Importance of Non-Covalent Interactions in Yeast Cell Wall Molecular Organization
Abstract
1. Introduction
2. Cell Wall Proteins—Evolution of Viewpoints
3. SEP Proteins Are a Heterogeneous Group of Proteins of the Cell Wall
4. Extracellular Vesicles as a Mode of Compartmentalization of NCAp
5. Polyfunctional Protein with Amyloid Properties, Bgl2, Is the Most Studied NCAp
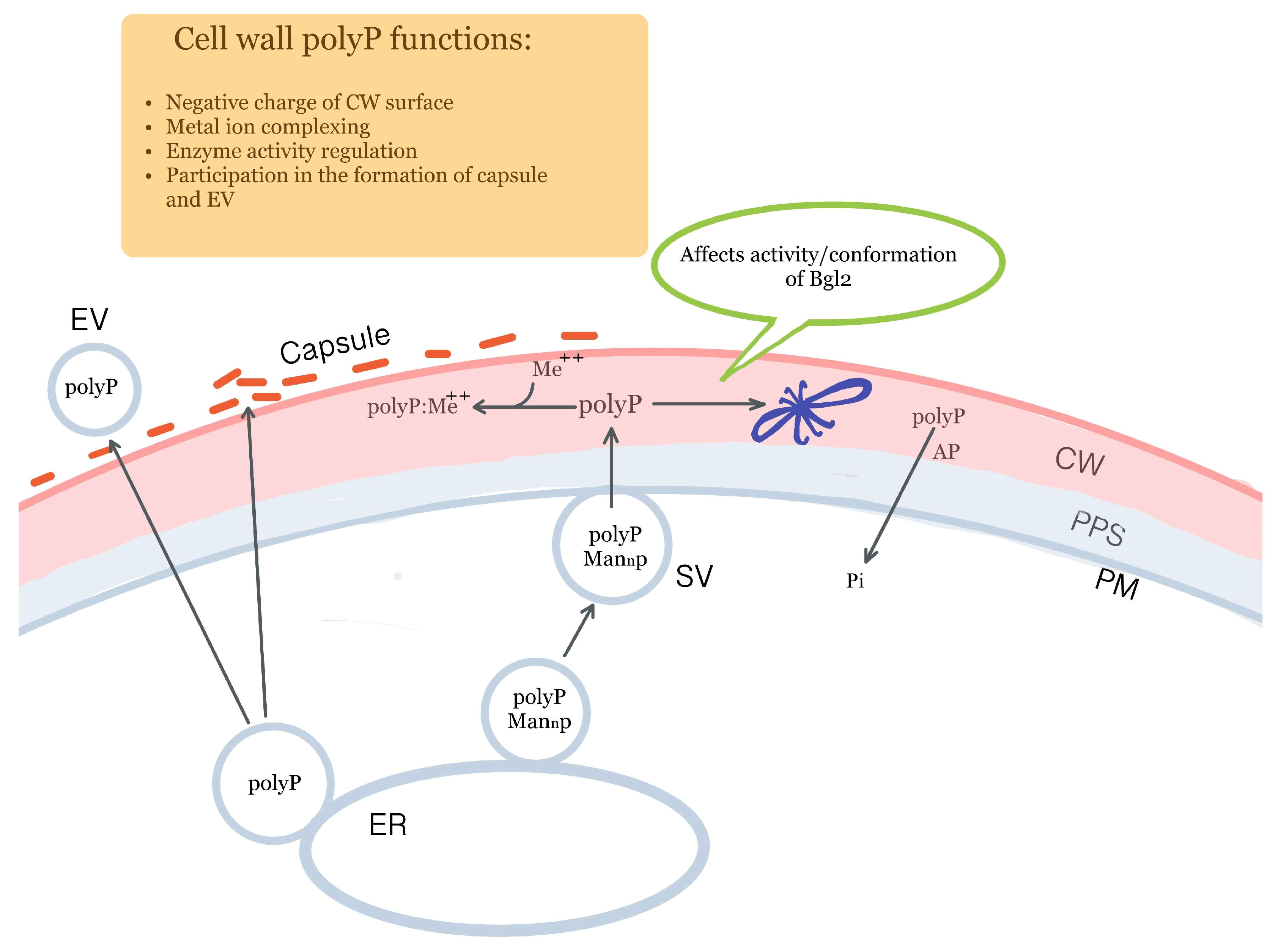
6. Non-Covalently Bound CW Polyphosphates Are a Necessary Component of the Cell Wall and Possible Participants in Enzyme Activity Regulation
7. Conclusions
8. Future Directions
- −
- Studying the properties (especially amyloidogenic properties) of NCAp which allow them to be incorporated into the yeast CW so strongly but flexibly;
- −
- Studying the localization and compartmentalization of NCAp in the yeast CW;
- −
- Studying of the role of polyphosphates in the regulation of the activity of enzymes with glucan-remodeling activity;
- −
- Studying of post-translational modifications of yeast CW proteins, primarily polysaccharide-modeling enzymes belong to NCAp in the CW.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Klis, F.M. Review: Cell wall assembly in yeast. Yeast 1994, 10, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; de Koster, C.G.; Brul, S. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 2014, 13, 2–9. [Google Scholar] [CrossRef]
- MacWilliam, I.C. The structure, synthesis and functions of the yeast cell wall—A review. J. Inst. Brew. 1970, 76, 513–571. [Google Scholar] [CrossRef]
- Kulaev, I.S.; Vagabov, V.M.; Tsiomenko, A.B. Correlation between cell wall polysaccharide accumulation and high-molecular polyphosphate fractions in yeasts. Dokl. Akad. Nauk. SSSR 1972, 204, 734–736. [Google Scholar] [PubMed]
- Tsiomenko, A.B.; Vagabov, V.M.; Avgustin, I.; Kulaev, I.S. Interrelation of metabolism of inorganic polyphosphates and mannan in yeasts. Dokl. Akad. Nauk. SSSR 1974, 215, 478–480. [Google Scholar] [PubMed]
- Casadevall, A.; Nosanchuk, J.D.; Williamson, P.; Rodrigues, M.L. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009, 17, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef]
- Vargas, G.; Rocha, J.D.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.; Medeiros, L.C.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 2015, 17, 389407. [Google Scholar] [CrossRef]
- Francois, J.M. Cell surface interference with plasma membrane and transport processes in yeasts. Adv. Exp. Med. Biol. 2016, 892, 11–31. [Google Scholar] [CrossRef]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.S.; et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2019, 2, 305. [Google Scholar] [CrossRef]
- Honorato, L.; de Araujo, J.F.D.; Ellis, C.C.; Piffer, A.C.; Pereira, Y.; Frases, S.; de Sousa Araújo, G.R.; Pontes, B.; Mendes, M.T.; Pereira, M.D.; et al. Extracellular vesicles regulate biofilm formation and yeast-to-hypha differentiation in Candida Albicans. Mbio 2022, 13, e0030122. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Lenardon, M.D. Architecture of the dynamic fungal cell wall. Nat. Rev. Microbiol. 2023, 21, 248–259. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI Pathway: Regulation of the transcriptional adaptive response to cell wall stress in yeast. J. Fungi 2017, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhao, F.; Lin, Y.; Zheng, S.; Liang, S.; Han, S. RNA-Seq analysis of global transcriptomic changes suggests a roles for the MAPK pathway and carbon metabolism in cell wall maintenance in a Saccharomyces cerevisiae FKS1 mutant. Biochem. Biophys. Res. Commun. 2018, 500, 603–608. [Google Scholar] [CrossRef]
- Liu, L.; Levin, D.E. Intracellular mechanism by which genotoxic stress activates yeast SAPK Mpk1. Mol. Biol. Cell 2018, 29, 2898–2909. [Google Scholar] [CrossRef]
- Molon, M.; Woznicka, O.; Zebrowski, J. Cell wall biosynthesis impairment affects the budding lifespan of the Saccharomyces cerevisiae yeast. Biogerontology 2018, 19, 67–79. [Google Scholar] [CrossRef]
