The Multienzyme Complex Nature of Dehydroepiandrosterone Sulfate Biosynthesis
Abstract
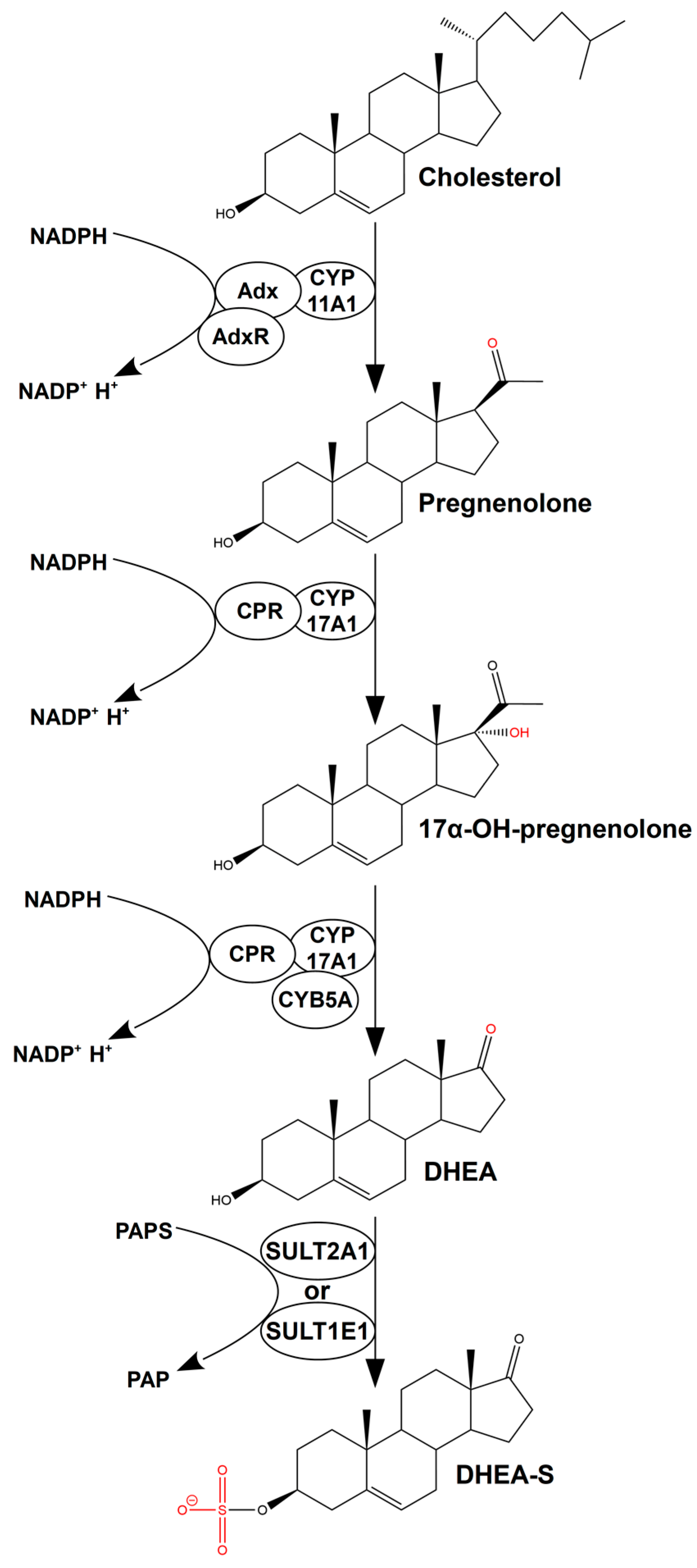
1. Introduction
2. Results
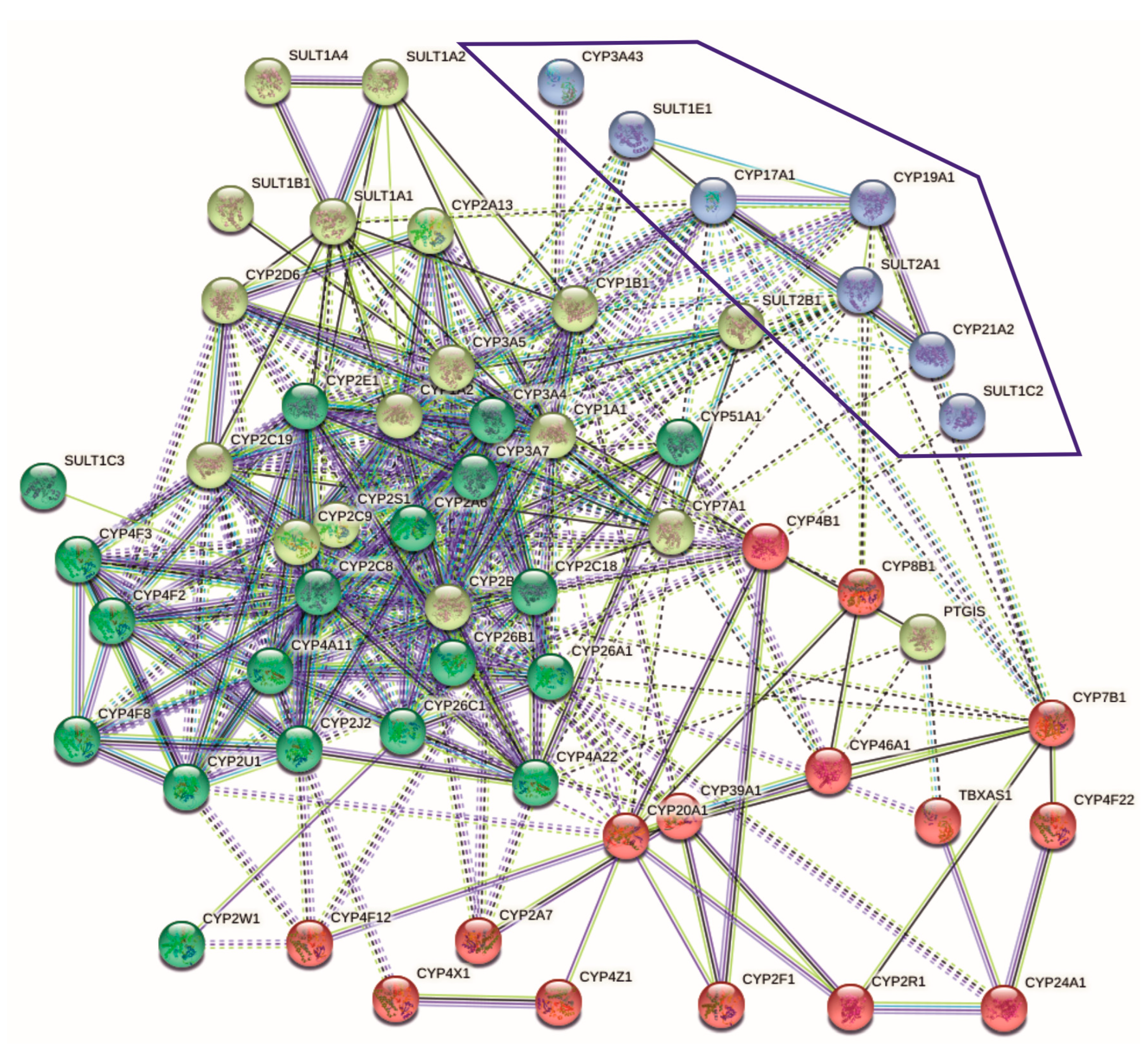
2.1. Relationships between SULTs and CYPs
2.2. SPR Analysis of Interactions of CYP17A1 and Its Redox Partners with SULT1E1 and SULT2A1
2.3. Mutual Effects of CYPs and SULTs on Their Enzymatic Activities
2.4. AP-MS Analysis
2.5. In Silico Modeling of Protein Complexes
3. Discussion
4. Materials and Methods
4.1. Instruments
4.2. Protein Preparations
4.3. Reagents
4.4. Bioinformatic Analysis
4.4.1. Protein–Protein Interaction Network Construction
4.4.2. Co-Expression Analysis
4.4.3. Text Mining Analysis
4.4.4. Structural Prediction of Protein–Protein Complexes In Silico
4.5. Surface Plasmon Resonance Analysis
4.6. Biochemical Tests
4.6.1. CYP17A1 Enzymatic Activity in the Presence of SULTs
4.6.2. SULT Enzymatic Activity in the Presence of CYP17A1 and CYB5A
4.7. Affinity Purification Combined with Mass Spectrometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vermeulen, A. Sex Hormone Status of the Postmenopausal Woman. Maturitas 1980, 2, 81–89. [Google Scholar] [CrossRef]
- Powrie, Y.S.L.; Smith, C. Central Intracrine DHEA Synthesis in Ageing-Related Neuroinflammation and Neurodegeneration: Therapeutic Potential? J. Neuroinflammation 2018, 15, 289. [Google Scholar] [CrossRef]
- Strushkevich, N.; MacKenzie, F.; Cherkesova, T.; Grabovec, I.; Usanov, S.; Park, H.-W. Structural Basis for Pregnenolone Biosynthesis by the Mitochondrial Monooxygenase System. PNAS 2011, 108, 10139–10143. [Google Scholar] [CrossRef]
- Auchus, R.J. Overview of Dehydroepiandrosterone Biosynthesis. Semin. Reprod. Med. 2004, 22, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.V.; Flück, C.E. NADPH P450 Oxidoreductase: Structure, Function, and Pathology of Diseases. Pharmacol. Ther. 2013, 138, 229–254. [Google Scholar] [CrossRef]
- Bhatt, M.R.; Khatri, Y.; Rodgers, R.J.; Martin, L.L. Role of Cytochrome B5 in the Modulation of the Enzymatic Activities of Cytochrome P450 17α-Hydroxylase/17,20-Lyase (P450 17A1). J. Steroid Biochem. Mol. Biol. 2017, 170, 2–18. [Google Scholar] [CrossRef]
- Akhtar, M.K.; Kelly, S.L.; Kaderbhai, M.A. Cytochrome B5 Modulation of 17α Hydroxylase and 17–20 Lyase (CYP17) Activities in Steroidogenesis. J. Endocrinol. 2005, 187, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Leowattana, W. DHEAS as a New Diagnostic Tool. Clin. Chim. Acta 2004, 341, 1–15. [Google Scholar] [CrossRef]
