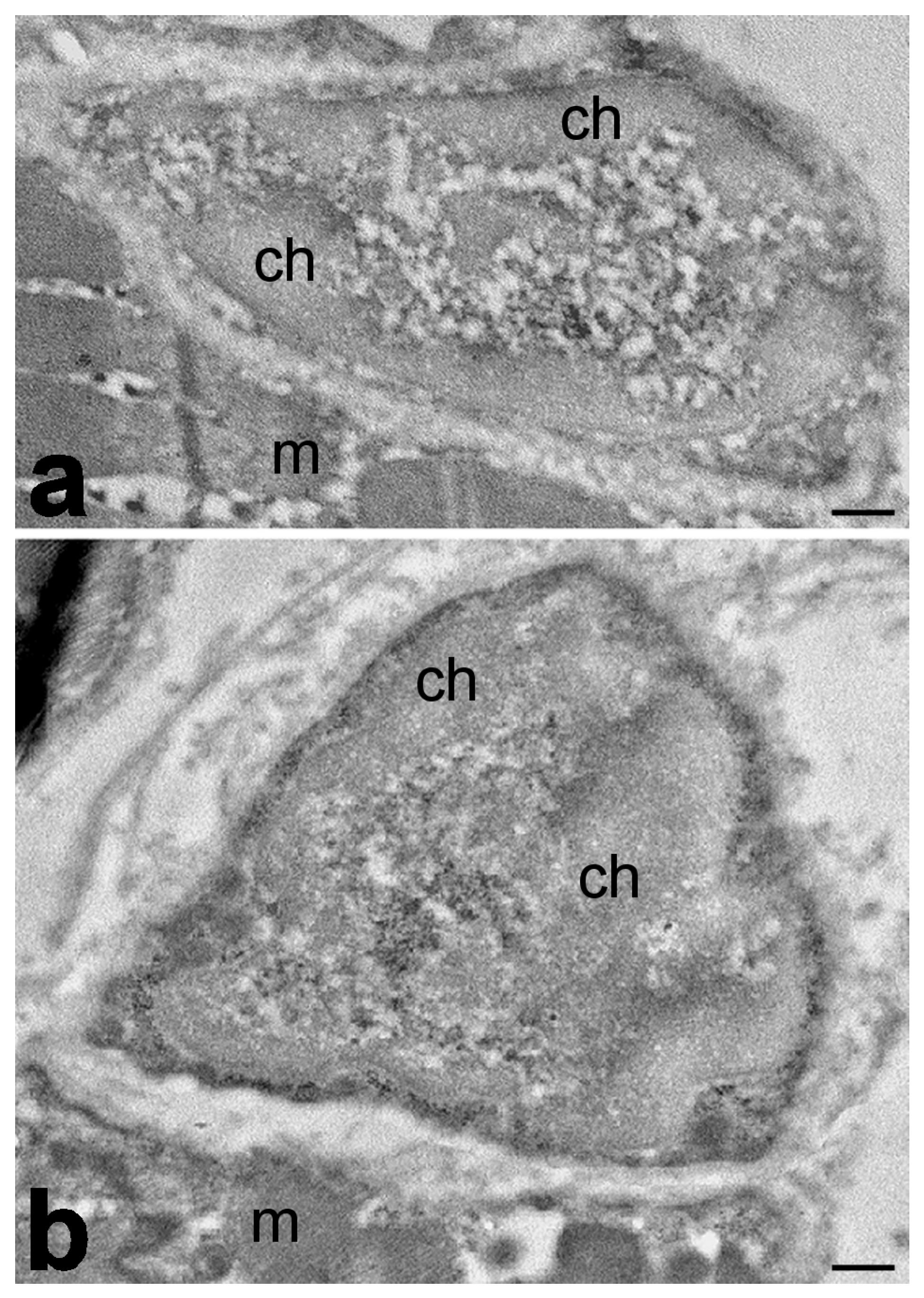
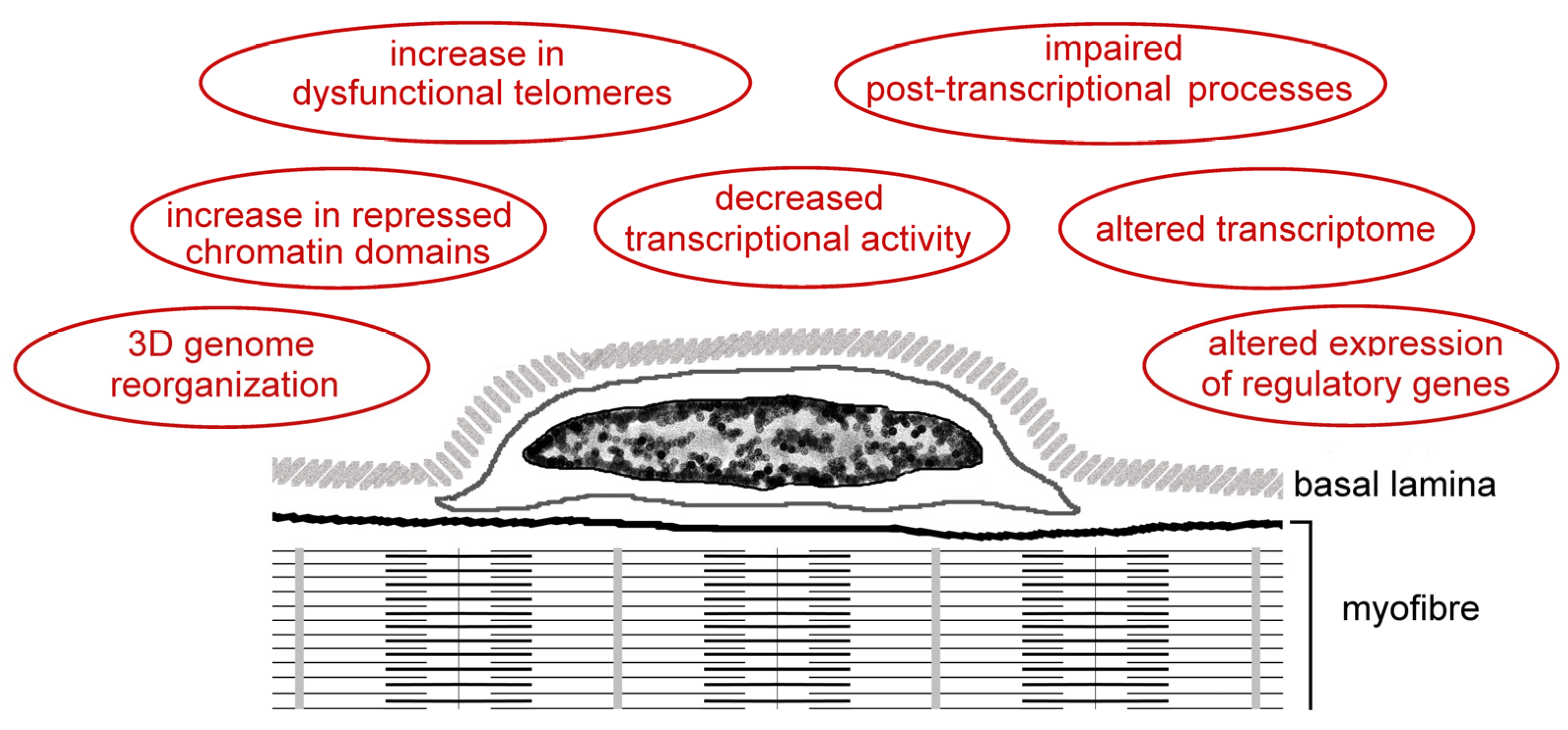
| Analyzed Features | Species | Main Findings | References |
|---|---|---|---|
| Number of myonuclei | human | Increases with aging | [76,77,78] |
| human | No change with aging | [80,81] | |
| rat | Increases with aging | [79] | |
| rat | No change with aging | [82] | |
| mouse | Decreases with aging | [70,83] | |
| Myonuclear spatial organization | human | Changes with aging | [78] |
| mouse | Changes with aging | [83] | |
| Myonuclear domain | human | Changes with aging (increases or decreases depending on the fiber type) | [78] |
| Myonuclear shape | human | Changes with aging | [78] |
| mouse | Changes with aging | [70,83,87] | |
| Myonuclear envelope | mouse | Changes with aging | [87] |
| Myonuclear area | rat | Decreases with aging | [79] |
| Condensed chromatin | rat | Increases in amount with aging | [79] |
| mouse | Increases in amount with aging | [98] | |
| Nuclear activity | rat | Decreased mRNA production with aging | [79] |
| mouse | Decreased mRNA production with aging | [100,119] | |
| mouse | Impairment of mRNA transport with aging | [119] | |
| mouse | Nucleolar changes with aging | [119] | |
| mouse | Change in pre-ribosome biogenesis and transport with aging | [79,100,120,121] | |
| mouse | Changes in transcriptional and post-transcriptional processes with aging | [120] | |
| Gene expression | mouse | Changes with aging | [69,125] |