- Davì, V.; Chevalier, L.; Guo, H.; Tanimoto, H.; Barrett, K.; Couturier, E.; Boudaoud, A.; Minc, N. Systematic mapping of cell wall mechanics in the regulation of cell morphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 13833–13838. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R.G. Adhesins of yeasts: Protein structure and interactions. J. Fungi 2018, 4, 119. [Google Scholar] [CrossRef]
- Jendretzki, A.; Wittland, J.; Wilk, S.; Straede, A.; Heinisch, J.J. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur. J. Cell Biol. 2011, 90, 740–744. [Google Scholar] [CrossRef]
- Kock, C.; Arlt, H.; Ungermann, C.; Heinisch, J.J. Yeast cell wall integrity sensors form specific plasma membrane microdomains important for signaling. Cell Microbiol. 2016, 18, 1251–1267. [Google Scholar] [CrossRef] [PubMed]
- Davì, V.; Tanimoto, H.; Ershov, D.; Haupt, A.; de Belly, H.; Le Borgne, R.; Couturier, E.; Boudaoud, A.; Minc, N. Mechanosensation Dynamically Coordinates Polar Growth and Cell Wall Assembly to Promote Cell Survival. Dev. Cell. 2018, 45, 170–182.e7. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Minc, N.; Peter, M. Cells under pressure: How yeast cells respond to mechanical forces. Trends Microbiol. 2022, 30, 495–510. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Díaz-Jiménez, D.F.; Jan Kullberg, B.; Brown, A.J.; Odds, F.C.; et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 2010, 285, 12087–12095. [Google Scholar] [CrossRef]
- Hoving, L.R.; van der Zande, H.J.P.; Pronk, A.; Guigas, B.; Willems van Dijk, K.; van Harmelen, V. Dietary yeast-derived mannan oligosaccharides have immune-modulatory properties but do not improve high fat diet-induced obesity and glucose intolerance. PLoS ONE 2018, 13, e0196165. [Google Scholar] [CrossRef]
- Kalebina, T.S.; Plotnikova, T.A.; Gorkovskii, A.A.; Selyakh, I.O.; Galzitskaya, O.V.; Bezsonov, E.E.; Gellissen, G.; Kulaev, I.S. Amyloid-like properties of Saccharomyces cerevisiae cell wall glucantransferase Bgl2p: Prediction and experimental evidences. Prion 2008, 2, 91–96. [Google Scholar] [CrossRef]
- Bezsonov, E.E.; Groenning, M.; Galzitskaya, O.V.; Gorkovskii, A.A.; Semisotnov, G.V.; Selyakh, I.O.; Ziganshin, R.H.; Rekstina, V.V.; Kudryashova, I.B.; Kuznetsov, S.A.; et al. Amyloidogenic peptides of yeast cell wall glucantransferase Bgl2p as a model for the investigation of its pH-dependent fibril formation. Prion 2013, 7, 175–184. [Google Scholar] [CrossRef]
- Mouyna, I.; Hartl, L.; Latge, J.P. β-1,3-glucan modifying enzymes in Aspergillus fumigatus. Front. Microbiol. 2013, 4, 81. [Google Scholar] [CrossRef]
- Klis, F.M.; Boorsma, A.; de Groot, P.W. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef]
- Teparic, R.; Lozancic, M.; Mrsa, V. Evolutionary Overview of Molecular Interactions and Enzymatic Activities in the Yeast Cell Walls. Int. J. Mol. Sci. 2020, 21, 8996. [Google Scholar] [CrossRef]
- Ene, I.V.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.A.; Munro, C.A.; Brown, A.J. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio 2015, 6, e00986. [Google Scholar] [CrossRef]
- Borovikova, D.; Teparić, R.; Mrša, V.; Rapoport, A. Anhydrobiosis in yeast: Cell wall mannoproteins are important for yeast Saccharomyces cerevisiae resistance to dehydration. Yeast 2016, 33, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, J.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. Differential Proteome Analysis of a Flor Yeast Strain under Biofilm Formation. Int. J. Mol. Sci. 2017, 18, 720. [Google Scholar] [CrossRef] [PubMed]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic Fungal Cell Wall Architecture in Stress Adaptation and Immune Evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Phaff, H.J. Cell wall of yeasts. Annu. Rev. Microbiol. 1963, 17, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E.; Farkas, V. The control of morphogenesis: An enzymatic mechanism for the initiation of septum formation in yeast. Proc. Nat. Acad. Sci. USA 1971, 68, 2052–2056. [Google Scholar] [CrossRef]
- Manners, D.J.; Masson, A.J.; Patterson, J.C. The structure of a β-(1→3)-d-glucan from yeast cell walls. Biochem. J. 1973, 135, 19–30. [Google Scholar] [CrossRef]