- Mueller, J.W.; Gilligan, L.C.; Idkowiak, J.; Arlt, W.; Foster, P.A. The Regulation of Steroid Action by Sulfation and Desulfation. Endocr. Rev. 2015, 36, 526–563. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.F. The Synthesis and Metabolism of Steroid Hormones. In Yen & Jaffe’s Reproductive Endocrinology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 66–92.e3. ISBN 978-1-4557-2758-2. [Google Scholar]
- Gamage, N.; Barnett, A.; Hempel, N.; Duggleby, R.G.; Windmill, K.F.; Martin, J.L.; McManus, M.E. Human Sulfotransferases and Their Role in Chemical Metabolism. Toxicol. Sci. 2006, 90, 5–22. [Google Scholar] [CrossRef]
- Frazier, M.N.; Pillon, M.C.; Kocaman, S.; Gordon, J.; Stanley, R.E. Structural Overview of Macromolecular Machines Involved in Ribosome Biogenesis. Curr. Opin. Struct. Biol. 2021, 67, 51–60. [Google Scholar] [CrossRef]
- Nassif, N.D.; Cambray, S.E.; Kraut, D.A. Slipping up: Partial Substrate Degradation by ATP-Dependent Proteases. IUBMB Life 2014, 66, 309–317. [Google Scholar] [CrossRef]
- Paiva, P.; Medina, F.E.; Viegas, M.; Ferreira, P.; Neves, R.P.P.; Sousa, J.P.M.; Ramos, M.J.; Fernandes, P.A. Animal Fatty Acid Synthase: A Chemical Nanofactory. Chem. Rev. 2021, 121, 9502–9553. [Google Scholar] [CrossRef]
- Smith, S.; Witkowski, A.; Joshi, A.K. Structural and Functional Organization of the Animal Fatty Acid Synthase. Prog. Lipid Res. 2003, 42, 289–317. [Google Scholar] [CrossRef]
- Ciszak, E.M.; Korotchkina, L.G.; Dominiak, P.M.; Sidhu, S.; Patel, M.S. Structural Basis for Flip-Flop Action of Thiamin Pyrophosphate-Dependent Enzymes Revealed by Human Pyruvate Dehydrogenase. J. Biol. Chem. 2003, 278, 21240–21246. [Google Scholar] [CrossRef] [PubMed]
- Vergères, G.; Winterhalter, K.H.; Richter, C. Localization of the N-Terminal Methionine of Rat Liver Cytochrome P-450 in the Lumen of the Endoplasmic Reticulum. Biochim. Biophys. Acta 1991, 1063, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Šrejber, M.; Navrátilová, V.; Paloncýová, M.; Bazgier, V.; Berka, K.; Anzenbacher, P.; Otyepka, M. Membrane-Attached Mammalian Cytochromes P450: An Overview of the Membrane’s Effects on Structure, Drug Binding, and Interactions with Redox Partners. J. Inorg. Biochem. 2018, 183, 117–136. [Google Scholar] [CrossRef]
- Sato, A.; Yamazaki, M.; Watanabe, H.; Sakurai, E.; Ebina, K. Human Estrogen Sulfotransferase and Its Related Fluorescently Labeled Decapeptides Specifically Interact with Oxidized Low-density Lipoprotein. J. Pept. Sci. 2020, 26, e3274. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Watanabe, H.; Yamazaki, M.; Sakurai, E.; Ebina, K. Interaction of Native- and Oxidized-Low-Density Lipoprotein with Human Estrogen Sulfotransferase. Protein J. 2021, 40, 192–204. [Google Scholar] [CrossRef]
