Iron–Imine Cocktail in Drug Development: A Contemporary Update
Abstract
1. Introduction
2. Bioactivity of Imine–Iron Complexes
2.1. Imine–Iron Complexes as Anticancer Agents
| Entry No. | Complex No. | Structures | Synthesis Condition | Complex and Positive Control | Cancer Cell Lines | Ref. | ||
|---|---|---|---|---|---|---|---|---|
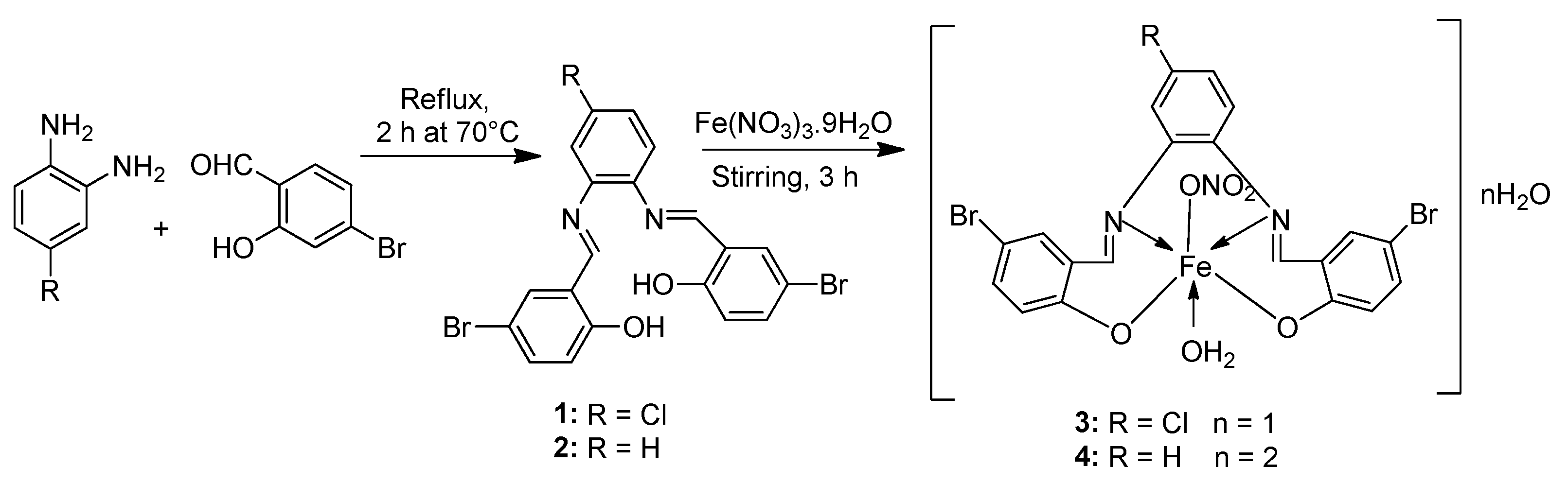
| 1. | 3, 4 | 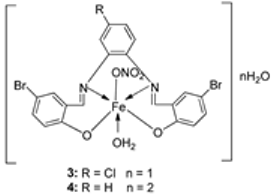 | EtOH Reflux, 2 h Stirring | MCF-7 | HepG-2 | HCT-116 | [21] | |
| 3 | 21.35 ± 0.12 | 27.70 ± 0.11 | 15.75 ± 0.07 | |||||
| 4 | 5.14 ± 0.05 | 6.75 ± 0.12 | 4.45 ± 0.14 | |||||
| Doxorubicin | 4.10 ± 0.13 | 5.15 ± 0.07 | 4.35 ± 0.15 | |||||
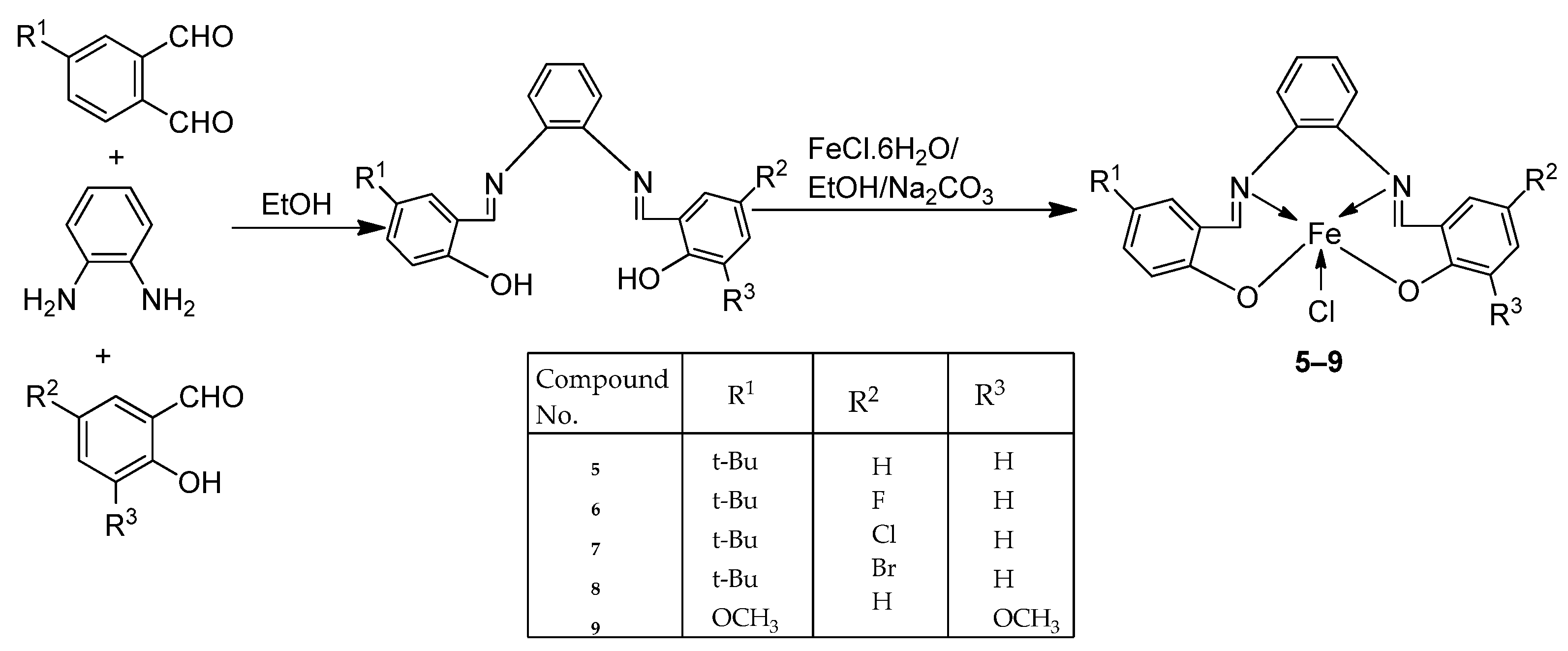
| 2. | 5–9 | 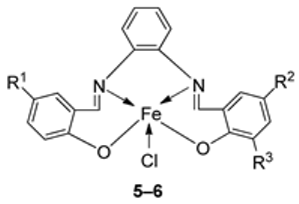 | EtOH Reflux, 3 h | KB | HepG-2 | [22] | ||
| 5 | 0.68 ± 0.05 | 0.83 ± 0.05 | ||||||
| 6 | 3.25 ± 0.16 | 7.05 ± 0.25 | ||||||
| 7 | 1.84 ± 0.10 | 6.07 ± 0.22 | ||||||
| 8 | 2.76 ± 0.17 | 19.78 ± 1.07 | ||||||
| 9 | 1.95 ± 0.13 | 2.38 ± 0.17 | ||||||
| Ellipticine | 1.14 ± 0.06 | 2.11 ± 0.12 | ||||||
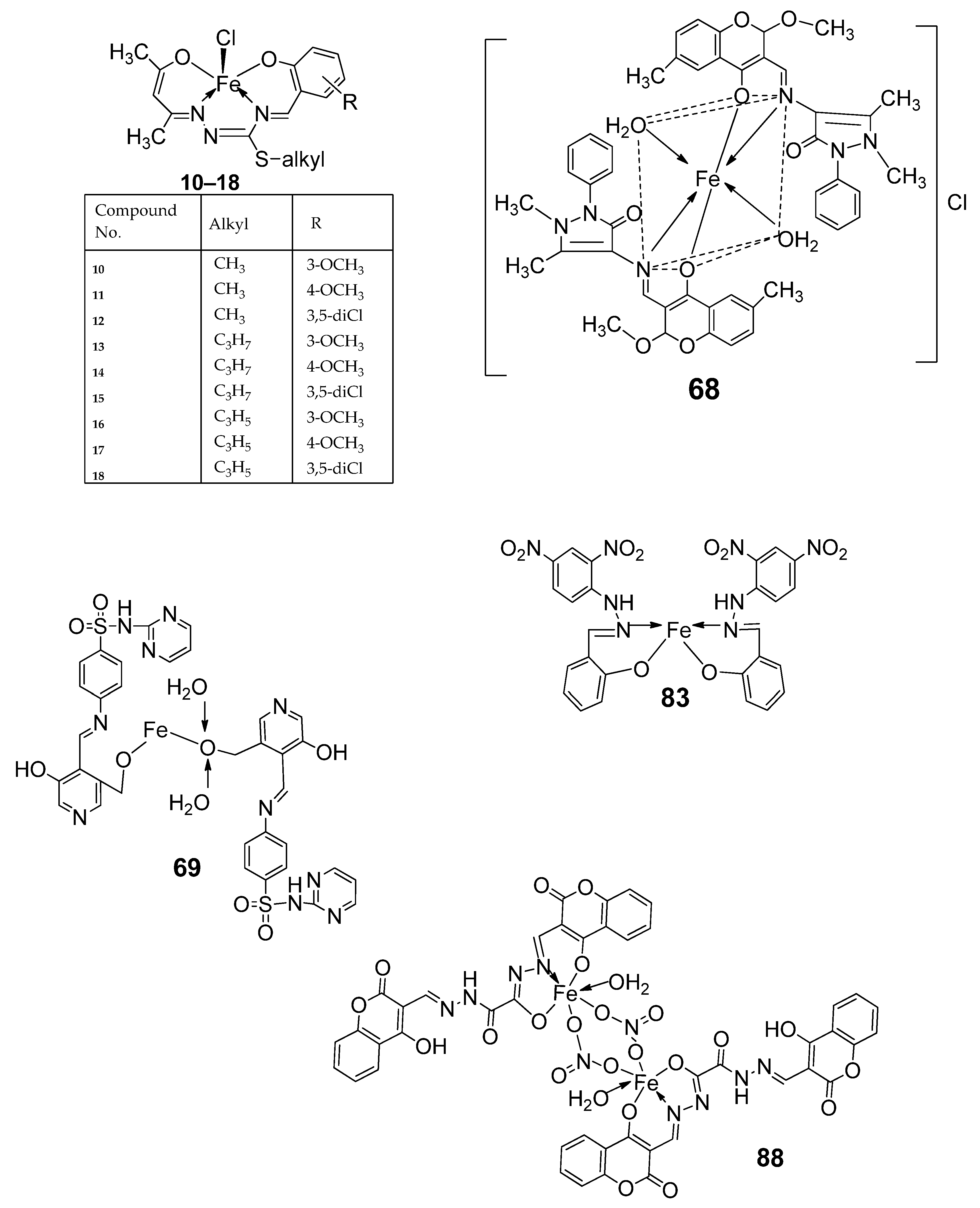
| 3. | 10–18 | 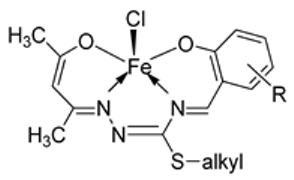 | Stirring, 30 min | K562 | P3HR1 | JURKAT | [27] | |
| 10 | >25 | >25 | >25 | |||||
| 11 | 9.25 ± 0.42 | 5.61 ± 0.19 | >25 | |||||
| 12 | 22.24 ± 0.06 | 8.09 ± 0.62 | >25 | |||||
| 13 | >25 | >25 | >25 | |||||
| 14 | 4.81 ± 0.15 | 11.98 ± 0.69 | 22.79 ± 0.54 | |||||
| 15 | >25 | 22.4 ± 0.47 | >25 | |||||
| 16 | 14.05 ± 0.31 | 5.72 ± 0.28 | >25 | |||||
| 17 | 5.04 ± 0.18 | 11.47 ± 0.42 | 22.0 ± 0.39 | |||||
| 18 | >25 | 21.03 ± 0.39 | >25 | |||||
| Imatinib | 9.67 ± 0.49 | 23.74 ± 1.02 | 3.73 ± 0.21 | |||||
