Isolation and Characterization of Paenibacillus polymyxa B7 and Inhibition of Aspergillus tubingensis A1 by Its Antifungal Substances
Abstract
:1. Introduction
2. Results
2.1. Dominant Molds
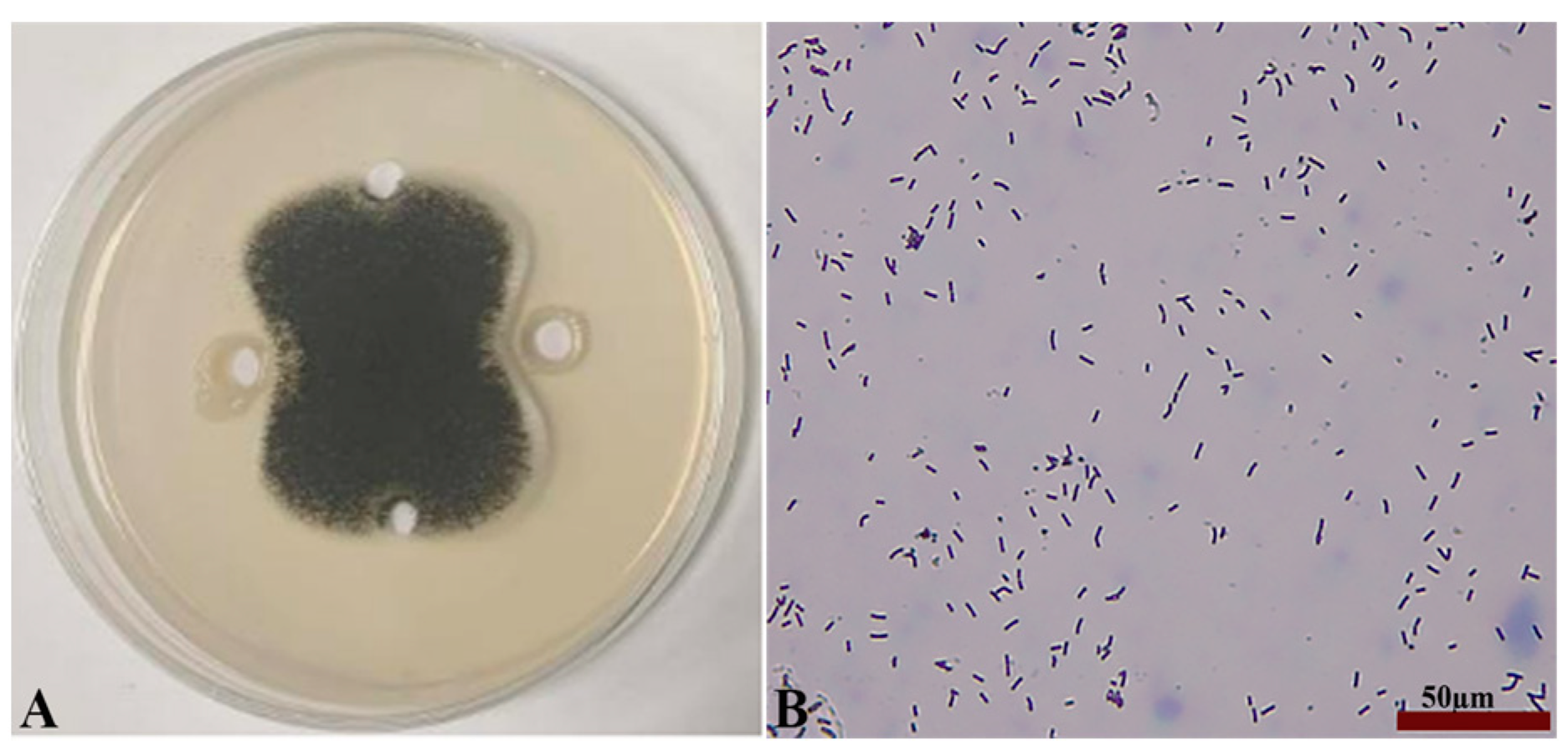
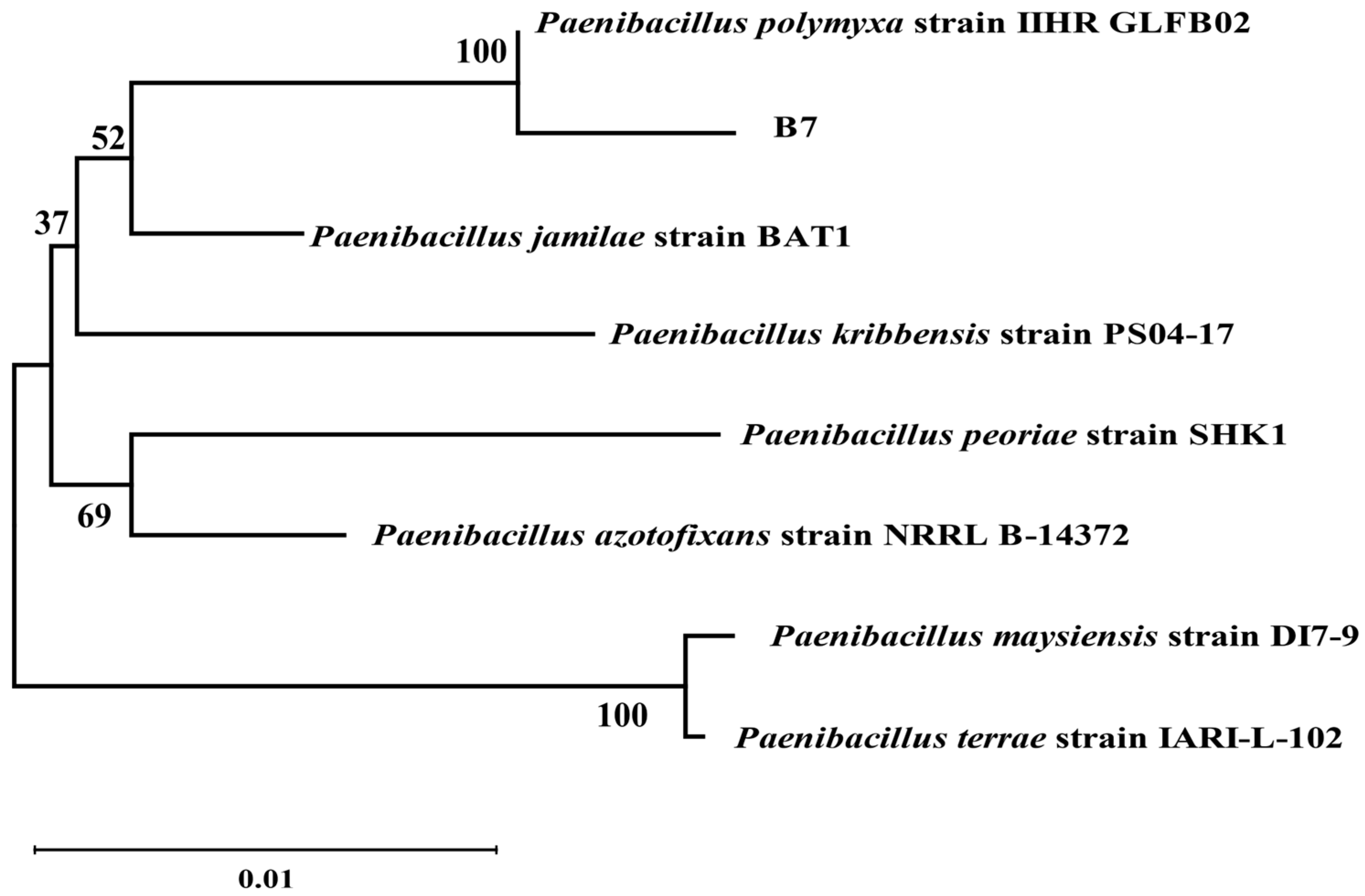
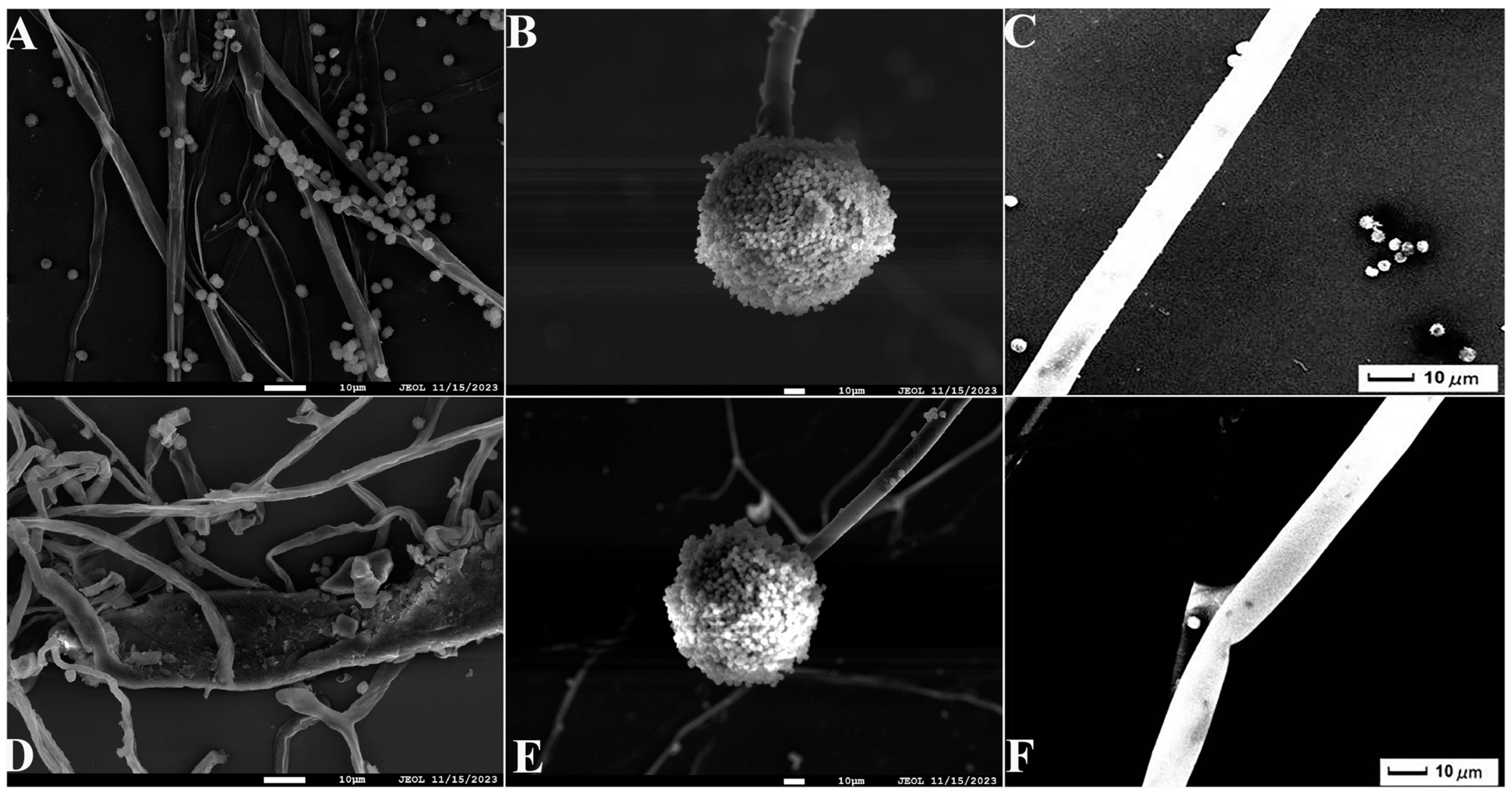
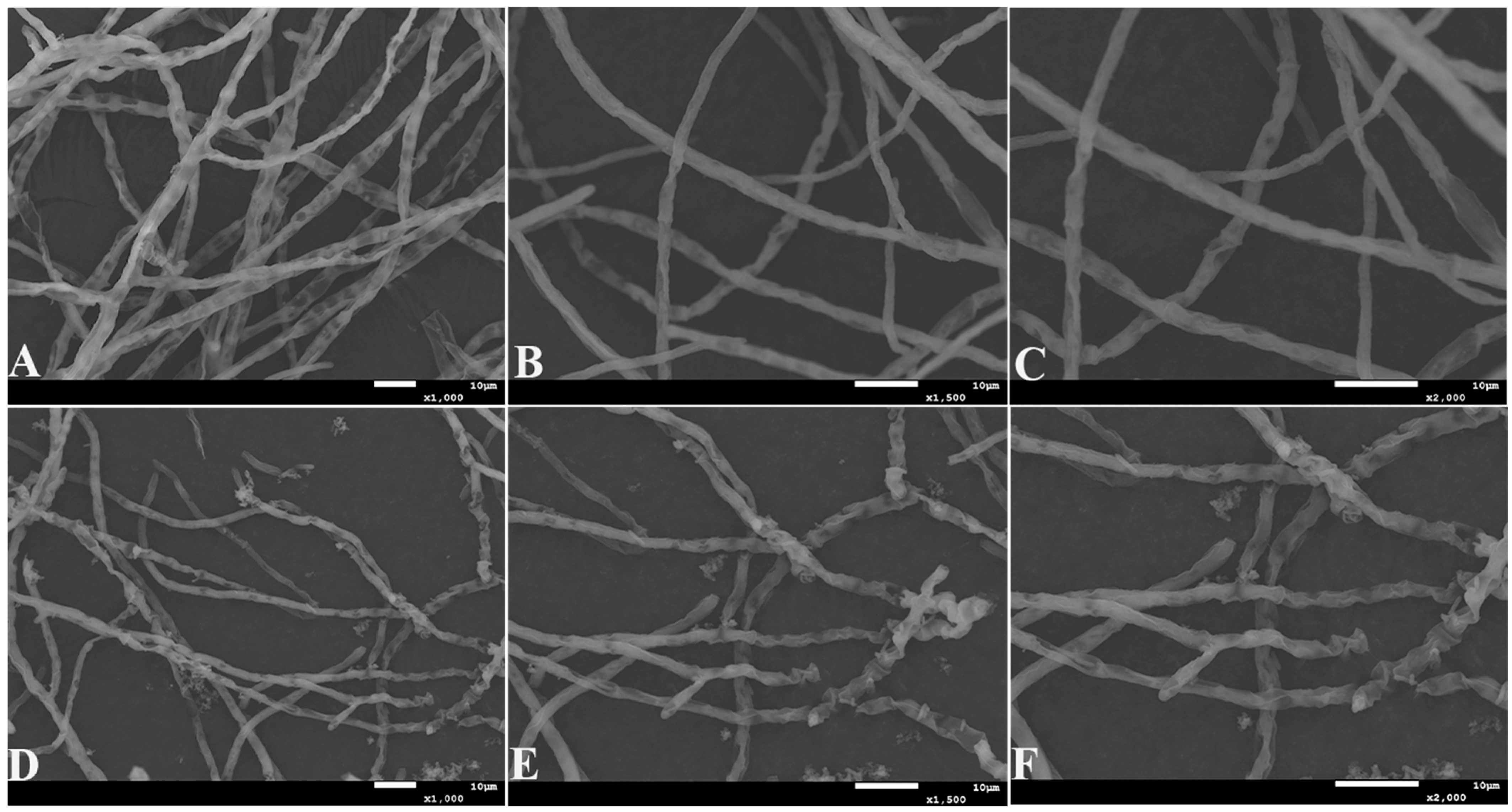
2.2. Identification of Antagonistic Bacteria Isolates and Inhibition of Mycelial Growth of A. tubingensis A1 with Scanning Electron Microscope (SEM) Evaluation
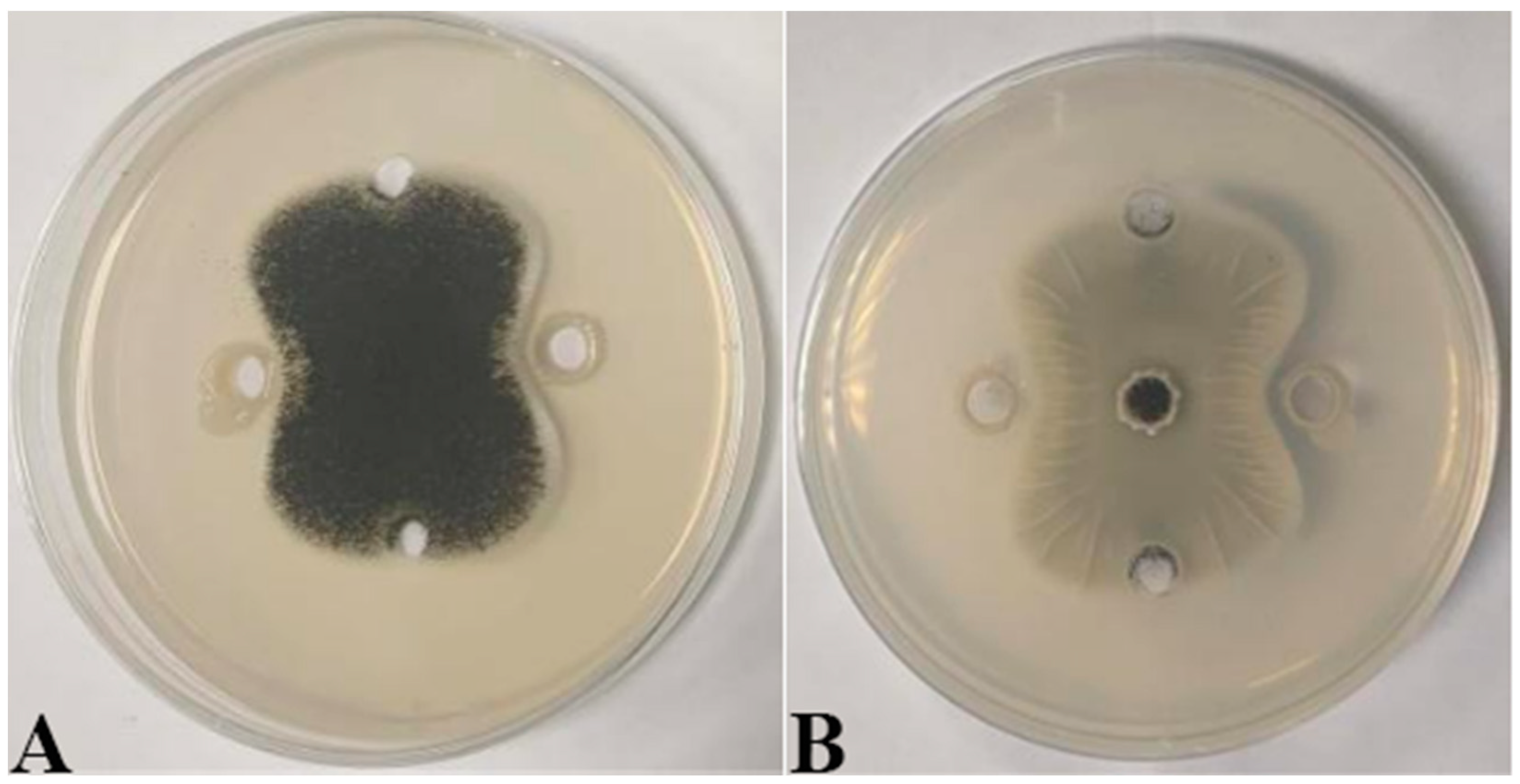
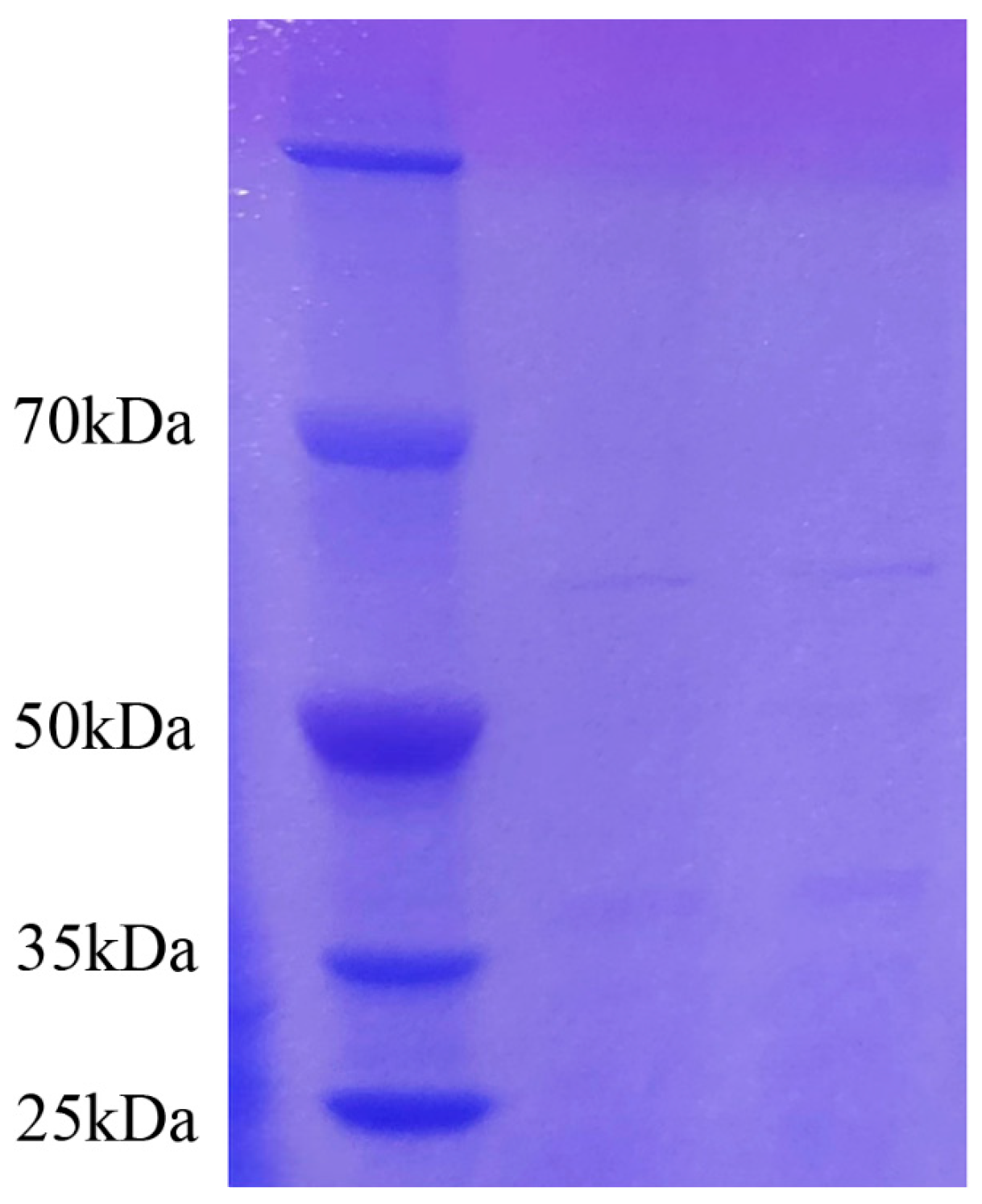