- Zimmer, B.; Tenbusch, L.; Klymiuk, M.C.; Dezhkam, Y.; Schuler, G. SULFATION PATHWAYS: Expression of SULT2A1, SULT2B1 and HSD3B1 in the Porcine Testis and Epididymis. J. Mol. Endocrinol. 2018, 61, M41–M55. [Google Scholar] [CrossRef]
- Kastrinou Lampou, V.; Poller, B.; Huth, F.; Fischer, A.; Kullak-Ublick, G.A.; Arand, M.; Schadt, H.S.; Camenisch, G. Novel Insights into Bile Acid Detoxification via CYP, UGT and SULT Enzymes. Toxicol. In Vitro 2023, 87, 105533. [Google Scholar] [CrossRef]
- Mellacheruvu, D.; Wright, Z.; Couzens, A.L.; Lambert, J.-P.; St-Denis, N.A.; Li, T.; Miteva, Y.V.; Hauri, S.; Sardiu, M.E.; Low, T.Y.; et al. The CRAPome: A Contaminant Repository for Affinity Purification-Mass Spectrometry Data. Nat. Methods 2013, 10, 730–736. [Google Scholar] [CrossRef]
- Idris, M.; Mitchell, D.J.; Gordon, R.; Sidharthan, N.P.; Butcher, N.J.; Minchin, R.F. Interaction of the Brain-Selective Sulfotransferase SULT4A1 with Other Cytosolic Sulfotransferases: Effects on Protein Expression and Function. Drug Metab. Dispos. 2020, 48, 337–344. [Google Scholar] [CrossRef]
- Pröschel, M.; Detsch, R.; Boccaccini, A.R.; Sonnewald, U. Engineering of Metabolic Pathways by Artificial Enzyme Channels. Front. Bioeng. Biotechnol. 2015, 3, 168. [Google Scholar] [CrossRef]
- Iyanagi, T. Molecular Mechanism of Phase I and Phase II Drug-Metabolizing Enzymes: Implications for Detoxification. Int. Rev. Cytol. 2007, 260, 35–112. [Google Scholar] [CrossRef]
- Liston, H.L.; Markowitz, J.S.; DeVane, C.L. Drug Glucuronidation in Clinical Psychopharmacology. J. Clin. Psychopharmacol. 2001, 21, 500–515. [Google Scholar] [CrossRef]
- Constantinescu, G.; Langton, K.; Conrad, C.; Amar, L.; Assié, G.; Gimenez-Roqueplo, A.-P.; Blanchard, A.; Larsen, C.K.; Mulatero, P.; Williams, T.A.; et al. Glucocorticoid Excess in Patients with Pheochromocytoma Compared with Paraganglioma and Other Forms of Hypertension. J. Clin. Endocrinol. Metab. 2020, 105, e3374–e3383. [Google Scholar] [CrossRef] [PubMed]
- Akwa, Y. Steroids and Alzheimer’s Disease: Changes Associated with Pathology and Therapeutic Potential. Int. J. Mol. Sci. 2020, 21, 4812. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Zárate, M.J.; Cittelly, D.M. Sex Steroid Hormone Function in the Brain Niche: Implications for Brain Metastatic Colonization and Progression. Cancer Rep. 2022, 5, e1241. [Google Scholar] [CrossRef] [PubMed]
- Blanco, L.Z., Jr.; Kuhn, E.; Morrison, J.C.; Bahadirli-Talbott, A.; Smith-Sehdev, A.; Kurman, R.J. Steroid Hormone Synthesis by the Ovarian Stroma Surrounding Epithelial Ovarian Tumors: A Potential Mechanism in Ovarian Tumorigenesis. Mod. Pathol. 2017, 30, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Eacker, S.M.; Agrawal, N.; Qian, K.; Dichek, H.L.; Gong, E.-Y.; Lee, K.; Braun, R.E. Hormonal Regulation of Testicular Steroid and Cholesterol Homeostasis. Mol. Endocrinol. 2008, 22, 623–635. [Google Scholar] [CrossRef]