- Manners, D.J.; Masson, A.J.; Patterson, J.C. The heterogeneity of glucan preparations from the walls of various yeasts. J. Gen. Microbiol. 1974, 80, 411–417. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ballou, C.E.; Lipke, P.N.; Rashke, W.C. Structure and immuno-chemistry of the cell wall mannans from S. chevalieri, S. italicus, S. diastaticus and S. carlsbergensis. J. Bacteriol. 1974, 117, 461–467. [Google Scholar] [CrossRef]
- Ballou, C.E.; Rashke, W.C. Polymorphism of the somatic antigen of yeast. Science 1974, 184, 127–134. [Google Scholar] [CrossRef]
- Fleet, G.H.; Phaff, H.J. Lysis of yeast cell walls: Glucanases from Bacillus circulans WL-12. J. Bacteriol. 1974, 119, 207–219. [Google Scholar] [CrossRef]
- Fleet, G.H.; Manners, D.J. The enzymic degradation of an alkali-soluble glucan from the cell walls of Saccharomyces cerevisiae. J. Gen. Microbiol. 1977, 98, 315–327. [Google Scholar] [CrossRef]
- Molano, J.; Bowers, B.; Cabib, E. Distribution of chitin in the yeast cell wall. An ultrastructural and chemical study. J. Cell Biol. 1980, 85, 199–212. [Google Scholar] [CrossRef]
- Holan, Z.; Pokorny, V.; Beran, K.; Gemperle, A.; Tuzar, Z.; Baldarian, J. The glucan-chitin complex in Saccharomyces cerevisiae. V. Precise location of chitin and glucan in bud scar and their physico-chemical characterization. Arch. Microbiol. 1981, 130, 312–319. [Google Scholar] [CrossRef]
- Fleet, G.H.; Phaff, H.J. Fungal Glucans—Structure and Metabolism. In Plant Carbohydrates II. Encyclopedia of Plant Physiology; Tanner, W., Loewus, F.A., Eds.; Springer: Berlin/Heidelberg, Germany, 1981; Volume 13, pp. 416–440. ISBN 978-3-642-68236-0. [Google Scholar]
- Kulaev, I.S. The Biochemistry of Inorganic Polyphosphates; John Wiley & Sons: New York, NY, USA, 1979; p. 255. ISBN 0-471-27574-3. [Google Scholar]
- Farkaš, V. The Fungal Cell Wall. In Fungal Protoplasts. Applications in Biochemistry and Genetics, 1st ed.; Peberdy, J.F., Ed.; CRC Press: Boca Raton, FL, USA, 1985; p. 27. ISBN 9781003065173. [Google Scholar]
- Lampen, J.O. External enzyme of yeast: Their nature and formuation. Ant. Van Leeuwenhoek 1968, 34, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kidby, D.K.; Davies, R. Invertase and disulphide bridges in the yeast wall. J. Gen. Microbiol. 1970, 61, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Iwata, H.; Nambap, Y. Proteolytic enzyme from Oerskovia sp. CK lysing viable yeast cells. Agric. Biol. Chem. 1977, 41, 2387–2394. [Google Scholar]
- Correa, J.U.; Elango, N.; Polacheck, I.; Cabib, E. Endochitinase, a mannan-associated enzyme from Saccharomyces cerevisiae. J. Biol. Chem. 1982, 257, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Kollar, R.; Petrakova, E.; Ashwell, G.; Robbins, P.W.; Cabib, E. Architecture of the yeast cell wall: The linkage between chitin and β-1,3–glucan. J. Biol. Chem. 1995, 270, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.C.; Montjin, R.C.; Vink, E.; de la Cruz, J.; Lobell, A.; Douwes, J.E.; Shimoi, H.; Lipke, P.R.; Klis, F.M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology 1996, 6, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.C.; Ram, A.F.J.; Groos, E.M.; Kollar, R.; Montijn, R.C.; Van Den Ende, H.; Llobell, A.; Cabib, E.; Klis, F.M. Altered extent of cross-linking of β-1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β-1,3–glucan content. J. Bacteriol. 1997, 179, 6279–6284. [Google Scholar] [CrossRef] [PubMed]
- Kollar, R.; Reinhold, B.B.; Petrakova, E.; Yeh, H.J.C.; Ash-well, G.; Drgonova, J.; Kapteyn, J.C.; Klis, F.M.; Cabib, E. Architecture of the yeast cell wall β-1,6–glucan interconnects mannoproteins, β-1,3–glucan, and chitin. J. Biol. Chem. 1997, 272, 17762–17775. [Google Scholar] [CrossRef]
- Lipke, P.N.; Ovalle, R. Cell wall architecture in yeast: New structure and new challenges. J. Bacteriol. 1998, 180, 3735–3740. [Google Scholar] [CrossRef]
- Kapteyn, J.C.; Van Den Ende, H.; Klis, F.M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1999, 1426, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Popolo, L.; Vai, M. The gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta 1999, 1426, 385–400. [Google Scholar] [CrossRef]