2.3. Analysis of Protein Concentration, SDS-PAGE Electrophoresis Result of Antifungal Proteins, and SEM Evaluation of the Inhibitory Effect of Antifungal Proteins on A. tubingensis Mycelial Growth
2.4. Effect of P. polymyxa on the Transcriptome of A. tubingensis
2.4.1. Analysis of Differentially Expressed Genes (DEGs)
2.4.2. GO Enrichment Analysis of DEGs
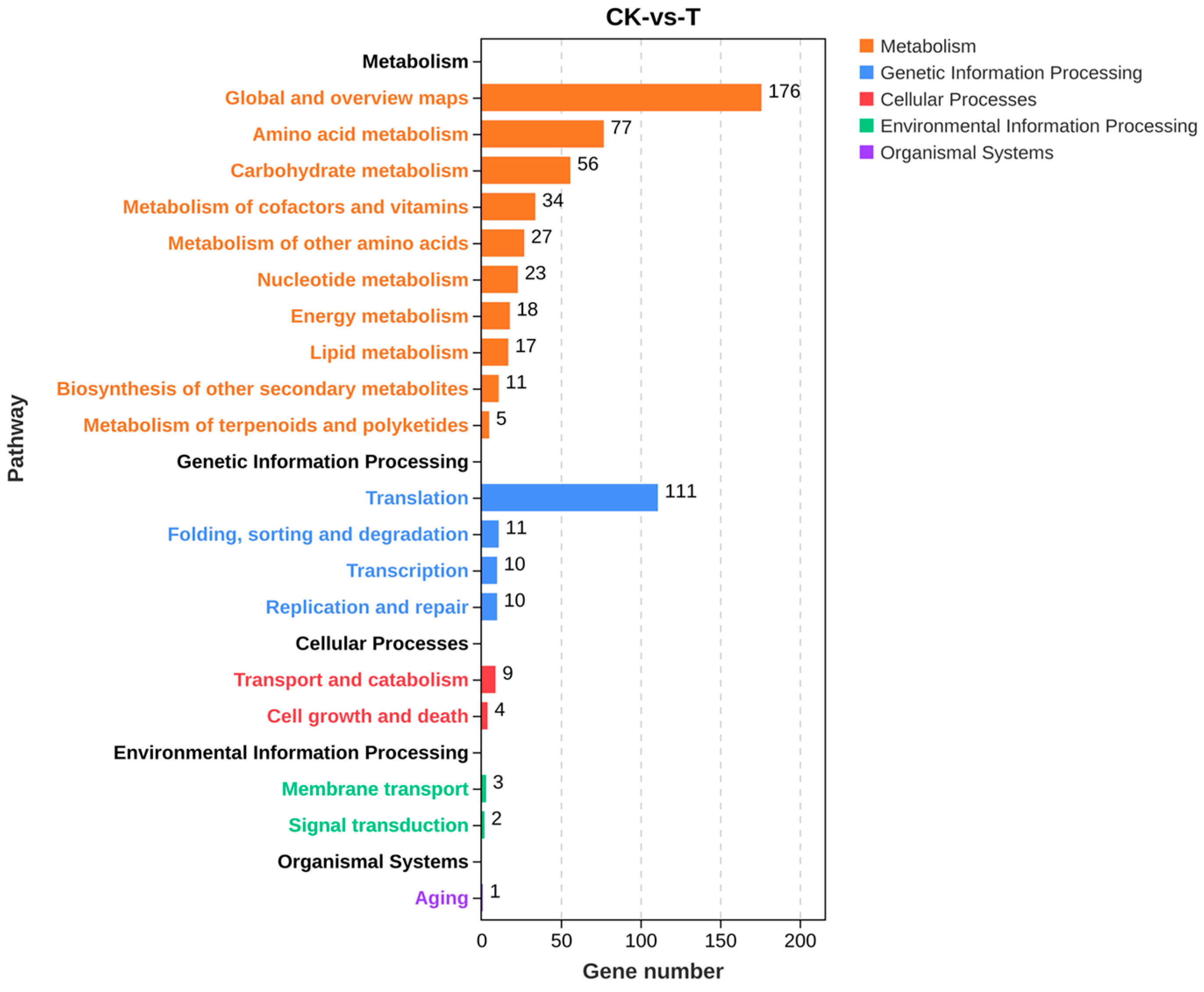
2.4.3. KEGG Enrichment Analysis of DEGs
2.4.4. Real-Time Quantitative PCR (RT-qPCR) Validation of DEGs
3. Discussion
4. Materials and Methods
4.1. Sample Source and Species
4.2. Isolation and Purification of Dominant Molds