- Mukudai, S.; Matsuda, K.I.; Nishio, T.; Sugiyama, Y.; Bando, H.; Hirota, R.; Sakaguchi, H.; Hisa, Y.; Kawata, M. Differential Responses to Steroid Hormones in Fibroblasts from the Vocal Fold, Trachea, and Esophagus. Endocrinology 2015, 156, 1000–1009. [Google Scholar] [CrossRef]
- Taves, M.D.; Ashwell, J.D. Glucocorticoids in T Cell Development, Differentiation and Function. Nat. Rev. Immunol. 2021, 21, 233–243. [Google Scholar] [CrossRef]
- Ershov, P.; Yablokov, O.; Florinskaya, A.; Mezentsev, Y.; Kaluzhskiy, L.; Tumilovich, A.; Gilep, A.; Usanov, S.; Ivanov, A.S. SPR-Based Study of Affinity of Cytochrome P450s / Redox Partners Interactions Modulated by Steroidal Substrates. J. Steroid Biochem. Mol. Biol. 2019, 187, 124–129. [Google Scholar] [CrossRef]
- Yoshinari, K.; Petrotchenko, E.V.; Pedersen, L.C.; Negishi, M. Crystal Structure-Based Studies of Cytosolic Sulfotransferase. J. Biochem. Mol. Toxicol. 2001, 15, 67–75. [Google Scholar] [CrossRef]
- Kakuta, Y.; Pedersen, L.G.; Carter, C.W.; Negishi, M.; Pedersen, L.C. Crystal Structure of Estrogen Sulphotransferase. Nat. Struct. Biol. 1997, 4, 904–908. [Google Scholar] [CrossRef]
- Petta, I.; Lievens, S.; Libert, C.; Tavernier, J.; De Bosscher, K. Modulation of Protein–Protein Interactions for the Development of Novel Therapeutics. Mol. Ther. 2016, 24, 707–718. [Google Scholar] [CrossRef]
- Dudas, B.; Toth, D.; Perahia, D.; Nicot, A.B.; Balog, E.; Miteva, M.A. Insights into the Substrate Binding Mechanism of SULT1A1 through Molecular Dynamics with Excited Normal Modes Simulations. Sci. Rep. 2021, 11, 13129. [Google Scholar] [CrossRef]
- Weagel, E.G.; Foulks, J.M.; Siddiqui, A.; Warner, S.L. Molecular Glues: Enhanced Protein-Protein Interactions and Cell Proteome Editing. Med. Chem. Res. 2022, 31, 1068–1087. [Google Scholar] [CrossRef]
- Liu, Y.; Denisov, I.G.; Sligar, S.G.; Kincaid, J.R. Substrate-Specific Allosteric Effects on the Enhancement of CYP17A1 Lyase Efficiency by Cytochrome b5. J. Am. Chem. Soc. 2021, 143, 3729–3733. [Google Scholar] [CrossRef]
- McDougle, D.R.; Palaria, A.; Magnetta, E.; Meling, D.D.; Das, A. Functional Studies of N-Terminally Modified CYP2J2 Epoxygenase in Model Lipid Bilayers. Protein Sci. 2013, 22, 964–979. [Google Scholar] [CrossRef]
- Yadav, R.; Scott, E.E. Endogenous Insertion of Non-Native Metalloporphyrins into Human Membrane Cytochrome P450 Enzymes. J. Biol. Chem. 2018, 293, 16623–16634. [Google Scholar] [CrossRef]