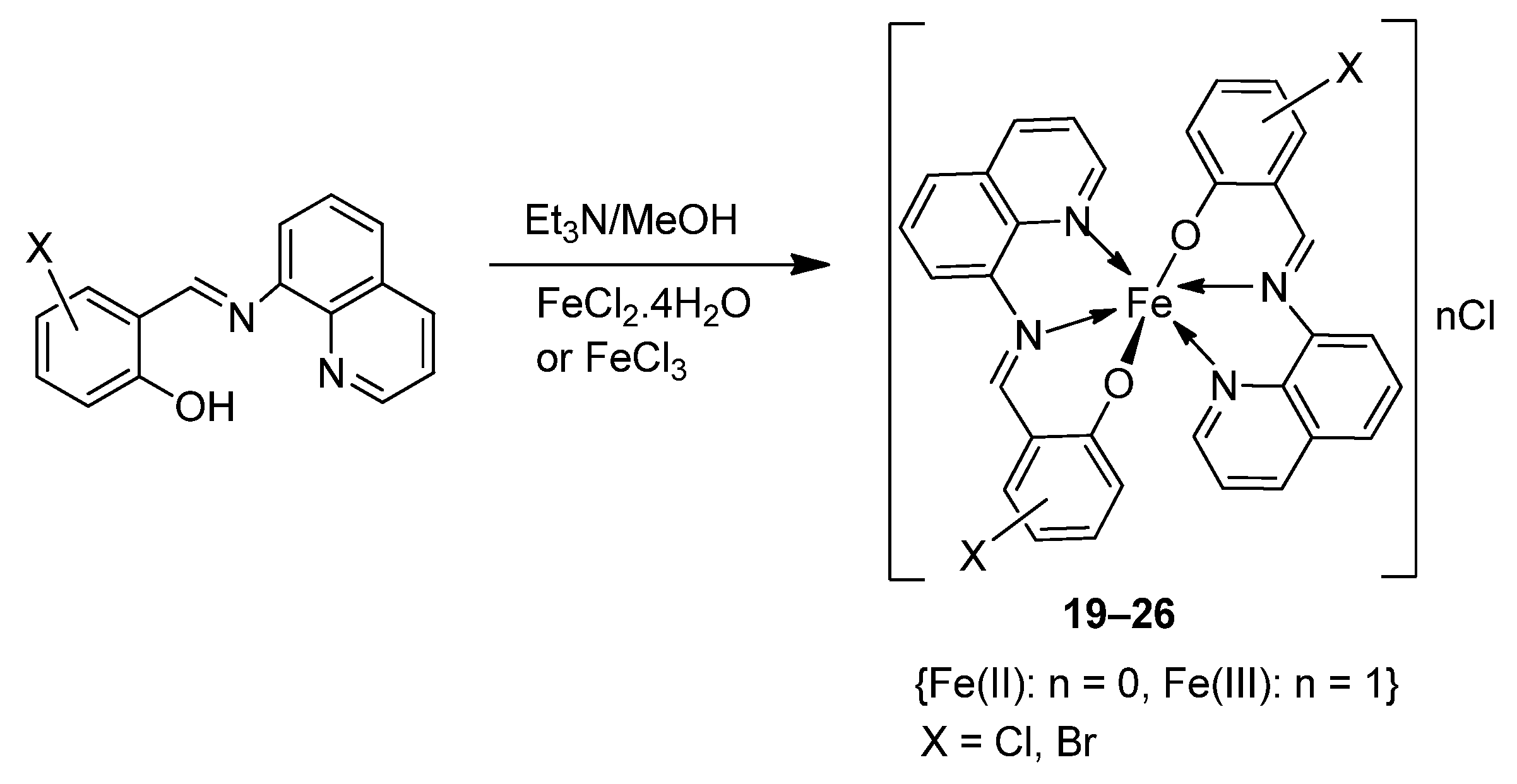
| 4. | 19–26 | 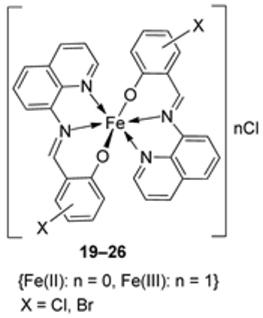 | 0 °C, 7 days | A549 | [28] | |||
| 19 | 30 ± 1.1 | |||||||
| 20 | 30 ± 7.7 | |||||||
| 21 | 28 ± 2.0 | |||||||
| 22 | 28 ± 2.0 | |||||||
| 23 | 28 ± 2.0 | |||||||
| 24 | 10 ± 2.1 | |||||||
| 25 | 34 ± 4.7 | |||||||
| 26 | 32 ± 1.5 | |||||||
| Etoposide | 19 ± 1.3 | |||||||
| Cisplatin | 16 ± 1.9 | |||||||
| 5. | 30 | 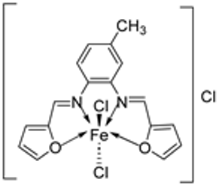 | Reflux, 3 h Stirring, 2 h | Hep-G2 | [29] | |||
| 30 | 7.31 | |||||||
| Vinblastine | 2.93 | |||||||
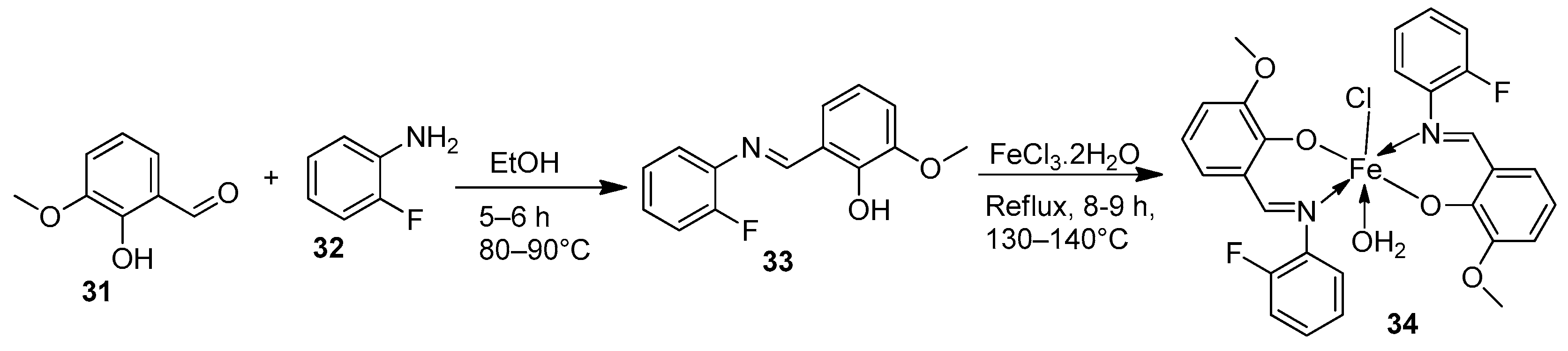
| 6. | 34 | 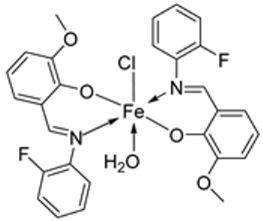 | Reflux, 8–9 h | Hela | MiaPaCa-2 | B16F10 | [30] | |
| 34 | 106.26 ± 0.5 | 112.13 ± 0.6 | 104.15 ± 1.2 | |||||
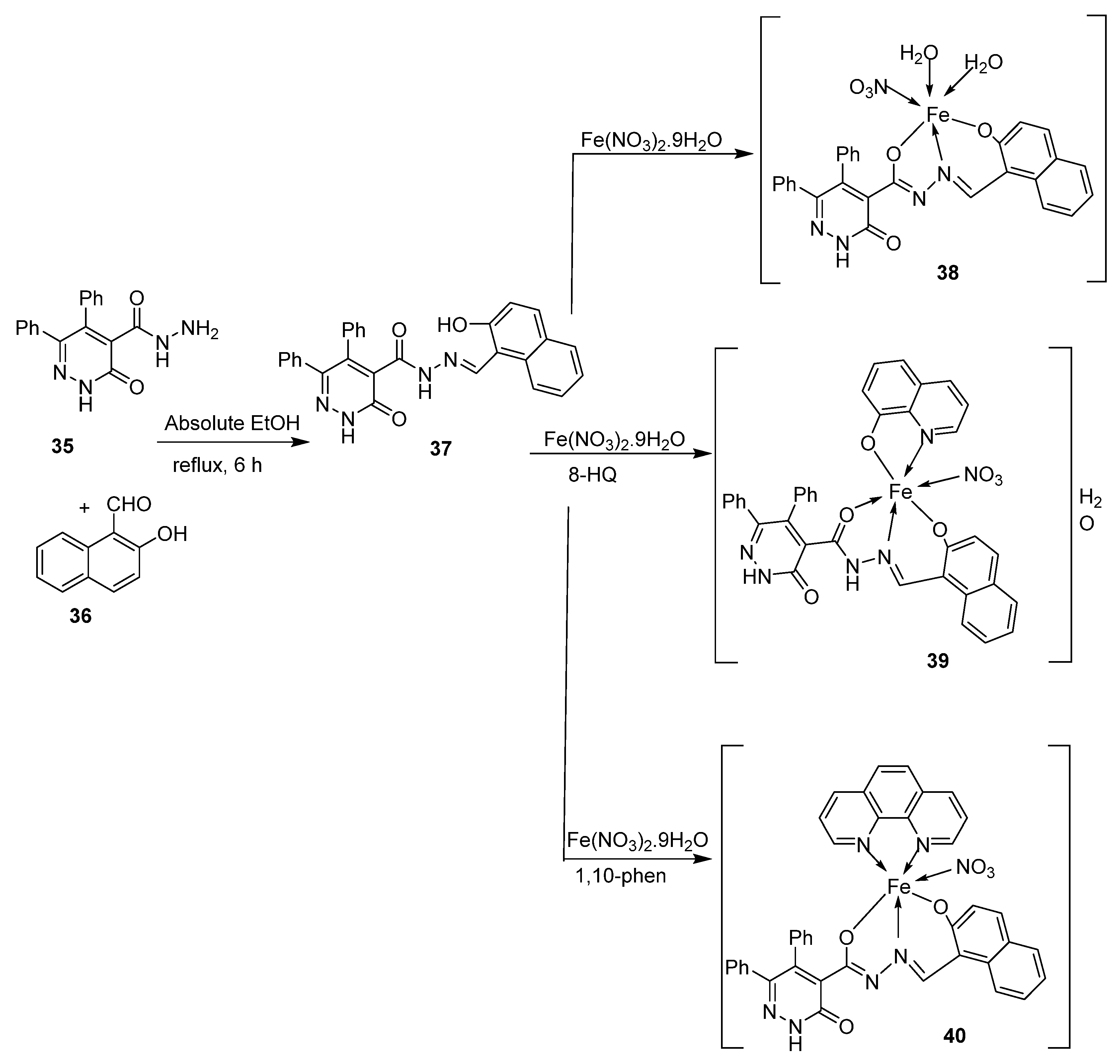
| 7. | 38–40 | 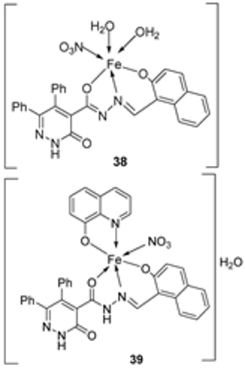 | Stir, 2 h Reflux, 12–15 h | Hep-G2 | [31] | |||
| 37 | 3.8058.00 | |||||||
| 38 | R | |||||||
| 39 | R | |||||||
| 40 | 3.27 | |||||||
| Cisplatin | ||||||||
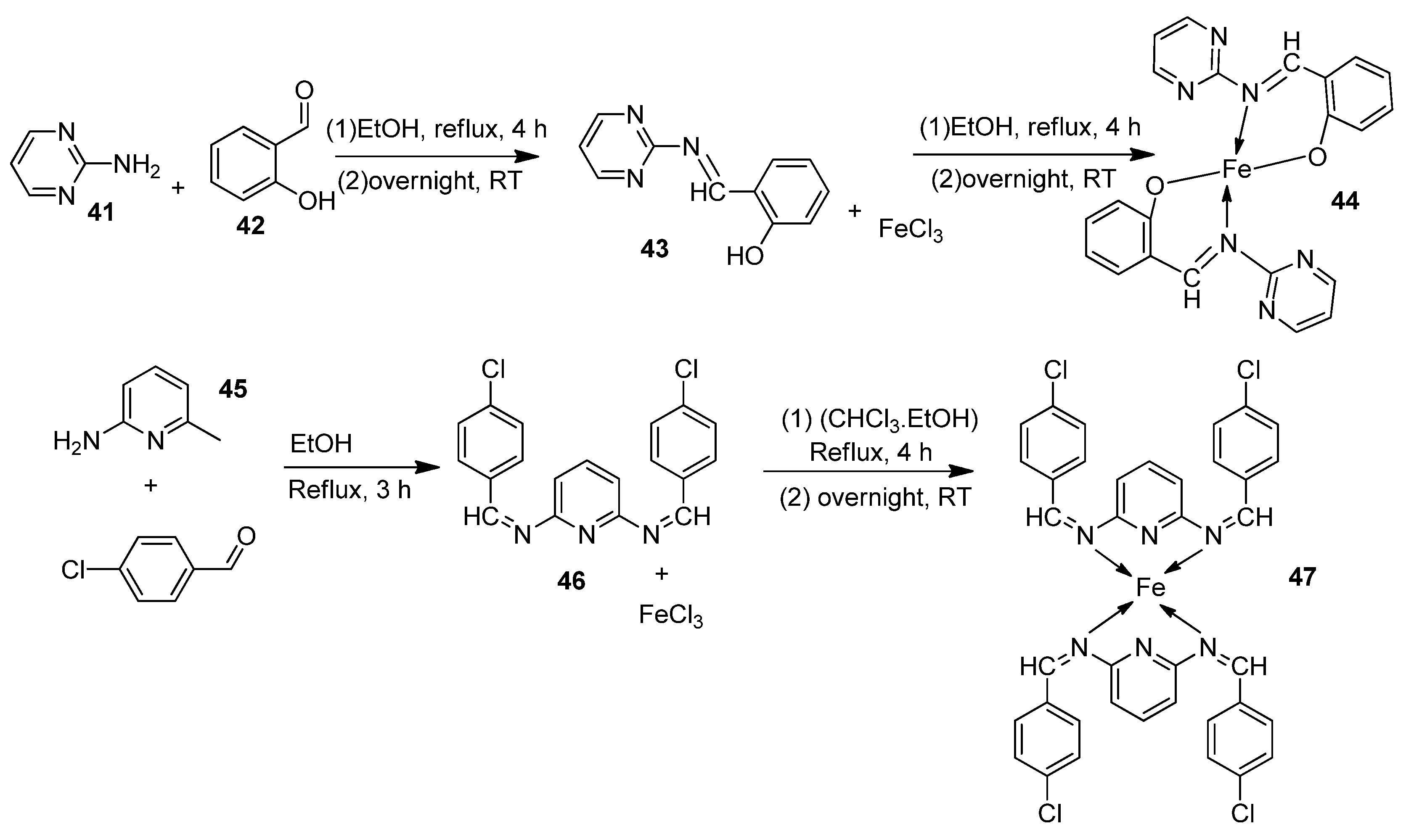
| 8. | 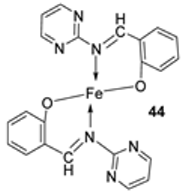 | Reflux, 3–4 h | L20B | [32] | ||||
| 44 | 44 | 8.70 | ||||||
| 45 | 45 | 13.20 | ||||||
| 46 | 46 | 18.4 | ||||||
| 47 | 47 | 22.9 | ||||||
2.2. Imine–Iron Complexes as Antimicrobial Agents
- Novel mechanism of action: Traditional antibiotics typically target specific bacterial functions like cell wall synthesis or protein translation, which can lead to resistance development as bacteria mutate those targets, imine–iron complexes employ diverse mechanisms, including iron starvation, DNA cleavage, and reactive oxygen species (ROS) generation, making it harder for bacteria to develop resistance.