4.3. Isolation and Characterization of Antagonist Strains
4.3.1. Isolation of Antagonist Bacteria (P. polymyxa B7)
4.3.2. Determination of Growth Curves of P. polymyxa B7
4.3.3. Physiological, Biochemical, and Molecular Biological Characterization of P. polymyxa B7
4.4. Inhibitory Effect of P. polymyxa B7 on A. tubingensis A1
4.5. Inhibition of A. tubingensis A1 Mycelial Growth Caused by P. polymyxa B7
4.6. Extraction of Antifungal Substances of P. polymyxa B7
4.6.1. Seed Fermentation Broth Preparation
4.6.2. Preparation of Bacteria-Free Culture Solution of the Strain
4.6.3. Crude Extraction of Antifungal Proteins
4.7. Determination of Inhibitory Protein Concentration and SDS-PAGE Electrophoresis
4.8. Effect of Antifungal Proteins on the Growth of A. tubingensis A1 Mycelia and SEM Observations
4.9. Transcriptomics of P. polymyxa B7 against A. tubingensis A1
4.9.1. Sample Preparation and RNA Extraction
4.9.2. Transcriptome Sequencing and Bioinformatics Analysis
4.9.3. RT-qPCR Validation
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Food and Agriculture Organization of the United Nations 2023. Available online: https://www.fao.org/faostat/en/#rankings/countries_by_commodity (accessed on 1 October 2023).