- Storbeck, K.-H.; Schiffer, L.; Baranowski, E.S.; Chortis, V.; Prete, A.; Barnard, L.; Gilligan, L.C.; Taylor, A.E.; Idkowiak, J.; Arlt, W.; et al. Steroid Metabolome Analysis in Disorders of Adrenal Steroid Biosynthesis and Metabolism. Endocr. Rev. 2019, 40, 1605–1625. [Google Scholar] [CrossRef]
- Fernández-Cancio, M.; Camats, N.; Flück, C.E.; Zalewski, A.; Dick, B.; Frey, B.M.; Monné, R.; Torán, N.; Audí, L.; Pandey, A.V. Mechanism of the Dual Activities of Human CYP17A1 and Binding to Anti-Prostate Cancer Drug Abiraterone Revealed by a Novel V366M Mutation Causing 17,20 Lyase Deficiency. Pharmaceuticals 2018, 11, 37. [Google Scholar] [CrossRef]
- Peng, H.-M.; Liu, J.; Forsberg, S.E.; Tran, H.T.; Anderson, S.M.; Auchus, R.J. Catalytically Relevant Electrostatic Interactions of Cytochrome P450c17 (CYP17A1) and Cytochrome B5. J. Biol. Chem. 2014, 289, 33838–33849. [Google Scholar] [CrossRef]
- Yablokov, E.; Florinskaya, A.; Medvedev, A.; Sergeev, G.; Strushkevich, N.; Luschik, A.; Shkel, T.; Haidukevich, I.; Gilep, A.; Usanov, S.; et al. Thermodynamics of Interactions between Mammalian Cytochromes P450 and B5. Arch. Biochem. Biophys. 2017, 619, 10–15. [Google Scholar] [CrossRef]
- Xu, D.; Tsai, C.J.; Nussinov, R. Hydrogen Bonds and Salt Bridges across Protein-Protein Interfaces. Protein Eng. Des. Sel. 1997, 10, 999–1012. [Google Scholar] [CrossRef]
- Conte, L.L.; Chothia, C.; Janin, J. The Atomic Structure of Protein-Protein Recognition Sites 1 1Edited by A. R. Fersht. J. Mol. Biol. 1999, 285, 2177–2198. [Google Scholar] [CrossRef]
- Deng, B.; Parthasarathy, S.; Wang, W.; Gibney, B.R.; Battaile, K.P.; Lovell, S.; Benson, D.R.; Zhu, H. Study of the Individual Cytochrome B5 and Cytochrome B5 Reductase Domains of Ncb5or Reveals a Unique Heme Pocket and a Possible Role of the CS Domain. J. Biol. Chem. 2010, 285, 30181–30191. [Google Scholar] [CrossRef]
- Esteves, F.; Campelo, D.; Gomes, B.C.; Urban, P.; Bozonnet, S.; Lautier, T.; Rueff, J.; Truan, G.; Kranendonk, M. The Role of the FMN-Domain of Human Cytochrome P450 Oxidoreductase in Its Promiscuous Interactions With Structurally Diverse Redox Partners. Front. Pharmacol. 2020, 11, 299. [Google Scholar] [CrossRef]
- Chapman, E.; Best, M.D.; Hanson, S.R.; Wong, C. Sulfotransferases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility. Angew. Chem. Int. Ed. 2004, 43, 3526–3548. [Google Scholar] [CrossRef]
- Thomas, M.P.; Potter, B.V.L. The Structural Biology of Oestrogen Metabolism. J. Steroid Biochem. Mol. Biol. 2013, 137, 27–49. [Google Scholar] [CrossRef]
- Otyepka, M.; Skopalík, J.; Anzenbacherová, E.; Anzenbacher, P. What Common Structural Features and Variations of Mammalian P450s Are Known to Date? Biochim. Biophys. Acta (BBA) Gen. Subj. 2007, 1770, 376–389. [Google Scholar] [CrossRef]