- Broad-spectrum activity: Traditional antibiotics often have specific targets, limiting their effectiveness against different types of bacteria, whilst imine–iron complexes can exhibit activity against a wider range of bacteria, including multi-drug-resistant strains, due to their multiple attacking mechanisms.
- Biofilm disruption: Traditional antibiotics may struggle to penetrate bacterial biofilms, protective communities are resistant to many drugs, whilst imine–iron complexes show potential to disrupt biofilms, exposing bacteria within to attack further.
- Reduced side effects: Traditional antibiotics can harm beneficial gut bacteria and other healthy cells due to their broad targeting, whilst imine–iron complexes can be designed to be more selective for bacterial targets, potentially reducing the side effects on human cells.
| No. | Complex No. | Structures of Synthesized Complexes | Reaction Conditions | Antimicrobial Biological Activity | Ref. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
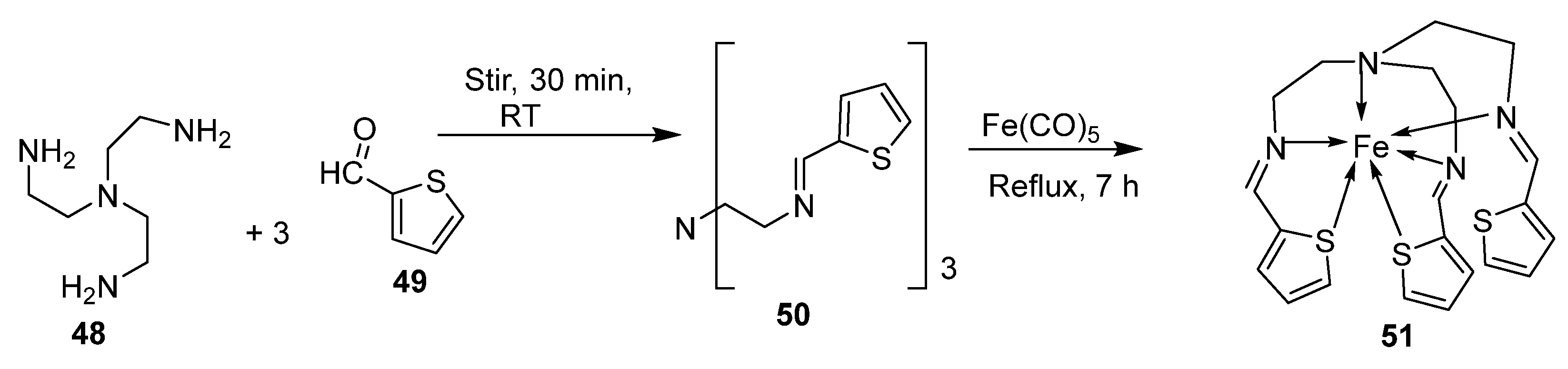
| 1. | 51 | 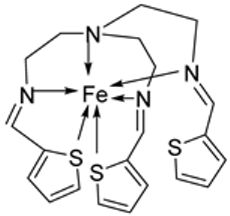 | Stirring, 30 min Reflux, 7 h | Zone of inhibition, mm | [40] | ||||||
| S. aureus | E. coli | P. aeruginosa | B. cereus | ||||||||
| 50 | 11 | 10 | 11 | 12 | |||||||
| 51 | 14 | 14 | 14 | 29 | |||||||
| Tetracycline | 9 | 10 | 12 | 11 | |||||||
| 2. | 55 | 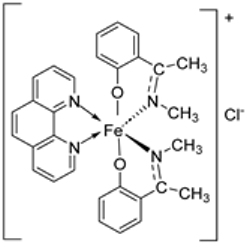 | Stirring, 1–2 h Reflux, 2–11 h | Zone of inhibition, mm | [42] | ||||||
| E. coli | |||||||||||
| 54 | 23 | ||||||||||
| 55 | 29 | ||||||||||
| Amoxicillin | 41 | ||||||||||
| Chloramphenicol | 39 | ||||||||||
| 3. | 34 | 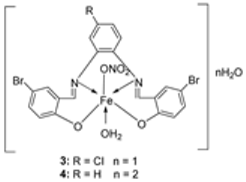 | Reflux, 2 h | Minimum Inhibitory Concentration (MIC)/µg/mL | [21] | ||||||
| Bacteria | Fungi | ||||||||||
| S. marcescence | E. coli | M. luteus | G. candidum | A. flavus | F. oxysporum | ||||||
| 1 | 7.25 | 7.25 | 6.25 | 6.75 | 8.00 | 7.50 | |||||
| 2 | 5.50 | 6.25 | 4.75 | 5.25 | 6.75 | 6.25 | |||||
| 3 | 3.75 | 4.25 | 3.00 | 4.00 | 4.50 | 4.25 | |||||
| 4 | 3.25 | 3.50 | 2.50 | 3.00 | 3.75 | 3.50 | |||||
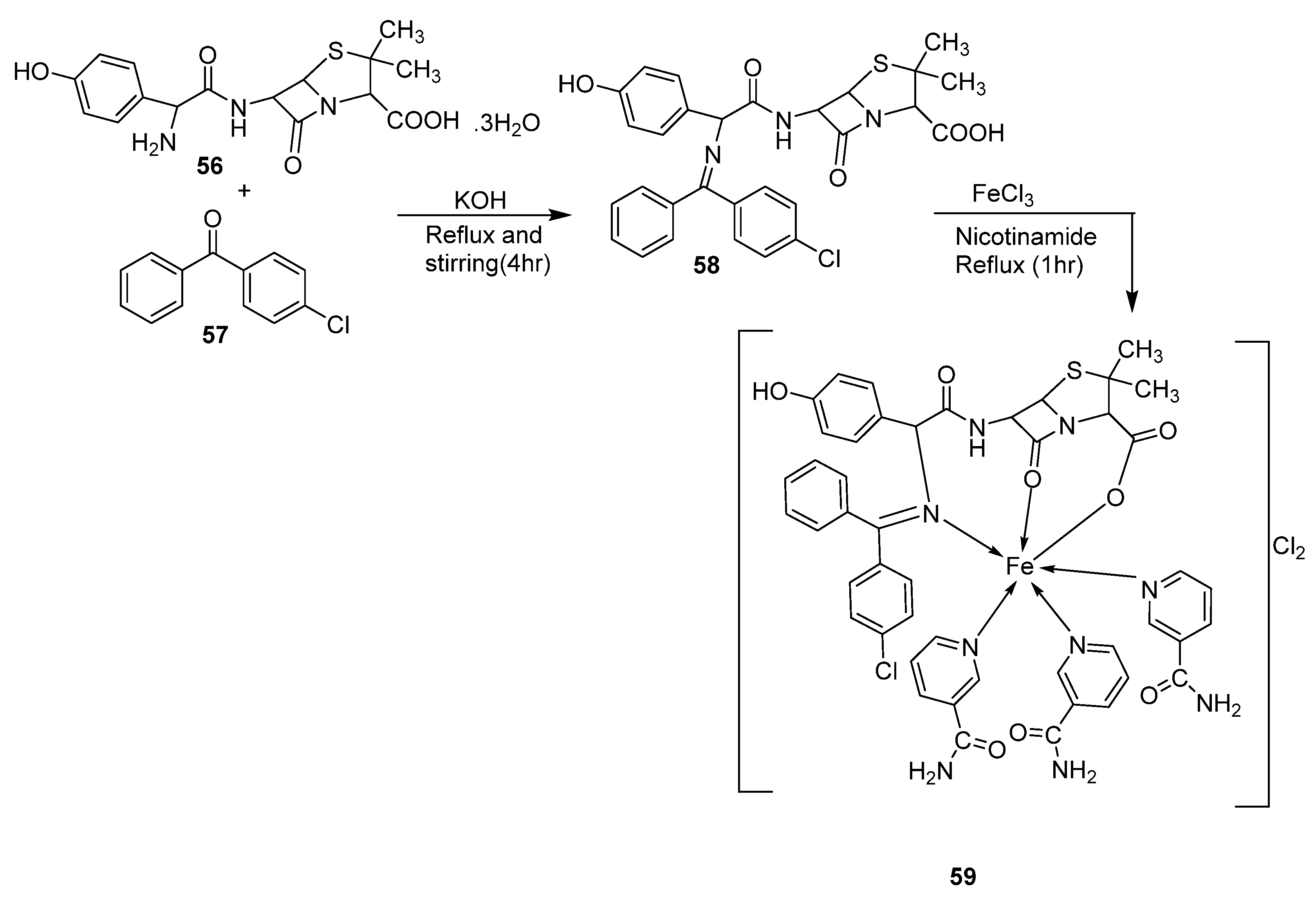
| 4. | 59 |  | Stirring Reflux, 1 h | Minimum Inhibitory Concentration (MIC)/µg/mL | [47] | ||||||
| E. coli | Pseudomonas | S. aureus | Bacillus | ||||||||
| 58 | 2.5 | 8 | 15 | 17 | |||||||
| 59 | 25 | R | R | ** R | |||||||
| 5. | 63 64 |  | Stirring, 1 h Reflux, 9 h | Zone of inhibition, mm | [48] | ||||||
| Bacillus subtilis | E. coli | ||||||||||
| 62 | 11 | 15 | |||||||||
| 63 | 12 | 11 | |||||||||
| 64 | 14 | 18 | |||||||||
| Amoxicillin | 16 | 20 | |||||||||
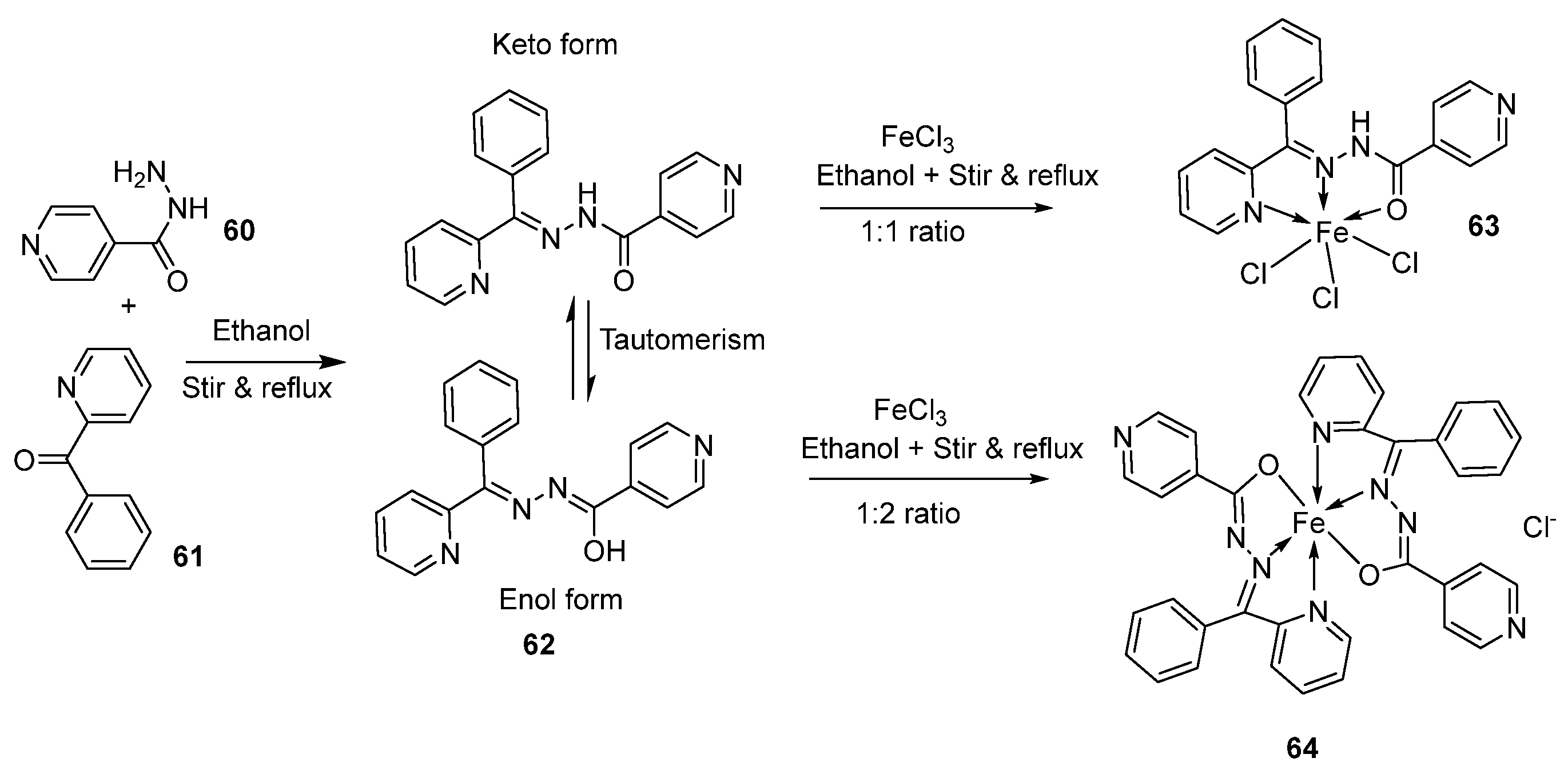
| 6. | 67 | 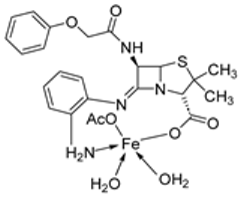 | Reflux, 5 h | Zone of inhibition, mm | [65] | ||||||
| S. v | E. sp | S. a | E. f | MRSA | |||||||
| 65 | 15 | 24 | 16 | 17 | R ** | ||||||
| 67 | 20 | 30 | 25 | 22 | 15 | ||||||
| Standard | 19 | 36 | 45 | 36 | R | ||||||
| 7. | 68 | 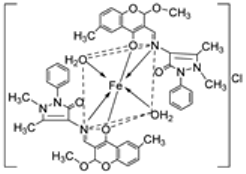 | Reflux, 4 h | Minimum Inhibitory Concentration (MIC)/µg/mL | [10] | ||||||
| C. albicans | C. neoformans | S. aureus | B. cereus | E. coli | |||||||
| 68 | 0.0156 | 0.0078 | 0.0625 | 0.0312 | 0.0625 | ||||||
| Nystatin | 0.032 | 0.032 | |||||||||
| Miconazole | 0.016 | 0.0162 | |||||||||
| Furacillinum | |||||||||||
| Ciprofloxacin | 0.0046 | 0.0046 | 0.0046 | ||||||||
| Amikacin | 0.001 | 0.0003 | 0.008 | ||||||||
| 8. | 69 |  | Reflux, 2 h | Zone of inhibition, mm | [51] | ||||||
| E. coli | E. aerogenes | C. butyrium | |||||||||
| Ligand | 14 | 12 | 12 | ||||||||
| 69 | 12 | 10 | 9 | ||||||||
| Standard | 11 | 7 | 9 | ||||||||
| 9. | 73 | 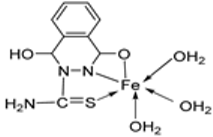 | Stir and reflux, 1 h | Zone of inhibition, mm | [52] | ||||||
| S. pneumonia | S. aureus | ||||||||||
| 72 | 7–10 | 1–3 | |||||||||
| 73 | 7–10 | 7–10 | |||||||||
| 10. | 76 | 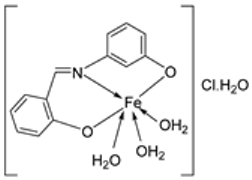 | Stir and reflux, 2 h | Zone of inhibition, mm | [4] | ||||||
| S. aureus | E. coli | P. aeruginosa | C. albicans | A. fumigatus | |||||||
| 75 | 15 | 14 | 16 | 13 | 15 | ||||||
| 76 | 16 | 14 | 15 | 16 | 18 | ||||||
| Ampicillin | 23 | 16 | |||||||||
| Gentamycin | |||||||||||
| Amphotericin | 19 | 25 | 23 | ||||||||
| 11. | 79 | 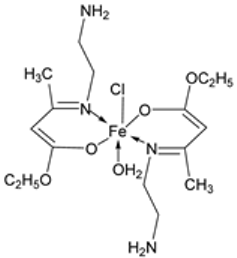 | Reflux, 4 h | Zone of inhibition, mm | [53] | ||||||
| S. aureus | P. aeruginosa | ||||||||||