- Perrone, G.; Susca, A.; Cozzi, G.; Ehrlich, K.; Varga, J.; Frisvad, J.C.; Meijer, M.; Noonim, P.; Mahakamchanakul, W.; Samson, R.A. Biodiversity of Aspergillus Species in Some Important Agricultural Products. Stud. Mycol. 2007, 59, 53–66. [Google Scholar] [CrossRef]
- Tan, S.; Ma, F.; Wu, Y.; Xu, Y.; Niu, A.; Chen, Y.; Wang, G.; Qiu, W. The Biodiversity of Aspergillus flavus in Stored Rice Grain Leads to a Decrease in the Overall Aflatoxin B1 Production in These Species. Int. J. Food Microbiol. 2023, 406, 110416. [Google Scholar] [CrossRef]
- Kumar, N.; Khurana, S.M.P.; Pandey, V.N. Deciphering of Seed Health of Common Food Grains (Wheat, Rice) of North Eastern UP and Gurgaon Haryana, India. Sci. Rep. 2023, 13, 8480. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, H.; Wang, G.; Wu, H.; Qiu, W. High-Throughput Sequencing to Analyze the Fungal Community Structure and Dominant Fungal Genera in Stored Rice. Food Sci. 2021, 42, 92–99. [Google Scholar]
- Aziz, N.H.; Mattar, Z.A.; Mahrous, S.R. Contamination of Grains by Mycotoxin-Producing Molds and Mycotoxins and Control by Gamma Irradiation. J. Food Saf. 2006, 26, 184–201. [Google Scholar] [CrossRef]
- Hua, C.; Kai, K.; Wang, X.; Shi, W.; Zhang, D.; Liu, Y. Curcumin Inhibits Gray Mold Development in Kiwifruit by Targeting Mitogen-Activated Protein Kinase (MAPK) Cascades in Botrytis cinerea. Postharvest Biol. Technol. 2019, 151, 152–159. [Google Scholar] [CrossRef]
- Alkuwari, A.; Ul Hassan, Z.; Zeidan, R.; Al-Thani, R.; Jaoua, S. Occurrence of Mycotoxins and Toxigenic Fungi in Cereals and Application of Yeast Volatiles for Their Biological Control. Toxins 2022, 14, 404. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Saba, M.K.; Ashengroph, M. Efficacy of Aquatic Yeasts to Control Gray Mold Decay, and Impacts on Strawberry Quality during Cold Storage and Shelf Life. Sci. Hortic. 2024, 323, 112503. [Google Scholar] [CrossRef]
- Zhang, Q.; Xing, C.; Li, S.; He, L.; Qu, T.; Chen, X. In Vitro Antagonism and Biocontrol Effects of Paenibacillus polymyxa JY1-5 against Botrytis cinerea in Tomato. Biol. Control 2021, 160, 104689. [Google Scholar] [CrossRef]
- Boukaew, S.; Petlamul, W.; Yossan, S.; Srinuanpan, S.; Nooprom, K.; Zhang, Z. Biofumigant Potential and Inhibition Mechanism of Trichoderma asperelloides SKRU-01 Volatile Organic Compounds for Controlling Aflatoxigenic Aspergillus parasiticus and Aspergillus flavus in Stored Peanuts. Food Control 2024, 157, 110194. [Google Scholar] [CrossRef]
- Godana, E.A.; Yang, Q.; Zhang, X.; Zhao, L.; Wang, K.; Dhanasekaran, S.; Mehari, T.G.; Zhang, H. Biotechnological and Biocontrol Approaches for Mitigating Postharvest Diseases Caused by Fungal Pathogens and Their Mycotoxins in Fruits: A Review. J. Agric. Food Chem. 2023, 71, 17584–17596. [Google Scholar] [CrossRef]
- Hossain, A.; Ali, M.A.; Lin, L.; Luo, J.; You, Y.; Masum, M.M.I.; Jiang, Y.; Wang, Y.; Li, B.; An, Q. Biocontrol of Soft Rot Dickeya and Pectobacterium Pathogens by Broad-Spectrum Antagonistic Bacteria within Paenibacillus polymyxa Complex. Microorganisms 2023, 11, 817. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, Y.; Yu, Z.; Zhuang, W.; Zeng, Z. Bacillus velezensis BV01 Has Broad-Spectrum Biocontrol Potential and the Ability to Promote Plant Growth. Microorganisms 2023, 11, 2627. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, Z.; Li, S.; Liu, T.; Song, J.; Li, J.; Zhang, X. Two Antimicrobial Peptides Derived from Bacillus and Their Properties. Molecules 2023, 28, 7899. [Google Scholar] [CrossRef] [PubMed]
- Hammad, M.; Ali, H.; Hassan, N.; Tawab, A.; Salman, M.; Jawad, I.; de Jong, A.; Moreno, C.M.; Kuipers, O.P.; Feroz, Y.; et al. Food Safety and Biological Control; Genomic Insights and Antimicrobial Potential of Bacillus velezensis FB2 against Agricultural Fungal Pathogens. PLoS ONE 2023, 18, e0291975. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current Knowledge and Perspectives of Paenibacillus: A Review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Bai, K.; Feng, C.; Jiang, L.; Zhang, L.; Zhang, J.; Zhang, L.; Wang, T. Dietary Effects of Bacillus subtilis Fmbj on Growth Performance, Small Intestinal Morphology, and Its Antioxidant Capacity of Broilers. Poult. Sci. 2018, 97, 2312–2321. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, X.; Zhao, T.; Ma, J.; Zhao, L.; Song, Q.; Sun, W. Screening of Bacillus velezensis E2 and the Inhibitory Effect of Its Antifungal Substances on Aspergillus flavus. Foods 2022, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Ran, J.; Wu, Y.; Zhang, B.; Su, Y.; Lu, N.; Li, Y.; Liang, X.; Zhou, H.; Shi, J. Paenibacillus polymyxa Antagonism towards Fusarium: Identification and Optimisation of Antibiotic Production. Toxins 2023, 15, 138. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Yue, Y.; Gu, G.; Deng, Y.; Jin, H.; Tao, K. Identification and Characterization of the Bacillus atrophaeus Strain J-1 as Biological Agent of Apple Ring Rot Disease. J. Plant Dis. Prot. 2020, 127, 367–378. [Google Scholar] [CrossRef]