- Anzenbacher, P.; Anzenbacherova, E.; Lange, R.; Skopalik, J.; Otyepka, M. Active Sites of Cytochromes P450: What Are They Like? Acta Chim. Slov. 2008, 55, 63–66. [Google Scholar]
- Urban, P.; Lautier, T.; Pompon, D.; Truan, G. Ligand Access Channels in Cytochrome P450 Enzymes: A Review. Int. J. Mol. Sci. 2018, 19, 1617. [Google Scholar] [CrossRef]
- Harford, S.; Weitzman, P.D. Evidence of Isosteric and Allosteric Nucleotide Inhibition of Citrate Synthease from Multiple-Inhibition Studies. Biochem. J. 1975, 151, 455–458. [Google Scholar] [CrossRef]
- Pechurskaya, T.A.; Lukashevich, O.P.; Gilep, A.A.; Usanov, S.A. Engineering, Expression, and Purification of “Soluble” Human Cytochrome P45017α and Its Functional Characterization. Biochemistry 2008, 73, 806–811. [Google Scholar] [CrossRef]
- Chudaev, M.V.; Usanov, S.A. Expression of Functionally Active Cytochrome B5 in Escherichia Coli: Isolation, Purification, and Use of the Immobilized Recombinant Heme Protein for Affinity Chromatography of Electron-Transfer Proteins. Biochemistry 1997, 62, 401–411. [Google Scholar]
- Bonina, T.A.; Gilep, A.A.; Estabrook, R.W.; Usanov, S.A. Engineering of Proteolytically Stable NADPH-Cytochrome P450 Reductase. Biochemistry 2005, 70, 357–365. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Allot, A.; Chen, Q.; Kim, S.; Vera Alvarez, R.; Comeau, D.C.; Wilbur, W.J.; Lu, Z. LitSense: Making Sense of Biomedical Literature at Sentence Level. Nucleic Acids Res. 2019, 47, W594–W599. [Google Scholar] [CrossRef]
- Wei, C.-H.; Allot, A.; Leaman, R.; Lu, Z. PubTator Central: Automated Concept Annotation for Biomedical Full Text Articles. Nucleic Acids Res. 2019, 47, W587–W593. [Google Scholar] [CrossRef]
- Tsuruoka, Y.; Tsujii, J.; Ananiadou, S. FACTA: A Text Search Engine for Finding Associated Biomedical Concepts. Bioinformatics 2008, 24, 2559–2560. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kwon, D.; Shin, S.-Y.; Wilbur, W.J. PIE the Search: Searching PubMed Literature for Protein Interaction Information. Bioinformatics 2012, 28, 597–598. [Google Scholar] [CrossRef]
- Arruda, H.; Silva, E.R.; Lessa, M.; Proença, D.; Bartholo, R. VOSviewer and Bibliometrix. J. Med. Libr. Assoc. 2022, 110, 392–395. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y. How Significant Is a Protein Structure Similarity with TM-Score = 0.5? Bioinformatics 2010, 26, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Davis, F.P.; Sali, A. PIBASE: A Comprehensive Database of Structurally Defined Protein Interfaces. Bioinformatics 2005, 21, 1901–1907. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wu, F.; Jernigan, R.L.; Dobbs, D.; Honavar, V. Characterization of Protein-Protein Interfaces. Protein J. 2008, 27, 59–70. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Jurcik, A.; Bednar, D.; Byska, J.; Marques, S.M.; Furmanova, K.; Daniel, L.; Kokkonen, P.; Brezovsky, J.; Strnad, O.; Stourac, J.; et al. CAVER Analyst 2.0: Analysis and Visualization of Channels and Tunnels in Protein Structures and Molecular Dynamics Trajectories. Bioinformatics 2018, 34, 3586–3588. [Google Scholar] [CrossRef]
- Müller, K.M.; Arndt, K.M.; Plückthun, A. Model and Simulation of Multivalent Binding to Fixed Ligands. Anal. Biochem. 1998, 261, 149–158. [Google Scholar] [CrossRef]
- Svirid, A.V.; Ershov, P.V.; Yablokov, E.O.; Kaluzhskiy, L.A.; Mezentsev, Y.V.; Florinskaya, A.V.; Sushko, T.A.; Strushkevich, N.V.; Gilep, A.A.; Usanov, S.A.; et al. Direct Molecular Fishing of New Protein Partners for Human Thromboxane Synthase. Acta Naturae 2017, 9, 92–100. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]

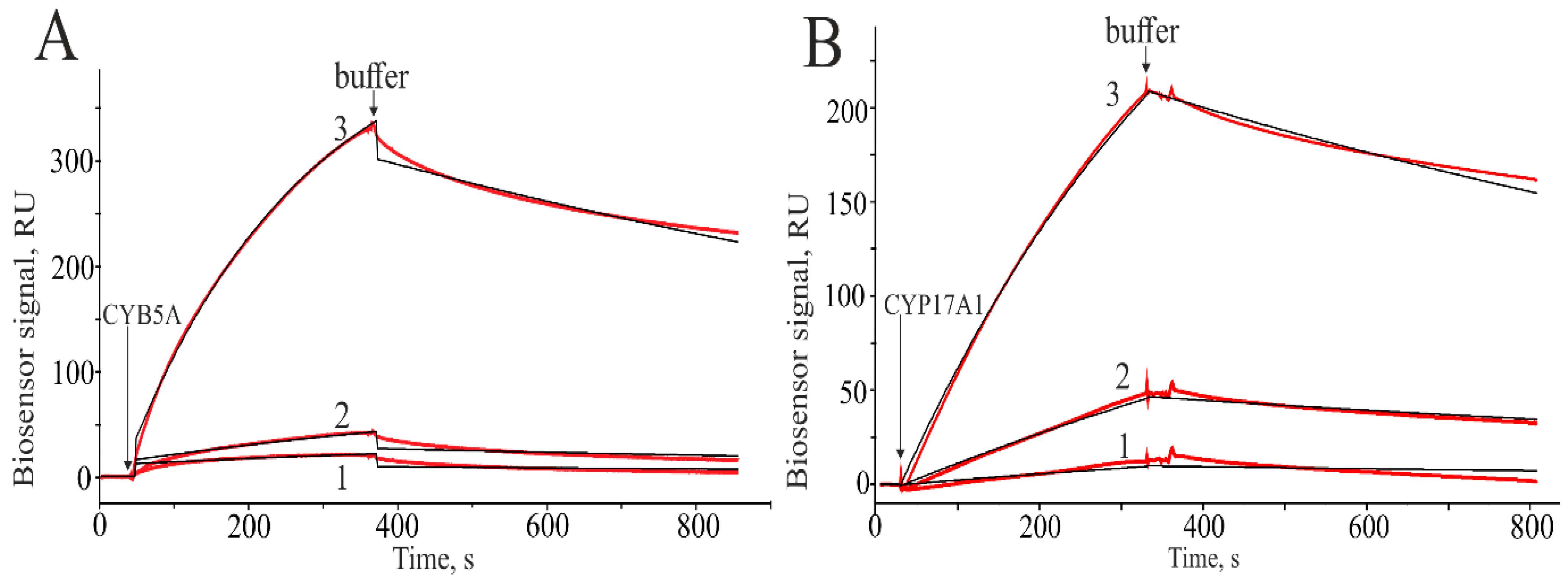
| Interacting Proteins (Ligand/Analyte) | kon (103 × M−1 s−1) | koff (10−4 × s−1) | Kd nM |
|---|---|---|---|
| SULT1E1/CYP17A1 | 2.96 ± 0.31 | 5.80 ± 0.49 | 196 ± 21 |
| SULT1E1/CYP17A1 (PAPS *) | 5.52 ± 0.52 | 7.60 ± 0.65 | 138 ± 15 |
| SULT2A1/CYP17A1 | 4.45 ± 0.35 | 5.50 ± 0.50 | 124 ± 11 |
| SULT2A1/CYP17A1 (PAPS) | 13.10 ± 1.28 | 13.70 ± 1.50 | 105 ± 12 |
| SULT1E1/CYB5A | 13.80 ± 1.12 | 9.80 ± 1.00 | 71 ± 8 |
| SULT1E1/CYB5A (PAPS) | 7.65 ± 0.60 | 6.70 ± 0.50 | 88 ± 8 |
| SULT2A1/CYB5A | n/d | n/d | n/d |
| SULT2A1/CYB5A (PAPS) | 19.90 ± 2.20 | 14.00 ± 1.20 | 71 ± 7 |
| CPR/SULT1E1 | n/d | n/d | n/d |
| CPR/SULT1E1 (PAPS) | n/d | n/d | n/d |
| CPR/SULT2A1 | 0.57 ± 0.48 | 7.80 ± 0.80 | 1370 ± 126 |
| CPR/SULT2A1 (PAPS) | 0.67 ± 0.55 | 5.10 ± 0.40 | 957 ± 89 |
| SULT1E1 Activity (min−1) | Resume | SULT2A1 Activity (min−1) | Resume | |
|---|---|---|---|---|
| Control | 0.09 ± 0.02 | - | 0.13 ± 0.06 | - |
| CYP17A1tr + CYB5A | 0.16 ± 0.03 | ↑ ↑ * | 0.30 ± 0.07 | ↑ ↑ |
| CYP17A1tr | 0.08 ± 0.02 | not changed | 0.23 ± 0.02 | ↑ ↑ |
| CYP17A1full + CYB5A | 0.1700 ± 0.0009 | ↑ ↑ | 0.13 ± 0.08 | not changed |
| CYP17A1full | 0.120 ± 0.004 | ↑ | 0.13 ± 0.04 | not changed |
| CYB5A | 0.050 ± 0.005 | ↓ | 0.080 ± 0.006 | ↓ |
| Gene Names | Protein Names | Protein IDs | Unique Peptides (Unmodified) | Sequence Coverage, % |
|---|---|---|---|---|
| Sult1b1 | Sulfotransferase family cytosolic 1B member 1 | P52847 | 4 | 13.7 |
| Sardh | Sarcosine dehydrogenase, mitochondrial | Q64380 | 4 | 3.2 |
| Sult1a1 | Sulfotransferase 1A1 | P17988 | 3 | 10.3 |