| 78 | 8 | 6 | |||||||||
| 79 | 14 | 11 | |||||||||
| Ampicillin | 14 | ||||||||||
| Choloramphenicol | 8 | ||||||||||
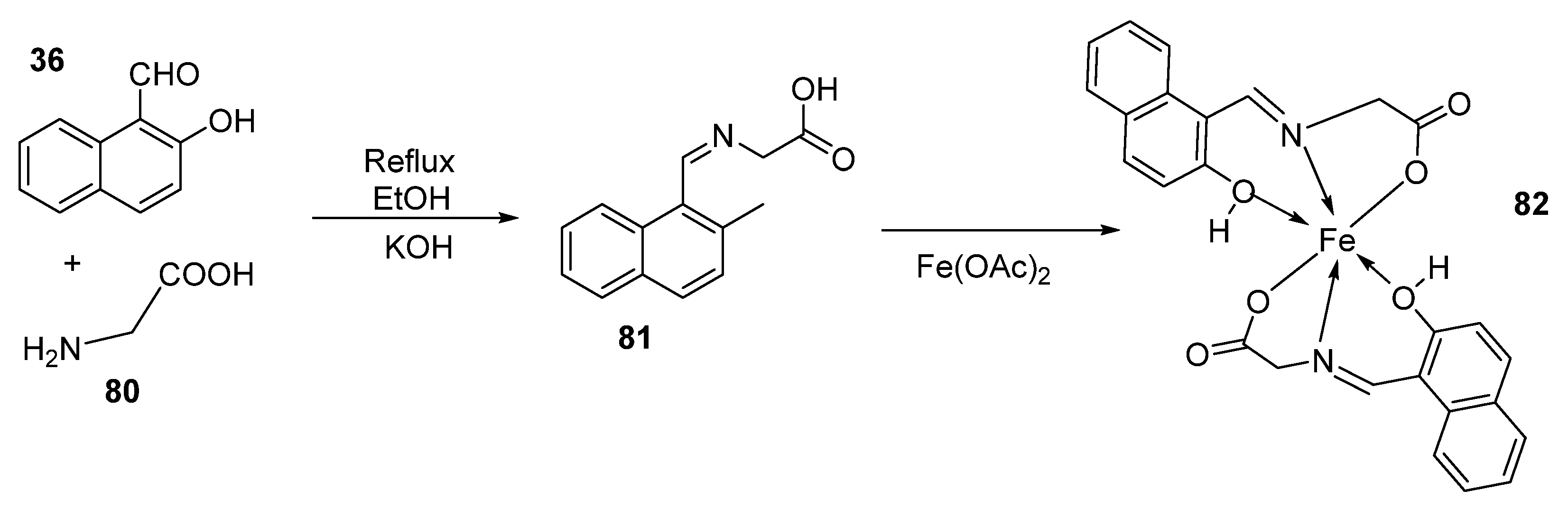
| 12. | 82 | 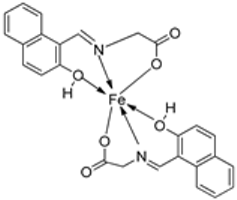 | Stir (overnight) | Minimum Inhibitory Concentration (MIC)/µg/mL | [2] | ||||||
| E. coli | S. aureus | C. albicans | A. niger | ||||||||
| 82 | 10 | 10 | 10 | 10 | |||||||
| Gentamicin | 10 | 10 | |||||||||
| Fluconazole | 20 | 20 | |||||||||
| 13. | 83 | 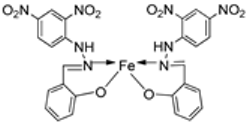 | Reflux and stirring, 3 h | Zone of inhibition, mm | [54] | ||||||
| E. coli | P. aeruginosa | S. aureus | C. albicans | F. solani | |||||||
| Ligand | R | R | 12 | ** R | ** R | ||||||
| 83 | 14 | 8 | 12 | 7 | 12 | ||||||
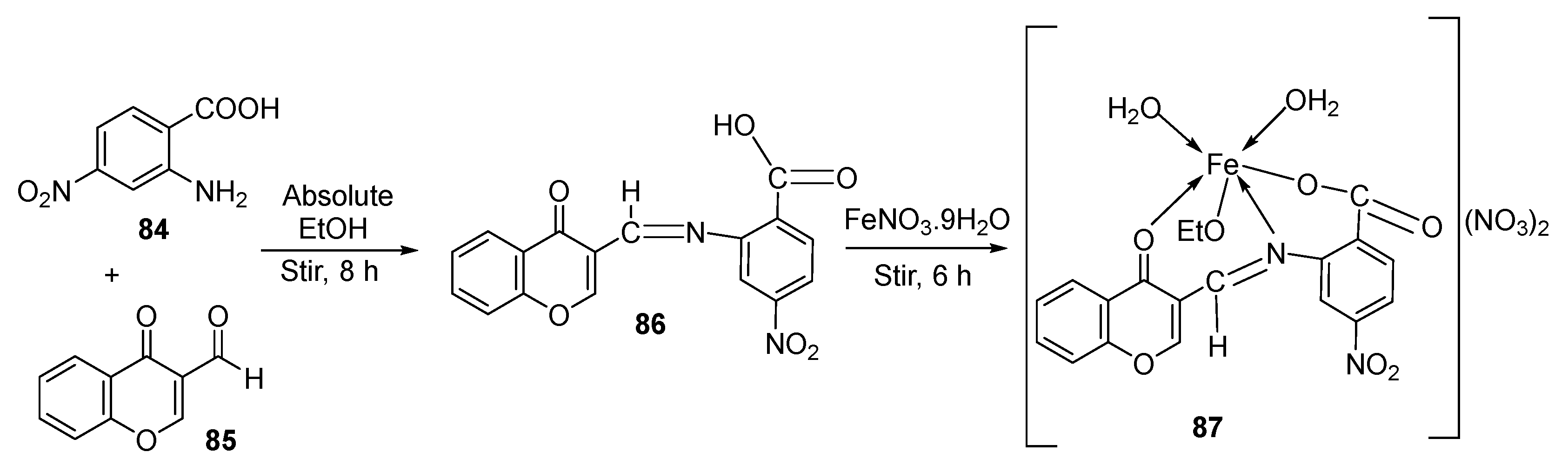
| 14. | 87 | 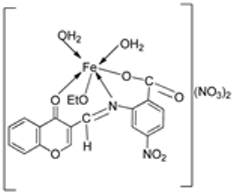 | Stirring, 6 h | Minimum Inhibitory Concentration (MIC)/µg/mL | [3] | ||||||
| E. coli | C. albicans | P. vulgaris | K. pneumonia | S. aureus | |||||||
| 86 | 12.5 | 4 | >50 | 1 | >50 | ||||||
| 87 | ˃50 | 8 | >50 | >50 | >50 | ||||||
| Doxymycin Fluconazole | 2 | 2 | 4 | 4 | |||||||
| 2 | |||||||||||
| 15. | 88 89 |  | Stir, 30 min Reflux, 6 h | Zone of inhibition, mm | [55] | ||||||
| S. aureus | P. phaseolicol | F. oxysporium | |||||||||
| Ligand | 22 ± 0.2 | 13 ± 0.1 | 17 ± 0.2 | ||||||||
| 88 | 37 ± 0.4 | 26 ± 0.1 | 31 ± 0.2 | ||||||||
| 89 | 32 ± 0.2 | 23 ± 0.1 | 30 ± 0.2 | ||||||||
| Cephalothin | 42 | ||||||||||
| Chloramphenicol | 36 | ||||||||||
| Cycloheximide | 40 | ||||||||||
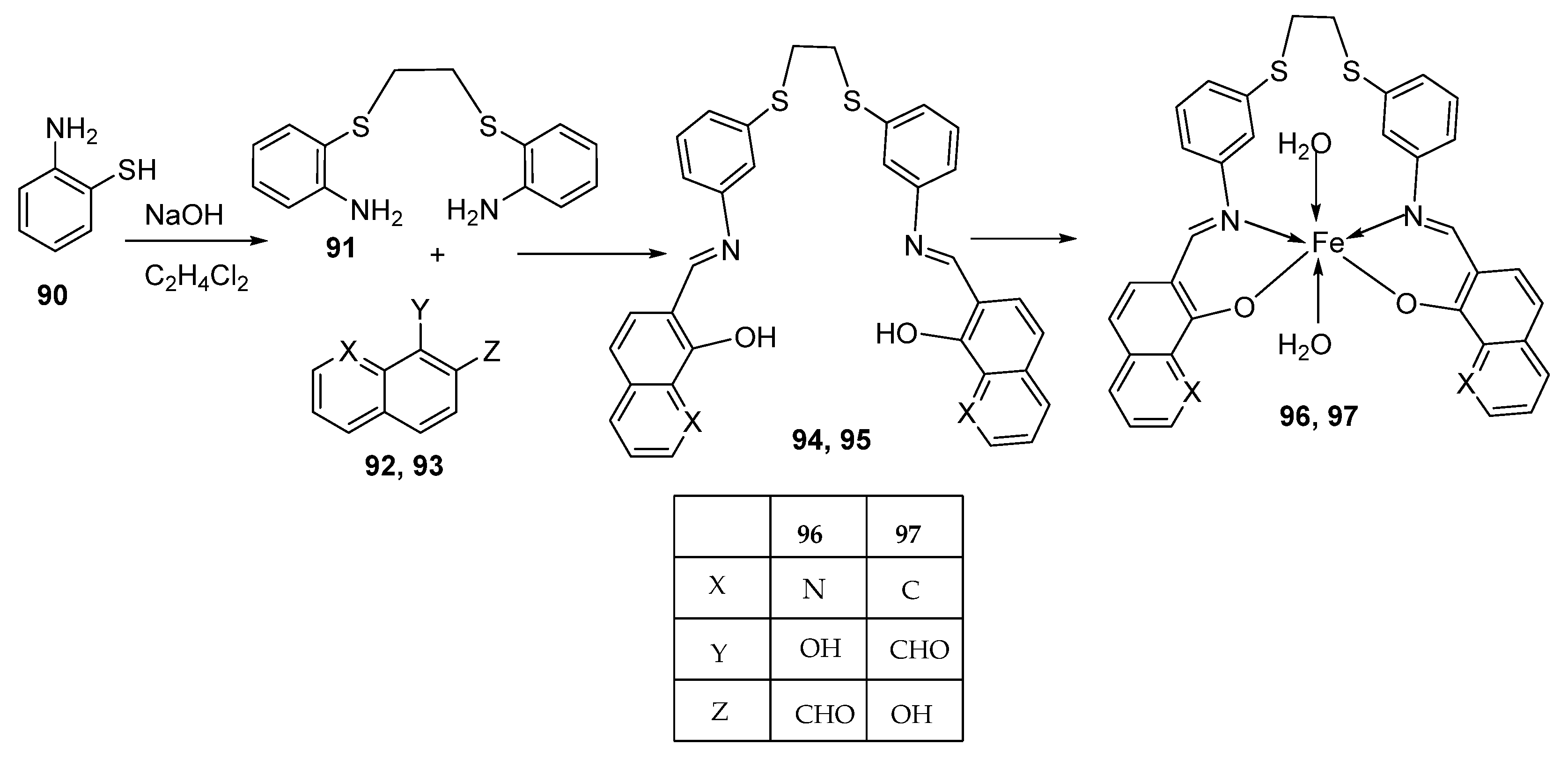
| 16. | 96 97 | 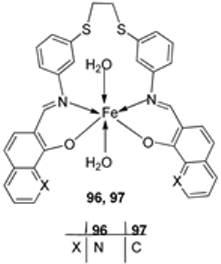 | Reflux and stirring, 4–5 h | Zone of inhibition, mm | [57] | ||||||
| S. epidermidis | E. faecalis | S. aureus | P. mirabilis | C. albicans | |||||||
| 94 | 5 | 9 | 7 | ** R | ** R | ||||||
| 95 | 6 | 8 | 9 | ** R | ** R | ||||||
| 96 | 14 | 15 | 12 | 8 | ** R | ||||||
| 97 | 12 | 8 | 7 | 22 | ** R | ||||||
| Amoxicillin | 28 | 26 | 27 | 44 | |||||||
| 17. | 100 101 |  | Reflux and stirring, 50 min | Zone of inhibition, mm | [59] | ||||||
| E. coli | P. aeruginosa | C. albicans | S. aureus | C. glabrata | |||||||
| 98 | 11 | 15 | 15 | 19 | 11 | ||||||
| 99 | 12 | 12 | 12 | 18 | 12 | ||||||
| 100 | 22 | 19 | 20 | 16 | <10 | ||||||
| 101 | 20 | 16 | 13 | 20 | 12 | ||||||
| Tetracycline | 25 | 20 | 23 | ||||||||
| Nystatin | 19 | 16 | |||||||||
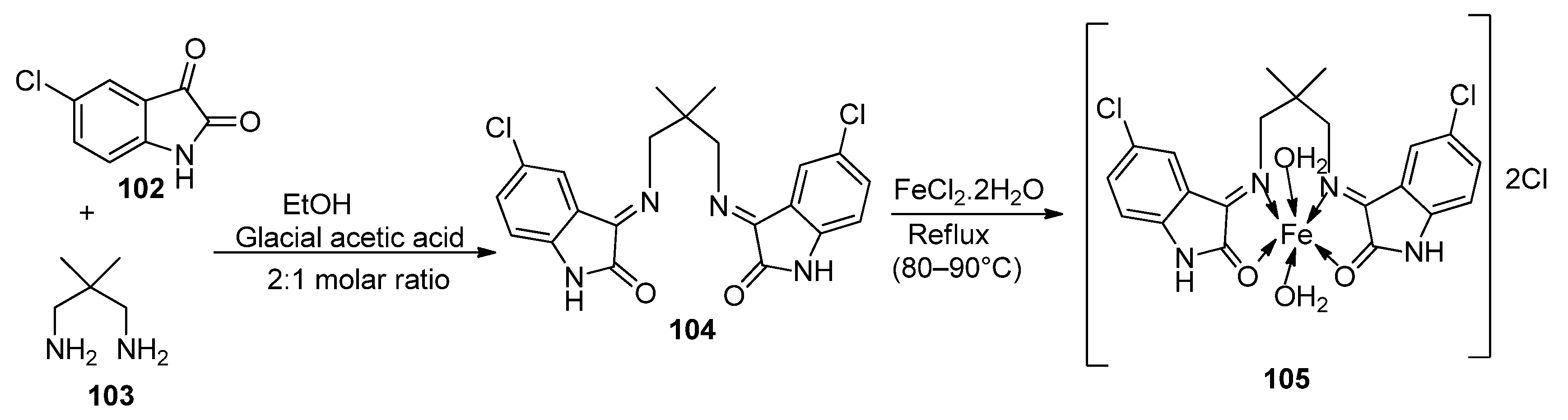
| 18. | 105 | 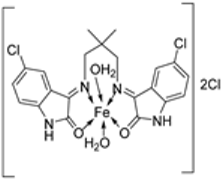 | Reflux, 15 h | Zone of inhibition, mm | [60] | ||||||
| E. coli | S. epidermidis | A. niger | A. flavus | C. lunata | |||||||
| 104 | ** R | 6 | 11 | 9 | 10 | ||||||
| 105 | 15 | 15 | 16 | 14 | 15 | ||||||
| 19. | 34 | 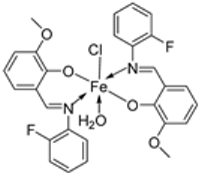 | Reflux, 8–9 h | Zone of inhibition, mm | [30] | ||||||
| Bacillus | Staphylococcus | E. coli | S. rolfsii | M. phaseolina | |||||||
| 33 | 1 | 1 | 1 | 2 | 8 | ||||||
| 34 | 4 | 3 | 4 | 6 | 14 | ||||||
| Streptomycin | 9 | 11 | 5 | ||||||||
| Mancozeb | 18 | 24 | |||||||||
| 20. | 109 | 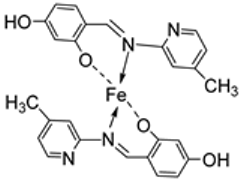 | Reflux, 4–5 h | Zone of inhibition, mm | [61] | ||||||
| S. aureus | E. coli | A. niger | C. albicans | F. moniliforme | |||||||
| 109 | 3.02 | ** R | 15.80 | 7.44 | ** R | ||||||
| Chloramphenicol | 15.11 | 25.44 | 23.23 | ||||||||
| Amphotericin | 15.78 | 12.58 | |||||||||
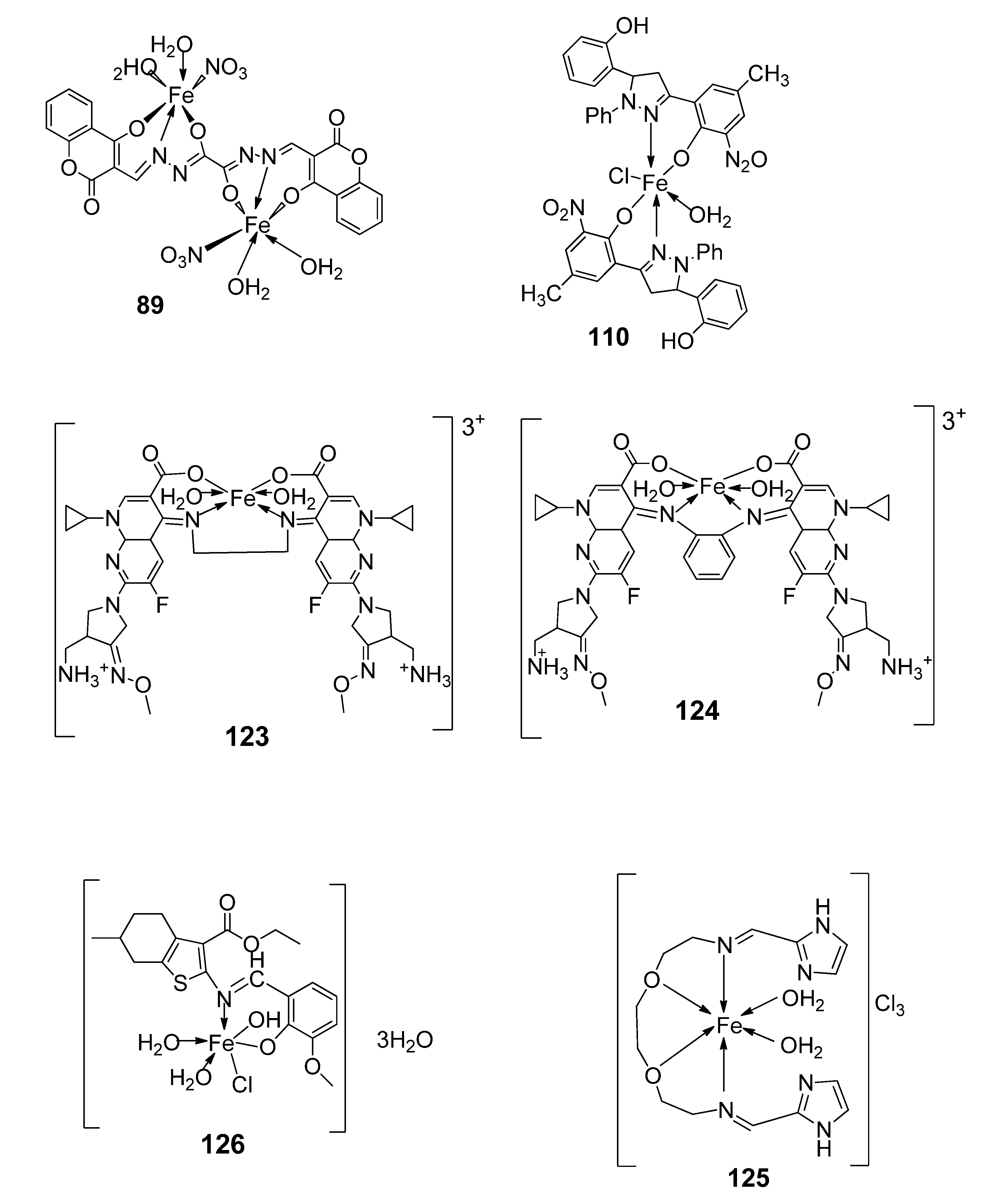
| 21. | 110 | 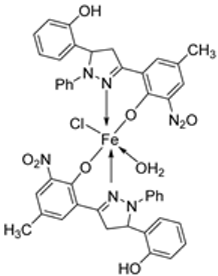 | Reflux, 15–16 h | Zone of inhibition, mm | [62] | ||||||
| S. pyrogenes | E. coli | S. typhi | |||||||||
| 110 | 25 | 16 | 19 | ||||||||
| 22. | 113 | 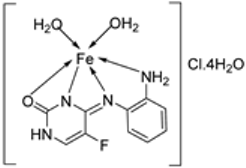 | Reflux, 6 h | Zone of inhibition, mm (concentration, mg/mL) | [64] | ||||||
| B. subtilis | B. megaterium | P. aeroginosa | K. pneumonia | E. aerogenes | |||||||
| 111 | 40±0.47(0.2) | 34±0.81(0.2) | 42±1.24(1) | 36±0.47(0.2) | 45 ± 0.00 | ||||||
| 112 | 30 ± 0.81(0.2) | 22±0.81(0.5) | 33±0.81(0.2) | ** R | 28 ± 0.00 | ||||||
| 113 | 21 ± 0.00(0.2) | ** R | 36±1.24(1) | ** R | ** R | ||||||
| Erythromycin | 20 ± 0.00 | 25±0.47 | 19±0.47 | 19±0.00 | 27 ± 1.24 | ||||||
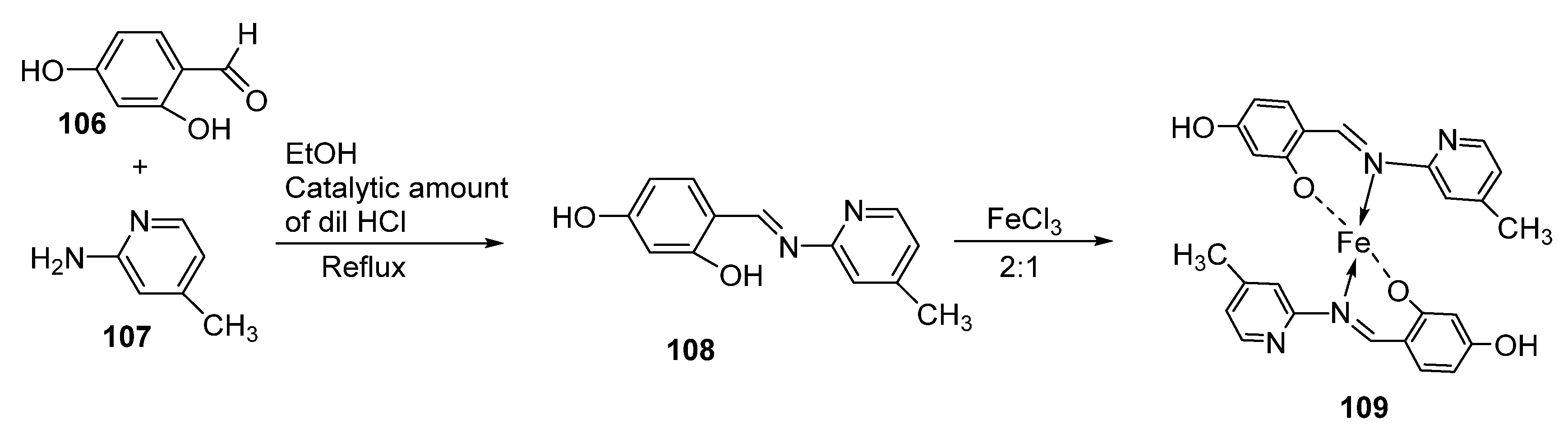
| 23. | 120 121 122 |  | Reflux, 8 h | Zone of inhibition, mm | [66] | ||||||
| S. aureus | P. aureginosa | E. coli | S. typhii | Aspergillus sp. | Penicillium sp. | ||||||
| 119 | 36 | 08 | 10 | 10 | 48 | 29 | |||||