- Yan, F.; Li, C.; Ye, X.; Lian, Y.; Wu, Y.; Wang, X. Antifungal Activity of Lipopeptides from Bacillus amyloliquefaciens MG3 against Colletotrichum gloeosporioides in Loquat Fruits. Biol. Control 2020, 146, 104281. [Google Scholar] [CrossRef]
- Chen, Y.; Yin, S.; Lin, H.; Zhang, G.; Cheng, H.; Zhang, L. Induced Resistance of Cucumber against Botrytis Cinerea by Bacillus subtilis BSD-2. North. Hortic. 2016, 12, 119–123. [Google Scholar] [CrossRef]
- Moreira, R.R.; Nesi, C.N.; May De Mio, L.L. Bacillus Spp. and Pseudomonas putida as Inhibitors of the Colletotrichum acutatum Group and Potential to Control Glomerella Leaf Spot. Biol. Control 2014, 72, 30–37. [Google Scholar] [CrossRef]
- Yu, X.; Ai, C.; Xin, L.; Zhou, G. The Siderophore-Producing Bacterium, Bacillus subtilis CAS15, has a Biocontrol Effect on Fusarium Wilt and Promotes the Growth of Pepper. Eur. J. Soil Biol. 2011, 47, 138–145. [Google Scholar] [CrossRef]
- Ling, L.; Zhao, Y.; Tu, Y.; Yang, C.; Ma, W.; Feng, S.; Lu, L.; Zhang, J. The Inhibitory Effect of Volatile Organic Compounds Produced by Bacillus subtilis CL2 on Pathogenic Fungi of Wolfberry. J. Basic Microbiol. 2021, 61, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Driss, F.; Rouis, S.; Azzouz, H.; Tounsi, S.; Zouari, N.; Jaoua, S. Integration of a Recombinant Chitinase into Bacillus thuringiensis Parasporal Insecticidal Crystal. Curr. Microbiol. 2011, 62, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, Y.; Hu, D.; Peng, J.; Jia, N.; Zhang, C.; Wang, X.; Wang, Z. Progress on Mechanism and Applications of Bacillus as a Biocontrol Microbe. Chin. J. Biol. Control 2018, 34, 639–648. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Wang, W. Bacillus amyloliquefaciens SN16-1-Induced Resistance System of the Tomato against Rhizoctonia solani. Pathogens 2022, 11, 35. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, R.; Tang, Y.; Cheng, Z.; Qian, M.; Li, W.; Shao, Y. Transcriptome Analysis Reveals the Inducing Effect of Bacillus siamensis on Disease Resistance in Postharvest Mango Fruit. Foods 2022, 11, 107. [Google Scholar] [CrossRef]
- Ran, J.; Jiao, L.; Zhao, R.; Zhu, M.; Shi, J.; Xu, B.; Pan, L. Characterization of a Novel Antifungal Protein Produced by Paenibacillus polymyxa Isolated from the Wheat Rhizosphere. J. Sci. Food Agric. 2021, 101, 1901–1909. [Google Scholar] [CrossRef]
- Yi, W.; Chen, C.; Gan, X. Polymyxin B1 and E2 From Paenibacillus polymyxa Y-1 for Controlling Rice Bacterial Disease. Front. Cell. Infect. Microbiol. 2022, 12, 866357. [Google Scholar] [CrossRef]
- Khan, M.S.; Gao, J.; Chen, X.; Zhang, M.; Yang, F.; Du, Y.; Moe, T.S.; Munir, I.; Xue, J.; Zhang, X. Isolation and Characterization of Plant Growth-Promoting Endophytic Bacteria Paenibacillus polymyxa SK1 from Lilium lancifolium. Biomed Res. Int. 2020, 2020, 8650957. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Choi, S.-K.; Ryu, C.-M.; Park, S.-H. Chronicle of a Soil Bacterium: Paenibacillus polymyxa E681 as a Tiny Guardian of Plant and Human Health. Front. Microbiol. 2019, 10, 467. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Yang, J.; Nie, Q.; Huang, D.; Yu, C.; Zheng, L.; Cai, M.; Thomashow, L.S.; Weller, D.M.; Yu, Z.; et al. Volatile Organic Compounds from Paenibacillus polymyxa KM2501-1 Control Meloidogyne incognita by Multiple Strategies. Sci. Rep. 2017, 7, 16213. [Google Scholar] [CrossRef]
- Ma, G.Z.; Fu, H.R.; Wu, S.J.; Bao, Z.H.; Wang, S.F.; Ge, P.H. Isolation and Structure Elucidation of Antifungal Metabolites from Marine Paenibacillus polymyxa Strain L1-9. Acta Phytopathol. Sin. 2014, 44, 486–496. [Google Scholar] [CrossRef]
- Kavitha, S.; Senthilkumar, S.; Gnanamanickam, S.; Inayathullah, M.; Jayakumar, R. Isolation and Partial Characterization of Antifungal Protein from Bacillus polymyxa Strain VLB16. Process Biochem. 2005, 40, 3236–3243. [Google Scholar] [CrossRef]
- Fu, H.; Li, Y.T.; Xiao, G.Q.; Peng, L.L.; Xia, J.M.; Shao, Z.Z.; Cao, J.W.; Lai, Q.L. Isolation and Purification of Bacillus velezensis MCCC 1A15695 Antibacterial Proteins and Application Studies. J. Appl. Oceanogr. 2023, 1–10. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.-X.; Ho, H.-H.; Chen, Z.-J.; Li, X.-Y.; He, Y.-Q. PBT1, a Novel Antimicrobial Protein from the Biocontrol Agent Bacillus subtilis XF-1 against Plasmodiophora brassicae. Eur. J. Plant Pathol. 2016, 145, 583–590. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Zheng, L.; Yang, X.; Liu, H.; Guo, J. Isolation and Characterization of an Antifungal Protein from Bacillus licheniformis HS10. Biochem. Biophys. Res. Commun. 2014, 454, 48–52. [Google Scholar] [CrossRef]
- Yehia, H.M.; Salmen, S.H.; Alharbi, S.A.; Khiyami, M.A.; Wainwright, M.; Alharbi, N.S.; Chinnathambi, A. Antifungal Protein ∼35kDa Produced by Bacillus cereus Inhibits the Growth of Some Molds and Yeasts. J. Pure Appl. Microbiol. 2013, 7, 401–406. [Google Scholar]