- Valentin, E.; Herrero, W.; Pastor, J.F.I.; Sentandreu, R. Solubilization and analysis of mannoprotein molecules from the cell wall of S. cerevisiae. J. Gen. Microbiol. 1984, 130, 1419–1428. [Google Scholar] [CrossRef]
- Mrsa, V.; Seidl, T.; Gentzsch, M.; Tanner, W. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast 1997, 30, 1145–1154. [Google Scholar] [CrossRef]
- Cappellaro, C.; Mrsa, V.; Tanner, W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 1998, 180, 5030–5037. [Google Scholar] [CrossRef]
- Lozancic, M.; Zunar, B.; Hrestak, D.; Lopandic, K.; Teparic, R.; Mrsa, V. Systematic comparison of cell wall-related proteins of different yeasts. J. Fungi 2021, 7, 128. [Google Scholar] [CrossRef]
- Cabib, E.; Durán, A. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 2005, 280, 9170–9179. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Sood, P.; Dorfmueller, H.C.; Brown, A.J.P.; Gow, N.A.R. Scalar nanostructure of the Candida albicans cell wall; a molecular, cellular and ultrastructural analysis and interpretation. Cell Surf. 2020, 6, 100047. [Google Scholar] [CrossRef]
- Ecker, M.; Deutzmann, R.; Lehle, L.; Mrsa, V.; Tanner, W. Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 2006, 281, 11523–11529. [Google Scholar] [CrossRef]
- Cezar, T.M.; Lozančić, M.; Novačić, A.; Matičević, A.; Matijević, D.; Vallée, B.; Mrša, V.; Teparić, R.; Žunar, B. Streamlining N-terminally anchored yeast surface display via structural insights into S. cerevisiae Pir proteins. Microb. Cell Fact. 2023, 22, 174. [Google Scholar] [CrossRef]
- Grbavac, A.; Čanak, I.; Stuparević, I.; Teparić, R.; Mrša, V. Proteolytic processing of the Saccharomyces cerevisiae cell wall protein Scw4 regulates its activity and influences its covalent binding to glucan. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 507–515. [Google Scholar] [CrossRef]
- Klebl, F.; Tanner, W. Molecular cloning of a cell wall exobeta-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 1989, 171, 6259–6264. [Google Scholar] [CrossRef]
- Goldman, R.C.; Sullivan, P.A.; Zakula, D.; Capobianco, O. Kinetics of beta-1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur. J. Biochem. 1995, 227, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Kalebina, T.S.; Egorov, N.S.; Arbatskyi, N.P.; Bezsonov, E.E.; Gorkovskyi, A.A.; Kulaev, I.S. The role of high-molecular-weight polyphosphates in activation of glucan transferase Bgl2p from Saccharomyces cerevisiae cell wall. Dokl. Biochem. Biophys. 2008, 420, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Aimanianda, V.; Simenel, C.; Garnaud, C.; Clavaud, C.; Tada, R.; Barbin, L.; Mouyna, I.; Heddergott, C.; Popolo, L.; Ohya, Y.; et al. The dual activity responsible for the elongation and branching of β-(1,3)-glucan in the fungal cell wall. Mbio 2017, 8, e00619-17. [Google Scholar] [CrossRef] [PubMed]
- Rekstina, V.V.; Bykova, A.A.; Ziganshin, R.H.; Kalebina, T.S. GPI-modified proteins non-covalently attached to Saccharomyces cerevisiae yeast cell wall. Biochemistry 2019, 84, 1513–1520. [Google Scholar] [CrossRef]
- Rekstina, V.V.; Sabirzyanova, T.A.; Sabirzyanov, F.A.; Adzhubei, A.A.; Tkachev, Y.V.; Kudryashova, I.B.; Snalina, N.E.; Bykova, A.A.; Alessenko, A.V.; Ziganshin, R.H.; et al. The post-translational modifications, localization, and mode of attachment of non-covalently bound glucanosyltransglycosylases of yeast cell wall as a key to understanding their functioning. Int. J. Mol. Sci. 2020, 21, 8304. [Google Scholar] [CrossRef]
- Sestak, S.; Hagen, I.; Tanner, W.; Strahl, S. Scw10p, a cell-wall glucanase/transglucosidase important for cell-wall stability in Saccharomyces cerevisiae. Microbiology 2004, 150, 3197–3208. [Google Scholar] [CrossRef]
- Kuznetsov, E.; Kučerová, H.; Váchová, L.; Palková, Z. SUN family proteins Sun4p, Uth1p and Sim1p are secreted from Saccharomyces cerevisiae and produced dependently on oxygen level. PLoS ONE 2013, 8, e73882. [Google Scholar] [CrossRef]