| 120 | 30 | 36 | 41 | 42 | 68 | 61 | |||||
| 121 | 24 | 25 | 22 | 28 | 51 | 54 | |||||
| 122 | 62 | 65 | 33 | 35 | 80 | 66 | |||||
| Imipenem | 100 | 100 | 100 | 100 | 57 | 65 | |||||
| Miconazole | |||||||||||
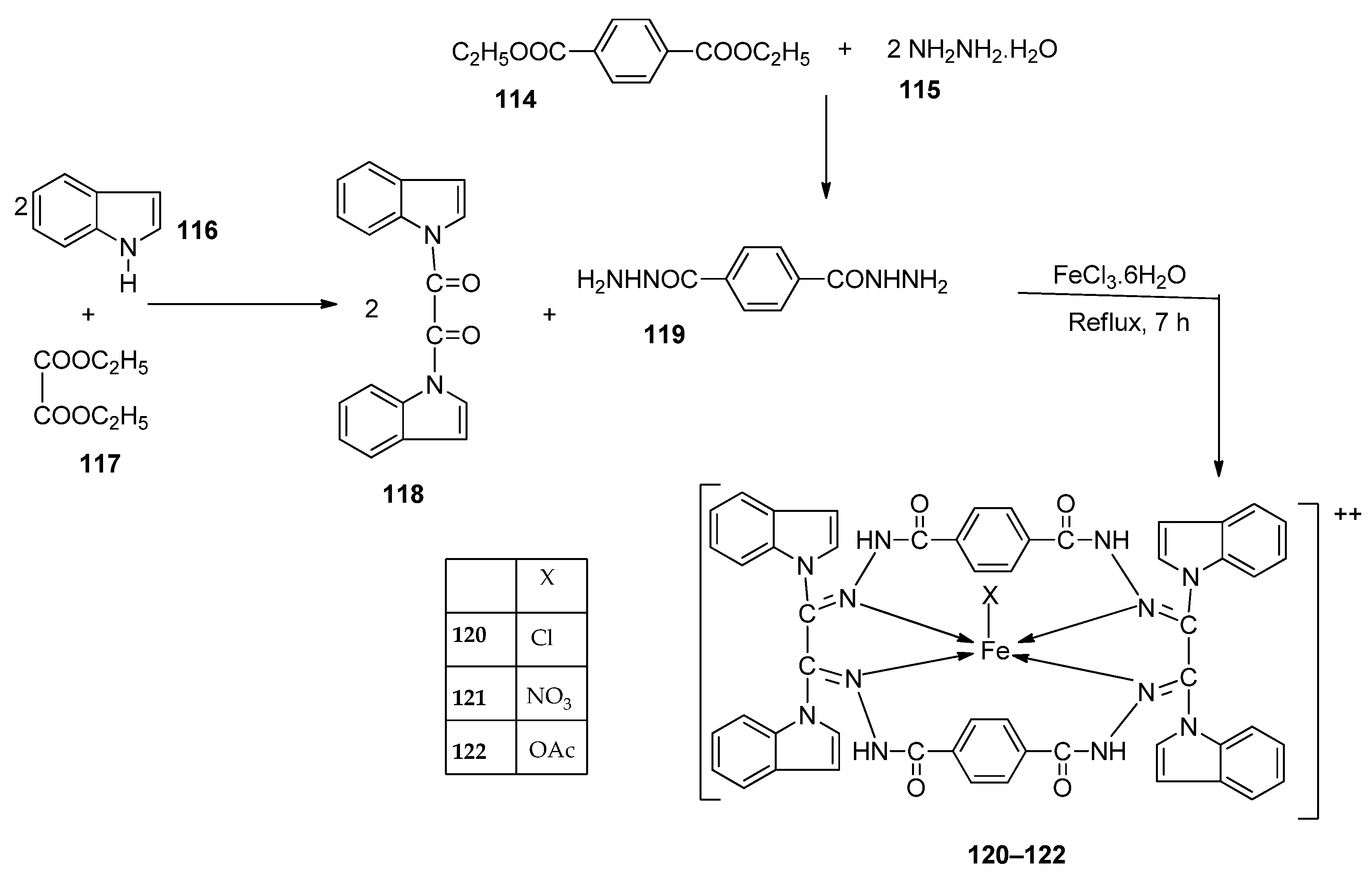
| 24. | 123 | 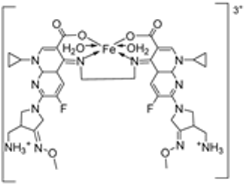 | Reflux, 3 h | Zone of inhibition, mm | [35] | ||||||
| X. campestris | B. megaterium | C. michiganensis | M. fructicola | P. digitatum | |||||||
| Ligand | 30 | 28 | 32 | 36.0 ± 3.1 | 28.0±3.5 | ||||||
| 123 | 26 | 19 | 20 | 62.5 ± 6.2 | 62.5 ± 8.2 | ||||||
| Tetracycline | 34 | 28 | 30 | ||||||||
| Azoxystrobin | 45.3 ± 2.1 | 58.1 ± 1.2 | |||||||||
| 25. | 124 |  | Reflux, 3 h | Zone of inhibition, mm (concentration, µg/mL) | [69] | ||||||
| E. coli | B. cereus | P. fluorescens | B. cinerea | A. flavus | |||||||
| Ligand | 20 | 12 | 11 | 0.0 ± 0.0 | 0.00 ±0.0 | ||||||
| 124 | 12 | 12 | 18 | 6.7 ± 2.3 | 6.7±2.6 | ||||||
| Tetracycline | 14 | 10 | 8 | ||||||||
| Cycloheximide | 42.2±2.6 | 9.7±3.0 | |||||||||
| 26. | 30 | 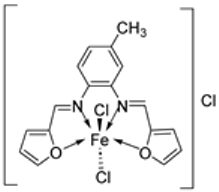 | Stir and reflux, 4 h | Zone of inhibition, mm | [29] | ||||||
| S. aureus | B. subtilis | E. coli | C. albicans | ||||||||
| Ligand | 19 | 25 | 24 | 25 | |||||||
| 30 | 17 | 16 | 19 | 15 | |||||||
| Gentamycin | 24 | 26 | 30 | ||||||||
| Ketoconazole | 20 | ||||||||||
| 27. | 125 | 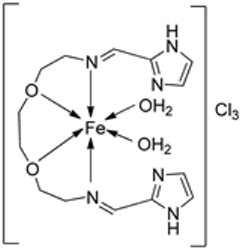 | Stirring and reflux, 1 h | Zone of inhibition, mm | [72] | ||||||
| S. aureus | E. coli | ||||||||||
| Ligand | 0.00 | 9 | |||||||||
| 125 | 10 | 10 | |||||||||
| Amikacin | 10 | 6 | |||||||||
| 28. | 38 39 40 | 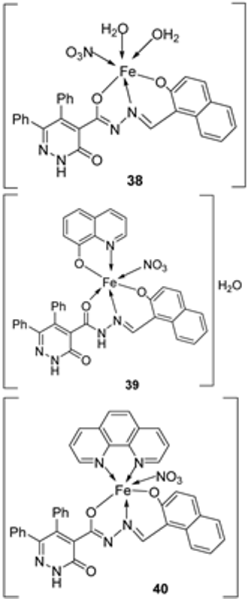 | Stirring, 2 h Reflux, 12–15 h | Zone of inhibition, mm | [31] | ||||||
| S. typhimurium | C. albicans | ||||||||||
| Ligand | ** R | 8 | |||||||||
| 38 | ** R | 14 | |||||||||
| 39 | 15 | 22 | |||||||||
| 40 | ** R | R | |||||||||
| Cephalothin | 36 | ||||||||||
| Cycloheximide | 35 | ||||||||||
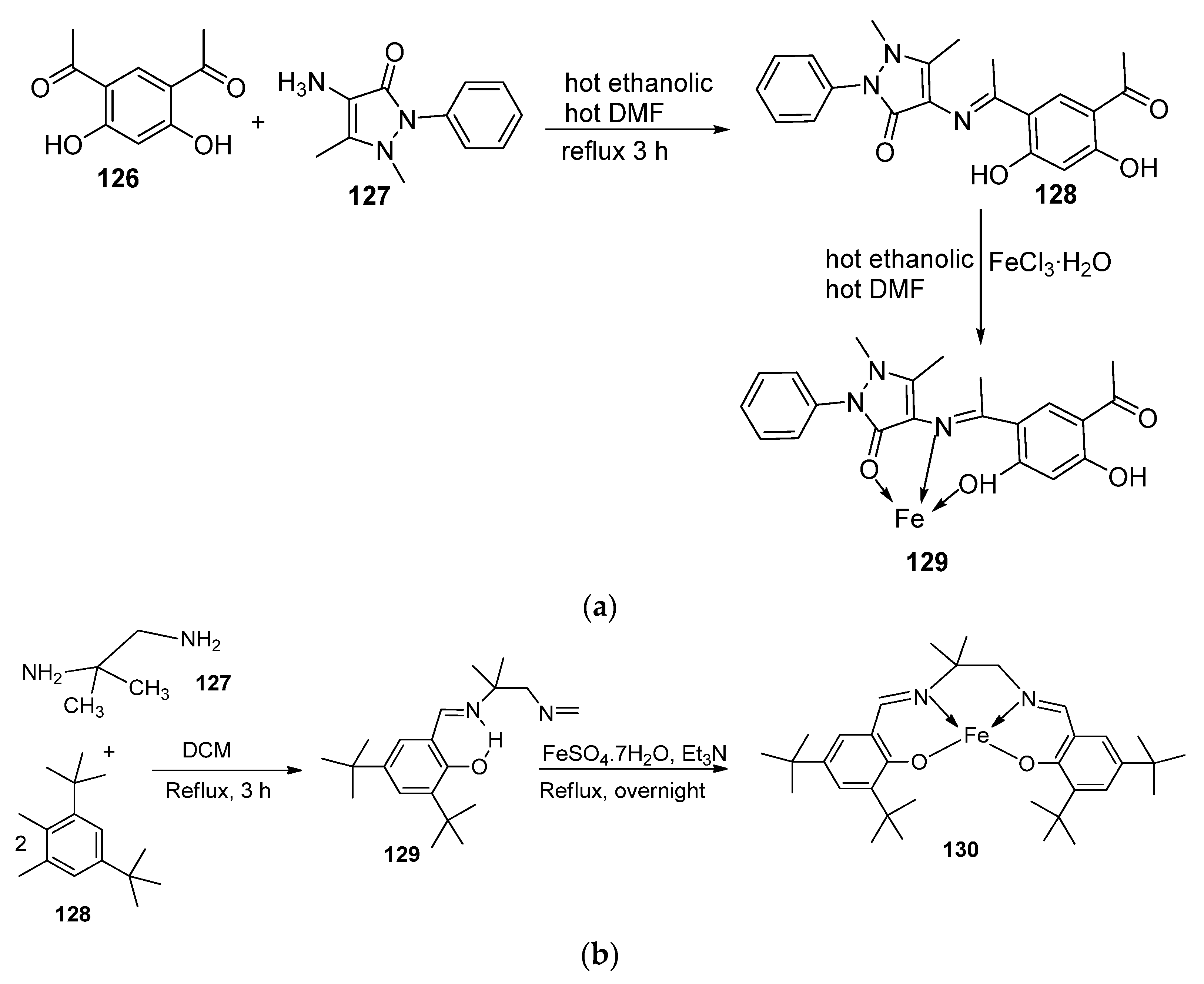
| 29. | 129 |  | Stirring and reflux, 1 h | Zone of inhibition, mm/mg | [75] | ||||||
| E. coli | S. aureus | C. albicans | A. flavus | ||||||||
| 128 | 14 | 12 | 10 | 0 | |||||||
| 129 | 13 | 11 | 12 | 14 | |||||||
| Amikacin | 6 | 10 | - | - | |||||||
| Ketoconazole | - | - | 9 | 8 | |||||||
2.3. Imine–Iron Complexes as Antioxidants
| Entry No. | Complex No. | Structures | Reaction Condition | Antioxidant Activity (IC50/µg/mL) | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 3 4 | 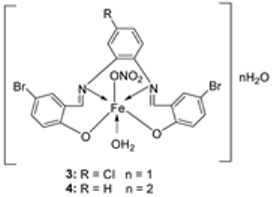 | Reflux (2 h) | DPPH | [21] | |||||
| 1 | 45 | |||||||||
| 3 | 22 | |||||||||
| 2 | 53 | |||||||||
| 4 | 32 | |||||||||
| Vit C | 55 | |||||||||
| 2. | 100 101 | 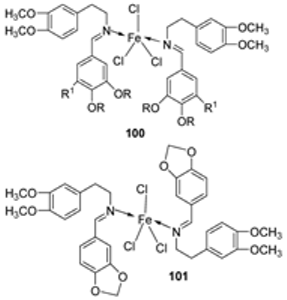 | Reflux and stirring (50 min) | DPPH | [59] | |||||
| 98 | 1.23 | |||||||||
| 100 | 1.70 | |||||||||
| 99 | 1.02 | |||||||||
| 101 | 1.41 | |||||||||
| Vit C | 1.14 | |||||||||
| 3. | 126 | 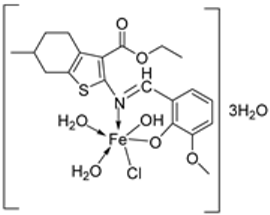 | Reflux (10 min) and stirring (24 h) | ABTS (734 nm) | DPPH (517 nm) | FRAP (700 nm) | [81] | |||
| Ligand | 1.90 | 1.35 | 0.50 | |||||||
| 126 | 0.60 | 1.25 | 0.40 | |||||||
| Ascorbic acid | 0.00 | 0.10 | 2.10 | |||||||
| BHA | 0.00 | 0.18 | 2.90 | |||||||
| BHT | 0.00 | 0.31 | 2.30 | |||||||
| 4. | 130 | 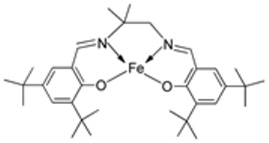 | Reflux and stir (overnight) | DPPH | H2O2 SA | FTC | HRSA | [82] | ||
| 129 | 53.55 ± 2.95 | 92.52 ± 0.07 | 48.81 ± 5.04 | 63.43 ± 5.66 | ||||||
| 130 | 44.65 ± 1.10 | 93.74 ± 0.43 | 9.47 ± 2.191 | 30.29 ± 0.81 | ||||||
| Trolox | 85.42 ± 0.04 | 91.80 ± 1.77 | 90.45 ± 6.70 | 57.72 ± 1.62 | ||||||
| BHA | 75.69 ± 0.11 | 92.97 ± 0.98 | 50.57 ± 5.42 | 10.00 ± 3.64 | ||||||
| 5. | 136 | 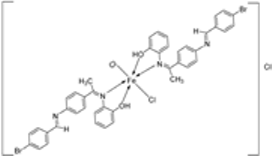 | Reflux (3 h) | DPPH (% scavenging) | [16] | |||||
| 135 | 24 | |||||||||
| 136 | 49 | |||||||||
| Ascorbic acid | 82 | |||||||||
| 6. | 124 |  | Reflux (3 h) | %RSA | [69] | |||||
| Ligand | 169.7 | |||||||||
| 124 | 164.6 | |||||||||
| 7. | 109 | 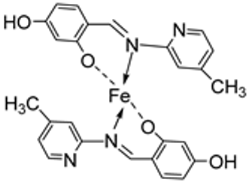 | Reflux (4–5 h) | DPPH | [61] | |||||
| 109 | 1615.22 | |||||||||
| 8. | 113 | 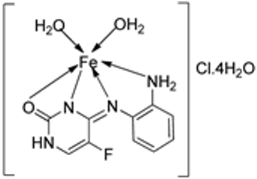 | Reflux (6 h) | DPPH | Total antioxidant | FRAP | CUPRAC | [64] | ||
| 111 | 1.9 | 0.64 | 0.06 | 0.30 | ||||||
| 112 | 0.8 | 0.62 | 0.38 | 3.50 | ||||||
| 113 | 0.70 | 0.61 | 0.11 | 1.20 | ||||||