- Chai, Q.; Li, Y.; Li, X.; Wu, W.; Peng, H.; Jia, R.; Sun, Q. Assessment of Variation in Paddy Microbial Communities under Different Storage Temperatures and Relative Humidity by Illumina Sequencing Analysis. Food Res. Int. 2019, 126, 108581. [Google Scholar] [CrossRef]
- Qi, Z.; Zhou, X.; Tian, L.; Zhang, H.; Cai, L.; Tang, F. Temporal and Spatial Variation of Microbial Communities in Stored Rice Grains from Two Major Depots in China. Food Res. Int. 2022, 152, 110876. [Google Scholar] [CrossRef]
- Katsurayama, A.M.; Martins, L.M.; Iamanaka, B.T.; Fungaro, M.H.P.; Silva, J.J.; Pitt, J.I.; Frisvad, J.C.; Taniwaki, M.H. Fungal Communities in Rice Cultivated in Different Brazilian Agroclimatic Zones: From Field to Market. Food Microbiol. 2020, 87, 103378. [Google Scholar] [CrossRef] [PubMed]
- Gil-Serna, J.; Garcia-Diaz, M.; Vazquez, C.; Gonzalez-Jaen, M.T.; Patino, B. Significance of Aspergillus niger Aggregate Species as Contaminants of Food Products in Spain Regarding Their Occurrence and Their Ability to Produce Mycotoxins. Food Microbiol. 2019, 82, 240–248. [Google Scholar] [CrossRef]
- Calvo, H.; Marco, P.; Blanco, D.; Oria, R.; Venturini, M.E. Potential of a New Strain of Bacillus amyloliquefaciens BUZ-14 as a Biocontrol Agent of Postharvest Fruit Diseases. Food Microbiol. 2017, 63, 101–110. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, X.; Wei, Y.; Shang, Q.; Liu, Z. Isolation and Identification of a Novel Antifungal Protein from a Rhizobacterium Bacillus subtilis Strain F3. J. Phytopathol. 2013, 161, 43–48. [Google Scholar] [CrossRef]
- Yu, C.; Yang, X.; Liang, X.; Song, Y.; Zhu, L.; Xing, S.; Yang, Y.; Gu, Q.; Borriss, R.; Dong, S.; et al. Fusaricidin Produced by the Rhizobacterium Paenibacillus polymyxa NX20 Is Involved in the Biocontrol of Postharvest Plant-Pathogenic Oomycete Phytophthora capsici. Postharvest Biol. Technol. 2023, 205, 112545. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, K.; Zhang, X.; Li, Z.; Li, J.; Qi, Y.; Tian, Y. Identification of a Biocontrol Strain of Oat Root Rot and Its Control Effect. Pratacultural Sci. 2021, 38, 2266–2273. [Google Scholar] [CrossRef]
- Lin, S.; Chen, X.; Xie, L.; Zhang, Y.; Zeng, F.; Long, Y.; Ren, L.; Qi, X.; Wei, J. Biocontrol Potential of Lipopeptides Produced by Paenibacillus polymyxa AF01 against Neoscytalidium dimidiatum in Pitaya. Front. Microbiol. 2023, 14, 1188722. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, Z.; Lu, F.; Wang, Y.; Bie, X. Study on an Antimicrobial Protein Produced by Paenibacillus polymyxa JSa-9 Isolated from Soil. World J. Microbiol. Biotechnol. 2011, 27, 1803–1807. [Google Scholar] [CrossRef]
- Castella, G.; Rosa Bragulat, M.; Aiese Cigliano, R.; Javier Cabanes, F. Transcriptome Analysis of Non-Ochratoxigenic Aspergillus carbonarius Strains and Interactions between Some Black Aspergilli Species. Int. J. Food Microbiol. 2020, 317, 108498. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Wu, F.; Liu, F.; Wang, Q.; Zhang, X.; Selvaraj, J.N.; Zhao, Y.; Xing, F.; Yin, W.-B.; et al. A Consensus Ochratoxin a Biosynthetic Pathway: Insights from the Genome Sequence of Aspergillus ochraceus and a Comparative Genomic Analysis. Appl. Environ. Microbiol. 2018, 84, e01009-18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Luo, W.; Chen, G.; Chen, J.; Lin, S.; Ren, T.; Lin, Z.; Zhao, C.; Wen, H.; Nie, Q.; et al. Chicken Muscle Antibody Array Reveals the Regulations of LDHA on Myoblast Differentiation through Energy Metabolism. Int. J. Biol. Macromol. 2024, 254, 127629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Meng, Y.; Li, Q.; Liang, Y. The Endosomal Sorting Complex Required for Transport Complex Negatively Regulates Erg6 Degradation under Specific Glucose Restriction Conditions. Traffic 2020, 21, 488–502. [Google Scholar] [CrossRef] [PubMed]
- Jahanshiri, Z.; Shams-Ghahfarokhi, M.; Asghari-Paskiabi, F.; Saghiri, R.; Razzaghi-Abyaneh, M. α-Bisabolol Inhibits Aspergillus Fumigatus Af239 Growth via Affecting Microsomal Δ24-Sterol Methyltransferase as a Crucial Enzyme in Ergosterol Biosynthesis Pathway. World J. Microbiol. Biotechnol. 2017, 33, 55. [Google Scholar] [CrossRef] [PubMed]
- Chi, F.; Zhao, J.; Yang, L.; Yang, X.; Zhao, X.; Zhao, S.; Zhan, J. Using Regular and Transcriptomic Analyses to Investigate the Biotransformation Mechanism and Phytotoxic Effects of 6:2 Fluorotelomer Carboxylic Acid (6:2 FTCA) in Pumpkin (Cucurbita maxima L.). Sci. Total Environ. 2024, 906, 167901. [Google Scholar] [CrossRef]
- Dong, X.; Cai, M. Handbook of Identification of Common Bacteria; Science Press: Beijing, China, 2001; pp. 370–384. [Google Scholar]