- Kuznetsov, E.; Vachova, L.; Palkova, Z. Cellular localization of Sun4p and its interaction with proteins in the yeast birth scar. Cell Cycle 2016, 15, 1898–1907. [Google Scholar] [CrossRef][Green Version]
- Nebreda, A.R.; Villa, T.G.; Villanueva, J.R.; del Rey, F. Cloning of genes related to exo-beta-glucanase production in Saccharomyces cerevisiae: Characterization of an exo-beta-glucanase structural gene. Gene 1986, 47, 245–259. [Google Scholar] [CrossRef]
- Nebreda, A.R.; Vazquez, C.R.; Villa, T.G.; Villanueva, J.R.; del Rey, F. Heterogeneous glycosylation of the EXG1 gene product accounts for the two extracellular exo-beta-glucanases of Saccharomyces cerevisiae. FEBS Lett. 1987, 220, 27–30. [Google Scholar] [CrossRef]
- Baladron, V.; Ufano, S.; Duenas, E.; Martin-Cuadrado, A.B.; del Rey, F.; Vazquez de Aldana, C.R. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell. 2002, 1, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, M.J.; Robbins, P.W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 1991, 266, 19758–19767. [Google Scholar] [CrossRef] [PubMed]
- Kalebina, T.S.; Kulakovskaya, E.V.; Rekstina, V.V.; Trilisenko, L.V.; Ziganshin, R.H.; Marmiy, N.V.; Esipov, D.S.; Kulakovskaya, T.V. Effect of deletions of the genes encoding Pho3p and Bgl2p on polyphosphate level, stress adaptation, and attachments of these proteins to Saccharomyces cerevisiae cell wall. Biochemistry 2023, 88, 152–161. [Google Scholar] [CrossRef]
- Russo, P.; Kalkkinen, N.; Sareneva, H.; Paakkola, J.; Makarow, M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc. Natl. Acad. Sci. USA 1992, 89, 3671–3675. [Google Scholar] [CrossRef] [PubMed]
- Moukadiri, I.; Zueco, J. Evidence for the attachment of Hsp150/Pir2 to the cell wall of Saccharomyces cerevisiae through disulfide bridges. FEMS Yeast Res. 2001, 1, 241–245. [Google Scholar] [CrossRef]
- Zarnowski, R.; Noll, A.; Chevrette, M.G.; Sanchez, H.; Jones, R.; Anhalt, H.; Fossen, J.; Jaromin, A.; Currie, C.; Nett, J.E.; et al. Coordination of fungal biofilm development by extracellular vesicle cargo. Nat. Commun. 2021, 12, 6235. [Google Scholar] [CrossRef]
- Kulig, K.; Karnas, E.; Woznicka, O.; Kuleta, P.; Zuba-Surma, E.; Pyza, E.; Osyczka, A.; Kozik, A.; Rapala-Kozik, M.; Karkowska-Kuleta, J. Insight into the properties and immunoregulatory effect of extracellular vesicles produced by Candida glabrata, Candida parapsilosis, and Candida tropicalis biofilms. Front. Cell Infect. Microbiol. 2022, 12, 879237. [Google Scholar] [CrossRef]
- Lai, Y.; Jiang, B.; Hou, F.; Huang, X.; Ling, B.; Lu, H.; Zhong, T.; Huang, J. The emerging role of extracellular vesicles in fungi: A double-edged sword. Front. Microbiol. 2023, 14, 1216895. [Google Scholar] [CrossRef]
- Morgan, A.E. Exocytosis. Essays Biochem. 1995, 30, 77–95. [Google Scholar] [PubMed]
- Prados-Rosales, R.; Brown, L.; Casadevall, A.; Montalvo-Quirós, S.; Luque-Garcia, J.L. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. Methods 2014, 1, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, M.C.; Matsuo, A.L.; Ganiko, L.; Medeiros, L.C.; Miranda, K.; Silva, L.S.; Freymüller-Haapalainen, E.; Sinigaglia-Coimbra, R.; Almeida, I.C.; Puccia, R. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic α-Galactosyl epitopes. Eukaryot. Cell. 2011, 10, 343–351. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Walker, L.; Sood, P.; Lenardon, M.D.; Milne, G.; Olson, J.; Jensen, G.; Wolf, J.; Casadevall, A.; Adler-Moore, J.; Gow, N.A.R. The viscoelastic properties of the fungal cell wall allow traffic of AmBisome as intact liposome vesicles. mBio 2018, 9, e02383-17. [Google Scholar] [CrossRef]
- Rizzo, J.; Rodrigues, M.L.; Janbon, G. Extracellular vesicles in fungi: Past, present, and future perspectives. Front. Cell Infect. Microbiol. 2020, 10, 346. [Google Scholar] [CrossRef]
- Winters, C.M.; Hong-Brown, L.Q.; Chiang, H.L. Intracellular vesicle clusters are organelles that synthesize extracellular vesicle-associated cargo proteins in yeast. J. Biol. Chem. 2020, 295, 2650–2663. [Google Scholar] [CrossRef]
- Wolf, J.M.; Casadevall, A. Challenges posed by extracellular vesicles from eukaryotic microbes. Curr. Opin. Microbiol. 2014, 22, 73–78. [Google Scholar] [CrossRef]