| BHT | 0.60 | 0.60 | 0.08 | 3.20 | ||||||
| BHA | 1.20 | 0.44 | 0.20 | 3.10 | ||||||
2.4. Other Pharmacological Activities of the Imine–Iron Complexes
| Entry No. | Complex No. | Structures | Reaction Conditions | Biological Activities | Ref | |
|---|---|---|---|---|---|---|
| 1. | 120 121 122 |  | Reflux, 8 h | Anti-inflammatory activity (%) | [66] | |
| 119 | 9.00 | |||||
| 120 | 28.50 | |||||
| 121 | 27.20 | |||||
| 122 | 31.10 | |||||
| Phenyl butazone | 18.20 | |||||
| 2. | 125 | 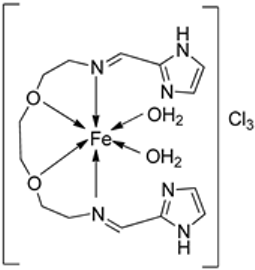 | Stirring and reflux, 1 h | Binding energy (kcal/mol) | [72] | |
| Ligand | −2.1 | |||||
| 125 | −8.5 | |||||
| 3. | 139 | 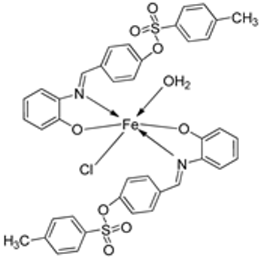 | Reflux, 12 h | % Inhibition of heat-induced denaturation of proteins | [84] | |
| 138 | 0.13 | |||||
| 139 | 0.70 | |||||
| Ibuprofen | 2.9 | |||||
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vija, K.J.; Abhishek, P.; Mohit, P.; Shweta, C.T.; Om, P.; Akansha, A.; Viveka, N. Schiff base metal complexes as a versatile catalyst: A review. J. Organomet. Chem. 2023, 999, 122825. [Google Scholar] [CrossRef]
- Manvatkar, V.D.; Patle, R.Y.; Meshram, P.H.; Dongre, R.S. Azomethine-functionalized organic–inorganic framework: An overview. Chem. Pap. 2023, 77, 5641–5662. [Google Scholar] [CrossRef]
- Ying, K.L.; Mohand, M.; Dominik, M.; Guy, B. An Air-Stable “Masked” Bis (imino) carbene: A Carbon-Based Dual Ambiphile. J. Am. Chem. Soc. 2023, 145, 2064–2069. [Google Scholar] [CrossRef]
- Chérifa, B.; Hana, F.; Amel, D.; Amel, D.; Abdesalem, K.; Abir, B.; Ahmad, S.D.; Tarek, R.; Rajesh, V.; Yacine, B. Schiff bases and their metal Complexes: A review on the history, synthesis, and applications. Inorg. Chem. Commun. 2023, 150, 110451. [Google Scholar] [CrossRef]
- El-Attar, M.S.; Elshafie, H.S.; Sadeek, S.A.; El-Farargy, A.F.; El-Desoky, S.I.; El-Shwiniy, W.H.; Camele, I. Biochemical Characterization and Antimicrobial Activity against Some Human or Phyto-Pathogens of New Diazonium Heterocyclic Metal Complexes. Chem. Biodivers. 2022, 19, 2. [Google Scholar] [CrossRef]
- Anjali Krishna, G.T.; Dhanya, M.; Shanty, A.A.; Raghu, K.G.; Mohanan, P.V. Transition metal complexes of imidazole derived Schiff bases: Antioxidant/anti-inflammatory/antimicrobial/enzyme inhibition and cytotoxicity properties. J. Mol. Struct. 2023, 1274, 134384. [Google Scholar] [CrossRef]
- Zhang, Z.; Song, Q.; Jin, Y.; Feng, Y.; Li, J.; Zhang, K. Advances in Schiff Base and Its Coating on Metal Biomaterials—A Review. Metals 2023, 13, 386. [Google Scholar] [CrossRef]
- Sohtun, W.P.; Khamrang, T.; Kannan, A.; Balakrishnan, G.; Saravanan, D.; Akhbarsha, M.A.; Velusamy, M.; Palaniandavar, M. Iron(III) bis-complexes of Schiff bases of S -methyldithiocarbazates: Synthesis, structure, spectral and redox properties and cytotoxicity. Appl. Organomet. Chem. 2020, 34, e5593. [Google Scholar] [CrossRef]
- Tukki, S.; Samya, B.; Akhtar, H. Significant photocytotoxic effect of an iron (iii) complex of a Schiff base ligand derived from vitamin B6 and thiosemicarbazide in visible light. RSC Adv. 2015, 5, 29276–29284. [Google Scholar] [CrossRef]
- Dubey, R.K.; Mariya, A.; Mishra, S.K. Synthesis and spectral (ir, nmr, fab-ms and xrd) characterization of lanthanide complexes containing bidentate schiff base derived from sulphadiazine and ovanillin. Int. J. Basic Appl. Sci. 2011, 1, 70–78. [Google Scholar]
- Basu, U.; Roy, M.; Chakravarty, A.R. Recent advances in the chemistry of iron-based chemotherapeutic agents. Coord. Chem. Rev. 2020, 417, 213339. [Google Scholar] [CrossRef]
- Satya, D.P.; Logesh, R.; Dhanabal, P.; Suresh, M.K. Importance of Iron Absorption in Human Health: An Overview. Curr. Nutr. Food Sci. 2021, 17, 293–301. [Google Scholar]
- Kargar, H.; Fallah-Mehrjardi, M.; Behjatmanesh-Ardakani, R.; Munawar, K.S.; Ashfaq, M.; Tahir, M.N. Diverse coordination of isoniazid hydrazone Schiff base ligand towards iron (III): Synthesis, characterization, SC-XRD, HSA, QTAIM, MEP, NCI, NBO and DFT study. J. Mol. Struct. 2022, 1250, 131691. [Google Scholar] [CrossRef]
- Lee, H.W.; Jeong, G.-U.; Kim, M.-C.; Kim, D.; Kim, S.; Han, S.S. Atomistic origin of mechanochemical NH3 synthesis on Fe catalysts. International Journal of Hydrogen Energy. Int. J. Hydrogen Energy 2023, 48, 3931–3941. [Google Scholar] [CrossRef]