| non-human primate | Changes with aging | [134] | |
| human | Changes with aging | [126] | |
| Epigenetic changes | human | Changes with aging | [102] |
| mouse | Changes with aging | [111] | |
| Telomeres | human | No change with aging | [149,150,151] |
| Analyzed Features | Species | Main Findings | References |
|---|---|---|---|
| Number of satellite cells | human | No changes with aging | [164,165] |
| human | Decreases with aging | [150,166,168,171] | |
| rat | No changes with aging | [167,170] | |
| rat | Decreases with aging | [169] | |
| mouse | Decreases with aging | [83,172,173] | |
| Myogenic potential | rat | No changes with aging | [176] |
| mouse | Decreases with aging but increase after physical exercise | [179,187] | |
| mouse | Decreased Notch signaling with aging | [181] | |
| Nuclear activity | rat | Decreased post-transcriptional activities with aging | [186] |
| mouse | Decreased transcriptional and post-transcriptional activities with aging but increases after physical exercise | [100] | |
| Telomeres | human | No changes in telomere length with aging | [189,190,191,192] |
| mouse | No changes in telomere length with aging | [188] | |
| human | Increases in dysfunctional telomeres with aging | [192] | |
| mouse | No changes in telomerase activity with aging | [194] | |
| Epigenetic changes | mouse | Decreased transcription of histone genes with aging | [198] |