| Sult1c1 | Sulfotransferase 1C1 | P50237 | 3 | 18.4 |
| Cyb5a | Cytochrome b5, microsomal | P00173 | 2 | 23.9 |
| Ass1 | Argininosuccinate synthase | P09034 | 2 | 4.9 |
| Aldob | Fructose-bisphosphate aldolase B | P00884 | 1 | 5.5 |
| Rab7a | Ras-related protein Rab-7a | P09527 | 1 | 6.8 |
| Cyp2d10 * | Cytochrome P450 2D10 | P12939 | 1 | 2.2 |
| Cyp2d1 * | Cytochrome P450 2D1 | P10633 | ||
| Bdh1 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial | P29147 | 1 | 5.8 |
| Pfkl | ATP-dependent 6-phosphofructokinase, liver type | P30835 | 1 | 1.2 |
| Aldh3a2 | Fatty aldehyde dehydrogenase | P30839 | 1 | 1.9 |
| Gnb2 * | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-2 | P54313 | 1 | 4.1 |
| Gnb4 * | Guanine nucleotide-binding protein subunit beta-4 | O35353 | ||
| Sult1c2a * | Sulfotransferase 1C2a | Q9WUW9 | 1 | 3.7 |
| Sult1c2 * | Sulfotransferase 1C2 | Q9WUW8 |
| Complexes | AlphaFold2 | PDBePISA | |||
|---|---|---|---|---|---|
| pLDDT | pTMscore | Interface Area (Å2) | Salt Bridges | H-Bonds | |
| CYP17A1/SULT1E1 (monomer) | 89.4 | 0.7 | 836 | 3 | 5 |
| CYP17A1/SULT1E1 (homodimer) | 90.6 | 0.6 | 1198 | 7 | 8 |
| CYB5A/SULT1E1 (monomer) | 88.1 | 0.8 | 699 | 6 | 7 |
| CYB5A/SULT1E1 (homodimer) | 90 | 0.8 | 702 | 0 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tumilovich, A.; Yablokov, E.; Mezentsev, Y.; Ershov, P.; Basina, V.; Gnedenko, O.; Kaluzhskiy, L.; Tsybruk, T.; Grabovec, I.; Kisel, M.; et al. The Multienzyme Complex Nature of Dehydroepiandrosterone Sulfate Biosynthesis. Int. J. Mol. Sci. 2024, 25, 2072. https://doi.org/10.3390/ijms25042072
Tumilovich A, Yablokov E, Mezentsev Y, Ershov P, Basina V, Gnedenko O, Kaluzhskiy L, Tsybruk T, Grabovec I, Kisel M, et al. The Multienzyme Complex Nature of Dehydroepiandrosterone Sulfate Biosynthesis. International Journal of Molecular Sciences. 2024; 25(4):2072. https://doi.org/10.3390/ijms25042072
Chicago/Turabian StyleTumilovich, Anastasiya, Evgeniy Yablokov, Yuri Mezentsev, Pavel Ershov, Viktoriia Basina, Oksana Gnedenko, Leonid Kaluzhskiy, Tatsiana Tsybruk, Irina Grabovec, Maryia Kisel, and et al. 2024. "The Multienzyme Complex Nature of Dehydroepiandrosterone Sulfate Biosynthesis" International Journal of Molecular Sciences 25, no. 4: 2072. https://doi.org/10.3390/ijms25042072
APA StyleTumilovich, A., Yablokov, E., Mezentsev, Y., Ershov, P., Basina, V., Gnedenko, O., Kaluzhskiy, L., Tsybruk, T., Grabovec, I., Kisel, M., Shabunya, P., Soloveva, N., Vavilov, N., Gilep, A., & Ivanov, A. (2024). The Multienzyme Complex Nature of Dehydroepiandrosterone Sulfate Biosynthesis. International Journal of Molecular Sciences, 25(4), 2072. https://doi.org/10.3390/ijms25042072