- Peterson, P.O.; Joannou, M.V.; Simmons, E.M.; Wisniewski, S.R.; Kim, J.; Chirik, P.J. Iron-Catalyzed C(sp2)-C(sp3) Suzuki-Miyaura Cross-Coupling Using an Alkoxide Base. ACS Catal. 2023, 13, 2443–2448. [Google Scholar] [CrossRef]
- Hayder, M.; Hayder, A.M. In vitro antioxidant activity of new Schiff base ligand and its metal ion complexes. J. Pharm. Sci. Res. 2019, 11, 2051–2061. [Google Scholar]
- Yahaya, N.P.; Mukhtar, M.S. Synthesis, Characterization and Antibacterial Activity of Mixed Ligands of Schiff Base and Its Metal (II) Complexes Derived from Ampicilin, 3-Aminophenol and Benzaldehyde. Sci. J. Chem. 2021, 9, 9–13. [Google Scholar] [CrossRef]
- Antolovich, M.; Prenzler, P.D.; Patsalides, E.; McDonald, S.; Robards, K. Methods for testing antioxidant activity. Anal. R. Soc. Chem. 2002, 27, 183–198. [Google Scholar] [CrossRef]
- Bharti, S.; Singh, S. Metal Based Drugs: Current Use and Future Potential. Der Pharm. Lett. 2009, 1, 39–51. [Google Scholar]
- Li, Y.; Qian, C.; Li, Y.; Yang, Y.; Lin, D.; Liu, X.; Chen, C. Syntheses, crystal structures of two Fe(III) Schiff base complexes with chelating o-vanillin aroylhydrazone and exploration of their bio-relevant activities. J. Inorg. Biochem. 2021, 218, 111405. [Google Scholar] [CrossRef]
- El-Lateef, H.M.A.; Khalaf, M.M.; Shehata, M.R.; Abu-Dief, A.M. Fabrication, DFT Calculation, and Molecular Docking of Two Fe(III) Imine Chelates as Anti-COVID-19 and Pharmaceutical Drug Candidate. Int. J. Mol. Sci. 2022, 23, 3994. [Google Scholar] [CrossRef] [PubMed]
- Bednarski, P.; Nguyen, Q.T.; Pham, T.P.N.; Nguyen, V.T. Synthesis, Characterization, and In Vitro Cytotoxicity of Unsymmetrical Tetradentate Schiff Base Cu(II) and Fe(III) Complexes. Bioinorg. Chem. Appl. 2021, 2021, 6696344. [Google Scholar]
- Zhao, W.L. Targeted therapy in T-cell malignancies: Dysregulation of the cellular signaling pathways. Leukemia 2010, 24, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.S.; Lotfi, M.; Soltani, N.; Farmani, E.; Fernandez, J.H.O.; Akhlaghitehrani, S.; Mohammed, S.H.; Yasamineh, S.; Kalajahi, H.G.; Gholizadeh, O. Recent advances on high-efficiency of microRNAs in different types of lung cancer: A comprehensive review. Cancer Cell Int. 2023, 23, 284. [Google Scholar] [CrossRef] [PubMed]
- Gujrati, H.; Ha, S.; Wang, B.D. Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms. Cancers 2023, 15, 3140. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.X.; Zheng, Z.; Xue, W.; Zhao, M.Z.; Fei, X.C.; Wu, L.L.; Huang, L.M.; Leboeuf, C.; Janin, A.; Wang, L.; et al. Is Overexpressed in T-Cell Leukemia/Lymphoma and Related to Chemoresistance. BioMed Res. Int. 2015, 2015, 197241. [Google Scholar] [CrossRef] [PubMed]
- Kalındemirtaş, F.D.; Kaya, B.; Bener, M.; Şahin, O.; Kuruca, S.; Demirci, T.B.; Ülküseven, B. Iron(III) complexes based on tetradentate thiosemicarbazones: Synthesis, characterization, radical scavenging activity and in vitro cytotoxicity on K562, P3HR1 and JURKAT cells. Appl. Organomet. Chem. 2021, 35, e6157. [Google Scholar] [CrossRef]
- Wongsuwan, S.; Chatwichien, J.; Pinchaipat, B. Synthesis, characterization and anticancer activity of Fe(II) and Fe(III) complexes containing N-(8-quinolyl)salicylaldimine Schiff base ligands. J. Biol. Inorg. Chem. 2021, 26, 327–339. [Google Scholar] [CrossRef]
- Ismail, B.A.; Nassar, D.A.; El–Wahab Abd, Z.H.; Ali, A.M.O. Synthesis, characterization, thermal, DFT computational studies and anticancer activity of furfural-type schiff base complexes. J. Mol. Struct. 2021, 1227, 129393. [Google Scholar] [CrossRef]
- Kavitha, B.; Sravanthi, M.; Reddy, P.S. Studies on DNA binding, cleavage, molecular docking, antimicrobial and anticancer activities of Cr(III), Fe(III), Co(II) and Cu(II) complexes of o-vanillin and fluorobenzamine Schiff base ligand. Appl. Organomet. Chem. 2022, 36, e6531. [Google Scholar] [CrossRef]
- Abdelrahman, M.S.A.; Omar, F.M.; Saleh, A.A.; El-ghamry, M.A. Synthesis, molecular modeling, and docking studies of a new pyridazinone-acid hydrazone ligand, and its nano metal complexes. Spectroscopy, thermal analysis, electrical properties, DNA cleavage, antitumor, and antimicrobial activities. J. Mol. Struct. 2022, 1251, 131947. [Google Scholar] [CrossRef]
- Farhan, L.K.; Awad, M.A.; Kshash, A.H. Synthesis, Characterization and Evaluation Anti-cancer Activity of Fe(III), Co(II), Ni(II) and Cu(II) Complexes Derived from Heterocyclic Schiff bases Ligands. J. Pharm. Sci. Res. 2019, 11, 1577–1581. [Google Scholar]