- de Souza, P.R.; Geibel, J. Direct observation of oxidative stress on the cell wall of Saccharomyces cerevisiae strains with atomic force microscopy. Mol. Cell Biochem. 1999, 201, 17–24. [Google Scholar] [CrossRef]
- Rodriguez, A.S.; Batac, J.; Killilea, A.N.; Filopei, J.; Simeonov, D.R.; Lin, I.; Paluh, J.L. Protein complexes at the microtubule organizing center regulate bipolar spindle assembly. Cell Cycle 2008, 7, 1246–1253. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.; Longo, L.V.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: Comparative analysis with other pathogenic fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Espadas-Moreno, J.; Luque-Garcia, J.L.; Casadevall, A. Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot. Cell. 2014, 13, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.J.; Chatterjee, V.; Swaidani, S.; Alluri, R.K.; Kundu, S.; Merkulova, A.; Angelini, D.; You, D.; Whitney, S.A.; Feener, E.P.; et al. Polyphosphate expression by cancer cell extracellular vesicles mediates binding of factor XII and contact activation. Blood Adv. 2021, 5, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Zvonarev, A.N.; Trilisenko, L.V.; Farofonova, V.V.; Kulakovskaya, E.V.; Abashina, T.N.; Dmitriev, V.V.; Kulakovskaya, T. The Extracellular Vesicles Containing Inorganic Polyphosphate of Candida Yeast upon Growth on Hexadecane. J. Xenobiot. 2023, 13, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Mrsa, V.; Klebl, F.; Tanner, W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J. Bacteriol. 1993, 175, 2102–2106. [Google Scholar] [CrossRef] [PubMed]
- Breidenbach, M.A.; Palaniappan, K.K.; Pitcher, A.A.; Bertozzi, C.R. Mapping yeast N-glycosites with isotopically recoded glycans. Mol. Cell. Proteom. 2012, 11, M111.015339. [Google Scholar] [CrossRef]
- Hartland, R.P.; Emerson, G.W.; Sullivan, P.A. A secreted beta-glucan-branching enzyme from Candida albicans. Proc. Biol. Sci. 1991, 246, 155–160. [Google Scholar] [CrossRef]
- Mouyna, I.; Hartland, R.P.; Fontaine, T.; Diaquin, M.; Simenel, C.; Delepierre, M.; Henrissat, B.; Latge, J.-P. A 1,3-beta-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast Bgl2p. Microbiology 1998, 144, 3171–3180. [Google Scholar] [CrossRef]
- Oro, L.; Zara, S.; Fancellu, F.; Mannazzu, I.; Budroni, M.; Ciani, M.; Comitini, F. TpBGL2 codes for a Tetrapisispora phaffii killer toxin active against wine spoilage yeasts. FEMS Yeast Res. 2014, 14, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Sabirzyanov, F.A.; Sabirzyanova, T.A.; Rekstina, V.V.; Adzhubei, A.A.; Kalebina, T.S. C-Terminal sequence is involved in the incorporation of Bgl2p glucanosyltransglycosylase in the cell wall of Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, fox093. [Google Scholar] [CrossRef]
- Adzhubei, A.A.; Sternberg, M.J.; Makarov, A.A. Polyproline-II Helix in proteins: Structure and function. J. Mol. Biol. 2013, 425, 2100–2132. [Google Scholar] [CrossRef]
- Kalebina, T.S.; Laurinavichiute, D.K.; Packeiser, A.N.; Morenkov, O.S.; Ter-Avanesyan, M.D.; Kulaev, I.S. Correct GPI-anchor synthesis is required for the incorporation of endoglucanase/glucanosyltransferase Bgl2p into the Saccharomyces cerevisiae cell wall. FEMS Microbiol. Lett. 2002, 210, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.B.; Yagi, H.; Manno, M.; Martorana, V.; Ban, T.; Christiansen, G.; Otzen, D.E.; Goto, Y.; Rischel, C. Branching in amyloid fibril growth. Biophys. J. 2009, 96, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.D.; Wong, S.S.; Lieber, C.M.; Lansbury, P.T. Observation of metastable Abeta amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997, 4, 119–125. [Google Scholar] [CrossRef]
- Moreno-García, J.; García-Martínez, T.; Moreno, J.; Millán, M.C.; Mauricio, J.C. A proteomic and metabolomic approach for understanding the role of the flor yeast mitochondria in the velum formation. Int. J. Food Microbiol. 2014, 172, 21–29. [Google Scholar] [CrossRef]