| mouse | Overexpression of Hoxa9 gene with aging | [199] | |
| mouse | Changes in genome compartmentalization with aging | [201] | |
| mouse | Changes in transcriptome, chromatin accessibility and DNA methylation with aging | [202] | |
| mouse | Increased DNA methylation with aging | [206] | |
| human | Increased DNA methylation with aging | [111,207,208] | |
| mouse | Decreased expression of Hairless gene with aging | [211] | |
| mouse | Decreased expression of Klotho gene with aging | [212] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cisterna, B.; Malatesta, M. Molecular and Structural Alterations of Skeletal Muscle Tissue Nuclei during Aging. Int. J. Mol. Sci. 2024, 25, 1833. https://doi.org/10.3390/ijms25031833
Cisterna B, Malatesta M. Molecular and Structural Alterations of Skeletal Muscle Tissue Nuclei during Aging. International Journal of Molecular Sciences. 2024; 25(3):1833. https://doi.org/10.3390/ijms25031833
Chicago/Turabian StyleCisterna, Barbara, and Manuela Malatesta. 2024. "Molecular and Structural Alterations of Skeletal Muscle Tissue Nuclei during Aging" International Journal of Molecular Sciences 25, no. 3: 1833. https://doi.org/10.3390/ijms25031833
APA StyleCisterna, B., & Malatesta, M. (2024). Molecular and Structural Alterations of Skeletal Muscle Tissue Nuclei during Aging. International Journal of Molecular Sciences, 25(3), 1833. https://doi.org/10.3390/ijms25031833