- Wang, L.; Hua, X.; Jing, N.; Ji, T.; Zhou, C.; Liu, W.; Lv, B.; Liu, L.; Chen, Y. Isolation and Characterization of Bacillus amyloliquefaciens YL-1 with Ochratoxin a Degradation Ability and Biocontrol Activity against Aspergillus westerdijkiae. Biol. Control 2022, 175, 105052. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, W.; Zhou, B.; Du, H.; Xi, L.; Zou, M.; Zou, H.; Xin, L.; Gao, Z.; Chen, Y. Variable Characteristics of Microbial Communities on the Surface of Sweet Cherries under Different Storage Conditions. Postharvest Biol. Technol. 2021, 173, 111408. [Google Scholar] [CrossRef]
- Ye, W.-Q.; Sun, Y.-F.; Tang, Y.-J.; Zhou, W.-W. Biocontrol Potential of a Broad-Spectrum Antifungal Strain Bacillus amyloliquefaciens B4 for Postharvest Loquat Fruit Storage. Postharvest Biol. Technol. 2021, 174, 111439. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhu, H.; Li, B.; Ngea, G.L.N.; Gu, X.; Zhang, X.; Dhanasekaran, S.; Zhang, H. Transcriptomic Analysis of the Disease-Resistance Response in Mandarins Induced by the Biocontrol Yeast, Yarrowia lipolytica. Biol. Control 2021, 163, 104607. [Google Scholar] [CrossRef]
- Xu, M.; Yang, Q.; Boateng, N.A.S.; Ahima, J.; Dou, Y.; Zhang, H. Ultrastructure Observation and Transcriptome Analysis of Penicillium expansum Invasion in Postharvest Pears. Postharvest Biol. Technol. 2020, 165, 111198. [Google Scholar] [CrossRef]




| Test Item | Result | Test Item | Result |
|---|---|---|---|
| Salt resistance | Glucose oxidative fermentation | Fermentation type | |
| 2% | + | Sodium citrate utilization | − |
| 5% | − | Sugar fermentation | |
| 10% | − | D-glucose | + |
| Exposure to enzymes | + | Mannitol | + |
| Methyl red | + | Lactose | + |
| Starch hydrolysis | + | Sucrose | + |
| V-P assay | + | Gram | + |
| Gene ID | Gene Description | Log2FC |
|---|---|---|
| ncbi_56006507 | Ornithine aminotransferase | −7.564785 |
| ncbi_56002670 | L-lactate/malate dehydrogenase | −7.494522 |
| ncbi_56004126 | Squalene synthetase | −7.400879 |
| ncbi_56006177 | Glutathione S-transferase | −6.853829 |
| ncbi_56007485 | Phytoene dehydrogenase | −5.403722 |
| ncbi_56006167 | Dihydrodipicolinate synthetase family protein | −5.357552 |
| ncbi_56001048 | Phytoene dehydrogenase | −4.459432 |
| ncbi_56003329 | Threonine aldolase | −4.058894 |
| ncbi_56005832 | Dihydroxy-acetone synthase | −4.058894 |
| ncbi_56002418 | Fungal chitosanase | −3.984418 |
| Gene ID | Gene Description | Log2FC |
|---|---|---|
| ncbi_56007446 | NAD binding Rossmann fold oxidoreductase | −3.529698 |
| ncbi_56002881 | Fructose-bisphosphate aldolase | −3.06848 |
| ncbi_56001061 | D-glycerate 3-kinase-like protein | −2.992715 |
| ncbi_56007372 | Cytochrome P450 | −2.907936 |
| ncbi_56006356 | Beta glucosidase | −2.898623 |
| ncbi_56008280 | Catabolic 3-dehydroquinase | −2.896164 |
| ncbi_56005213 | Isocitrate lyase | −2.069498 |
| Gene ID | Symbol | Primers (5′ to 3′) |
|---|---|---|
| ncbi_56000074 | UFD1 | F: CGGGTGATGTCCAGTGTAAG |
| R: GGTGCCATAAGCCGAGTC | ||
| ncbi_56002166 | UEF-CFP | F: GCGAAGAAAAGGGTCAAAG |
| R: GGTAATGTCAAGGCTGGTCA | ||
| ncbi_56000480 | PepA | F: GTTGAGCGGCTACTCTTGG |
| R: AGGTGGTCTGTGCCTTGG | ||
| ncbi_56000968 | EcdD | F: GGCTATCATCTTGGGAGGTG |
| R: AGTCCGAGTCTTGGGGCT | ||
| ncbi_56001045 | RntA | F: GAGCATCATCGCCCTTCT |
| R: TGCGGGTAGTCGTCAATAGTA | ||
| ncbi_56002623 | Blh | F: TCAACAGAATCAAGCCCCA |
| R: CGAGTCGCCCGTGAATAC | ||
| ncbi_56002624 | CDO1 | F: CAACTTTCCCTGGCTCTGC |
| R: AACCTTGTGGTCAACATTTTCT | ||
| ncbi_56008178 | YAT1 | F: CATTGAATCGCCCAAGTCT |
| R: CGAAGCCAAAGTGCCGTA | ||
| ncbi_56004358 | RPS9 | F: CGCAACAAGCGTGAGGTG |
| R: CAGACGGCGAATCAAAGC | ||
| ncbi_56004736 | RPS8A | F: AACTTCTCGTGGGGTTCTGA |
| R: GGTCTCGGTCTTCTGCTGG | ||
| ncbi_56008317 | RPL6 | F: ATGTCGGACTCTACTGTTGGC |
| R: CGGACTTTCTTGGGCTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Ma, J.; Lin, M.; Gao, C.; Zhao, Y.; Li, X.; Sun, W. Isolation and Characterization of Paenibacillus polymyxa B7 and Inhibition of Aspergillus tubingensis A1 by Its Antifungal Substances. Int. J. Mol. Sci. 2024, 25, 2195. https://doi.org/10.3390/ijms25042195
Zhao T, Ma J, Lin M, Gao C, Zhao Y, Li X, Sun W. Isolation and Characterization of Paenibacillus polymyxa B7 and Inhibition of Aspergillus tubingensis A1 by Its Antifungal Substances. International Journal of Molecular Sciences. 2024; 25(4):2195. https://doi.org/10.3390/ijms25042195
Chicago/Turabian StyleZhao, Tianyuan, Jianing Ma, Meiwei Lin, Chen Gao, Yuhao Zhao, Xin Li, and Weihong Sun. 2024. "Isolation and Characterization of Paenibacillus polymyxa B7 and Inhibition of Aspergillus tubingensis A1 by Its Antifungal Substances" International Journal of Molecular Sciences 25, no. 4: 2195. https://doi.org/10.3390/ijms25042195
APA StyleZhao, T., Ma, J., Lin, M., Gao, C., Zhao, Y., Li, X., & Sun, W. (2024). Isolation and Characterization of Paenibacillus polymyxa B7 and Inhibition of Aspergillus tubingensis A1 by Its Antifungal Substances. International Journal of Molecular Sciences, 25(4), 2195. https://doi.org/10.3390/ijms25042195