- Moreno-García, J.; García-Martínez, T.; Moreno, J.; Mauricio, J.C. Proteins involved in flor yeast carbon metabolism under biofilm formation conditions. Food Microbiol. 2015, 46, 25–33. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, R.; Takada, A.; Iwatani, S.; Oka, C.; Kitamoto, T.; Kajiwara, S. The role of Bgl2p in the transition to filamentous cells during biofilm formation by Candida Albicans. Mycoses 2017, 60, 96–103. [Google Scholar] [CrossRef]
- Jeng, H.W.; Holmes, A.R.; Cannon, R.D. Characterization of two Candida albicans surface mannoprotein adhesins that bind immobilized saliva components. Med. Mycol. 2005, 43, 209–217. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Zhang, R.; Inokawa, N.; Oura, T.; Chen, X.; Iwatani, S.; Niimi, K.; Niimi, M.; Holmes, A.R.; Cannon, R.D.; et al. Candida albicans Bgl2p, Ecm33p, and Als1p proteins are involved in adhesion to saliva-coated hydroxyapatite. J. Oral. Microbiol. 2021, 13, 1879497. [Google Scholar] [CrossRef]
- Weimberg, R.; Orton, W.L. Evidence for an exocellular site for the acid phosphatase of Saccharomyces mellis. J. Bacteriol. 1964, 88, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Kulaev, I.S.; Krasheninikov, I.A.; Kokurina, I.A. On Localization of inorganic polyphosphates and nucleotides in Neurospora crassa. Biokhimiya 1966, 31, 850–858. (In Russian) [Google Scholar]
- Tijssen, J.P.F.; Beekes, H.W.; Van Steveninck, J. Localization of polyphosphate in Saccharomyces fragilis, as revealed by 4′6′-diamidino-2-phenylindole fluorescence. Biochem. Biophys. Acta 1982, 721, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Vagabov, V.M.; Chemodanova, O.V.; Kulaev, I.S. Effect of inorganic polyphosphates on the value of negative charge of yeast cell wall. Dokl. AN SSSR 1990, 313, 989–992. [Google Scholar]
- Ivanov, A.J.; Vagabov, V.M.; Fomchenkov, V.M.; Kulaev, I.S. Study of the influence of polyphosphates of cell envelope on the sensitivity of yeast Saccharomyces carlsbergensis to the cytyl-3-methylammonium bromide. Microbiologiia 1996, 65, 611–616. [Google Scholar]
- Tijssen, J.P.F.; Van Steveninck, J. Cytochemical staining of a yeast polyphosphate fraction, localized outside the plasma membrane. Protoplasma 1985, 125, 124–128. [Google Scholar] [CrossRef]
- Shabalin, Y.A.; Vagabov, V.M.; Kulaev, I.S. Mechanism of coupling of biosynthesis of high-molecular-weight polyphosphates and mannan in yeast Saccharomyces carlsbergensis. Dokl. AN SSSR 1979, 249, 243–246. (In Russian) [Google Scholar]
- Shabalin, Y.A.; Vagabov, V.M.; Kulaev, I.S. Dolichyl phosphate mannose: Intermediate of glycoprotein synthesis in yeast? Dokl. AN SSSR 1985, 283, 720–723. [Google Scholar]
- Gerasimaitė, R.; Mayer, A. Enzymes of yeast polyphosphate metabolism: Structure, enzymology and biological roles. Biochem. Soc. Trans. 2016, 44, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, F.; Rush, J.S.; Toke, D.A.; Han, G.; Quinn, J.E.; Carman, G.M.; Chio, J.-Y.; Voelker, D.R.; Aebi, M.; Weachter, C.J. The CWH8 gene encodes a dolichyl pyrophosphate phosphatase with luminal orientated active site in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 41455–41464. [Google Scholar] [CrossRef] [PubMed]
- Neville, N.; Lehotsky, K.; Yang, Z.; Klupt, K.A.; Denoncourt, A.; Downey, M.; Jia, Z. Modification of histidine repeat proteins by inorganic polyphosphate. Cell Rep. 2023, 42, 113082. [Google Scholar] [CrossRef] [PubMed]
- Lempart, J.; Jakob, U. Role of polyphosphate in amyloidogenic processes. Cold Spring Harb. Perspect. Biol. 2019, 11, a034041. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Jakob, U. Inorganic polyphosphate, a multifunctional polyanionic protein scaffold. J. Biol. Chem. 2019, 294, 2180–2190. [Google Scholar] [CrossRef] [PubMed]
- Cremers, C.M.; Knoefler, D.; Gates, S.; Martin, N.; Dahl, J.U.; Lempart, J.; Xie, L.; Chapman, M.R.; Galvan, V.; Southworth, D.R.; et al. Polyphosphate: A conserved modifier of amyloidogenic processes. Mol. Cell. 2016, 63, 768–780. [Google Scholar] [CrossRef]
- Lempart, J.; Tse, E.; Lauer, J.A.; Ivanova, M.I.; Sutter, A.; Yoo, N.; Huettemann, P.; Southworth, D.; Jakob, U. Mechanistic insights into the protective roles of polyphosphate against amyloid cytotoxicity. Life Sci. Alliance 2019, 2, e201900486. [Google Scholar] [CrossRef]
- Brady, D.; Duncan, J.R. Binding of heavy metals by the cell walls of Saccharomyces cerevisiae. Enzym. Microb. Technol. 1994, 16, 633–638. [Google Scholar] [CrossRef]
- Park, Y.; Malliakas, C.D.; Zhou, Q.; Gu, A.Z.; Aristilde, L. Molecular coordination, structure, and stability of metal-polyphosphate complexes resolved by molecular modeling and x-ray scattering: Structural insights on the biological fate of polyphosphate. Environ. Sci. Technol. 2021, 55, 14185–14193. [Google Scholar] [CrossRef]
- Kulakovskaya, T.; Ryazanova, L.; Zvonarev, A.; Khokhlova, G.; Ostroumov, V.; Vainshtein, M. The biosorption of cadmium and cobalt and iron ions by yeast Cryptococcus humicola at nitrogen starvation. Folia Microbiol. 2018, 63, 507–510. [Google Scholar] [CrossRef]
- Zvonarev, A.N.; Crowley, D.E.; Ryazanova, L.P.; Lichko, L.P.; Rusakova, T.G.; Kulakovskaya, T.V.; Dmitriev, V.V. Cell wall canals formed upon growth of Candida maltosa in the presence of hexadecane are associated with polyphosphates. FEMS Yeast Res. 2017, 17, fox026. [Google Scholar] [CrossRef]
- Ramos, C.L.; Gomes, F.M.; Girard-Dias, W.; Almeida, F.P.; Albuquerque, P.C.; Kretschmer, M.; Kronstad, J.W.; Frases, S.; de Souza, W.; Rodrigues, M.L.; et al. Phosphorus-Rich Structures and Capsular Architecture in Cryptococcus Neoformans. Future Microbiol. 2017, 12, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Kulaev, I.S.; Vagabov, V.M.; Kulakovskaya, T.V. High Molecular Inorganic Polyphosphates: Biochemistry, Cell Biology, Biotechnology; Scientific World: Moscow, Russia, 2005; p. 215. ISBN 5-89176-326-5. [Google Scholar]
- Utama, G.M.; Oktaviani, L.; Balia, R.L.; Rialita, T. Potential Application of Yeast Cell Wall Biopolymers as Probiotic Encapsulants. Polymers 2023, 15, 3481. [Google Scholar] [CrossRef] [PubMed]
- Yammine, M.; Bray, F.; Flament, S.; Picavet, A.; Lacroix, J.-M.; Poilpré, E.; Mouly, I.; Rolando, C. Reliable Approach for Pure Yeast Cell Wall Protein Isolation from Saccharomyces cerevisiae Yeast Cells. ACS Omega 2022, 7, 29702–29713. [Google Scholar] [CrossRef] [PubMed]
- Kalebina, T.; Rekstina, V. Molecular organization of yeast cell envelope. Mol. Biol. 2019, 53, 968–981. [Google Scholar] [CrossRef]
| Name of Protein | Participating in CW Remodeling | References |
|---|---|---|
| Bgl2 | + | [26,27,69,70,71,72,73,74] |
| Scw4 | + | [62,68,74,75] |
| Scw10 | + | [62,74,75] |
| Scw11 | + | [62,75] |
| Sun4 | + | [76,77] |
| Uth1 | + | [76] |
| Sim1 | + | [76] |
| Exg1 | + | [26,78,79] |
| Dse2 | + | [77] |
| Eng1/Dse4 | + | [74,77,80] |
| Cts1 | + | [80,81] |
| Pho3 | hypothetically | [71,74,82] |
| Ygp1 | − | [74] |
| Tos1 * | − | [74] |
| Pry3 | − | [74] |
| Hsp150 | − | [74,83,84] |
| Gas1 | + | [59,72,73,74] |
| Gas3 | − | [8,73,74] |
| Gas5 | + | [8,73,74] |
| Cwp1 | − | [73,74] |
| Sag1 | − | [73,74] |
| Crh1 | + | [8,73,74] |
| Crh2 | + | [8,73,74] |
| Ecm33 * | − | [73,74] |
| Tir1 | − | [73,74] |
| Tip1 | − | [73,74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalebina, T.S.; Rekstina, V.V.; Pogarskaia, E.E.; Kulakovskaya, T. Importance of Non-Covalent Interactions in Yeast Cell Wall Molecular Organization. Int. J. Mol. Sci. 2024, 25, 2496. https://doi.org/10.3390/ijms25052496
Kalebina TS, Rekstina VV, Pogarskaia EE, Kulakovskaya T. Importance of Non-Covalent Interactions in Yeast Cell Wall Molecular Organization. International Journal of Molecular Sciences. 2024; 25(5):2496. https://doi.org/10.3390/ijms25052496
Chicago/Turabian StyleKalebina, Tatyana S., Valentina V. Rekstina, Elizaveta E. Pogarskaia, and Tatiana Kulakovskaya. 2024. "Importance of Non-Covalent Interactions in Yeast Cell Wall Molecular Organization" International Journal of Molecular Sciences 25, no. 5: 2496. https://doi.org/10.3390/ijms25052496
APA StyleKalebina, T. S., Rekstina, V. V., Pogarskaia, E. E., & Kulakovskaya, T. (2024). Importance of Non-Covalent Interactions in Yeast Cell Wall Molecular Organization. International Journal of Molecular Sciences, 25(5), 2496. https://doi.org/10.3390/ijms25052496