- Claudel, M.; Schwarte, J.V.; Fromm, K.M. New Antimicrobial Strategies Based on Metal Complexes. Chemistry 2020, 2, 849–899. [Google Scholar] [CrossRef]
- Angelo, F.; Alysha, G.E.; Alex, K.; Hue, D.; Stefan, B.; Alice, E.B.; Mitchell, R.B.; Feng, C.; Dhirgam, H.; Nicole Jung, A.P.K.; et al. Metal complexes as antifungals? From a crowd-sourced compound library to the first in vivo experiments. JACS Au 2022, 2, 2277–2294. [Google Scholar] [CrossRef]
- Mohamed, A.A.; Elshafie, H.S.; Sadeek, S.A.; Camele, I. Biochemical Characterization, Phytotoxic Effect and Antimicrobial Activity against Some Phytopathogens of New Gemifloxacin Schiff Base Metal Complexes. Chem. Biodivers. 2021, 18, 9. [Google Scholar] [CrossRef]
- Raman, S.R.N.; Johnson, A.S. Transition metal complexes with Schiff-base ligands: 4-aminoantipyrine based derivatives—A review. J. Coord. Chem. 2009, 62, 691–709. [Google Scholar] [CrossRef]
- Chohan, Z.H.; Shaikh, A.U.; Naseer, M.M.; Supran, C.T. In-vitro antibacterial, antifungal and cytotoxic properties of metal-based furanyl derived sulfonamides. J. Enzym. Inhib. Med. Chem. 2006, 21, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Chohan, Z.H.; Arif, M.; Akhtar, M.A.; Supuran, C.T. Metal-Based Antibacterial and Antifungal Agents: Synthesis, Characterization, and In Vitro Biological Evaluation of Co(II), Cu(II), Ni(II), and Zn(II) Complexes with Amino Acid-Derived Compounds. Bioinorg. Chem. Appl. 2006, 2006, 83131. [Google Scholar] [CrossRef]
- Tsacheva, I.; Todorova, Z.; Momekova, D.; Momekov, G.; Koseva, N. Pharmacological Activities of Schiff Bases and Their Derivatives with Low and High Molecular Phosphonates. Pharmaceuticals 2023, 16, 938. [Google Scholar] [CrossRef]
- Rahmatabadi, F.D.; Khojasteh, R.R.; Fard, H.K. New Cr, Mo, W, and Fe Metal Complexes with Potentially Heptadentate (S3N4) Tripodal Schiff Base Ligand: Synthesis, Characterization, and Antibacterial Activity. Russ. J. Gen. Chem. 2020, 90, 1317–1321. [Google Scholar] [CrossRef]
- Parekh, J.; Inamdhar, P.; Nair, R.; Baluja, S.; Chanda, S. Synthesis and antibacterial activity of some Schiff bases derived from 4-aminobenzoic acid. J. Serbian Chem. Soc. 2005, 70, 1155–1162. [Google Scholar] [CrossRef]
- Shukla, S.N.; Gaur, P.; Vaidya, P.; Chaurasia, B.; Jhariya, S. Biomimetic complexes of Mn(II), Fe(III), Co(II), and Ni(II) with 1,10-phenanthroline and a salen type ligand: Tailored synthesis, characterization, DFT, enzyme kinetics, and antibacterial screening. J. Coord. Chem. 2018, 71, 3912–3933. [Google Scholar] [CrossRef]
- Gehad, G.M.; Carmen, M.S. Metal complexes of Schiff base derived from sulphametrole and o-vanilin: Synthesis, spectral, thermal characterization and biological activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2007, 66, 949–958. [Google Scholar] [CrossRef]
- Gowda, K.R.S.; Naik, H.S.B.; Kumar, B.V.; Sudhamani, C.N.; Sudeep, H.V.; Naik, T.R.R.; Krishnamurthy, G. Synthesis, antimicrobial, DNA-binding and photonuclease studies of Cobalt(III) and Nickel(II) Schiff base complexes. Spectrochim. Acta Part A 2013, 105, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Abu-Dief, A.M.; Nassr, L.A.M.E. Tailoring, physicochemical characterization, anti-bacterial and DNA binding mode studies of Cu (II) Schiff bases amino acid bioactive agents incorporating 5-bromo-2-hydroxybenzaldehyde. J. Iran. Chem. Soc. 2015, 12, 943–955. [Google Scholar] [CrossRef]
- Tweedy, B.G. Plant extracts with metal ions as potential antimicrobial agents. Phytopathology 1964, 55, 910–914. [Google Scholar]
- Karem, L.K.A.; Al-Noor, T.H. Mixed Ligand Complexes of Schiff Base and Nicotinamide: Synthesis, Characterization and Antimicrobial Activities. J. Phys. Conf. 2020, 1660, 012094. [Google Scholar] [CrossRef]
- Shukla, S.N.; Gaur, P.; Raidas, M.L.; Chaurasia, B.; Bagri, S.S. Novel NNO pincer type Schiff base ligand and its complexes of Fe(IIl), Co(II) and Ni(II): Synthesis, spectroscopic characterization, DFT, antibacterial and anticorrosion study. J. Mol. Struct. 2021, 1240, 130582. [Google Scholar] [CrossRef]
- Ibrahim, M.; Khan, A.; Ikram, M.; Rehman, S.; Shah, M.; Un Nabi, H.; Ahuchaogu, A.A. Ahuchaogu, in vitro Antioxidant Properties of Novel Schiff Base Complexes. Asian J. Chem. Sci. 2017, 2, 1–12. [Google Scholar] [CrossRef]
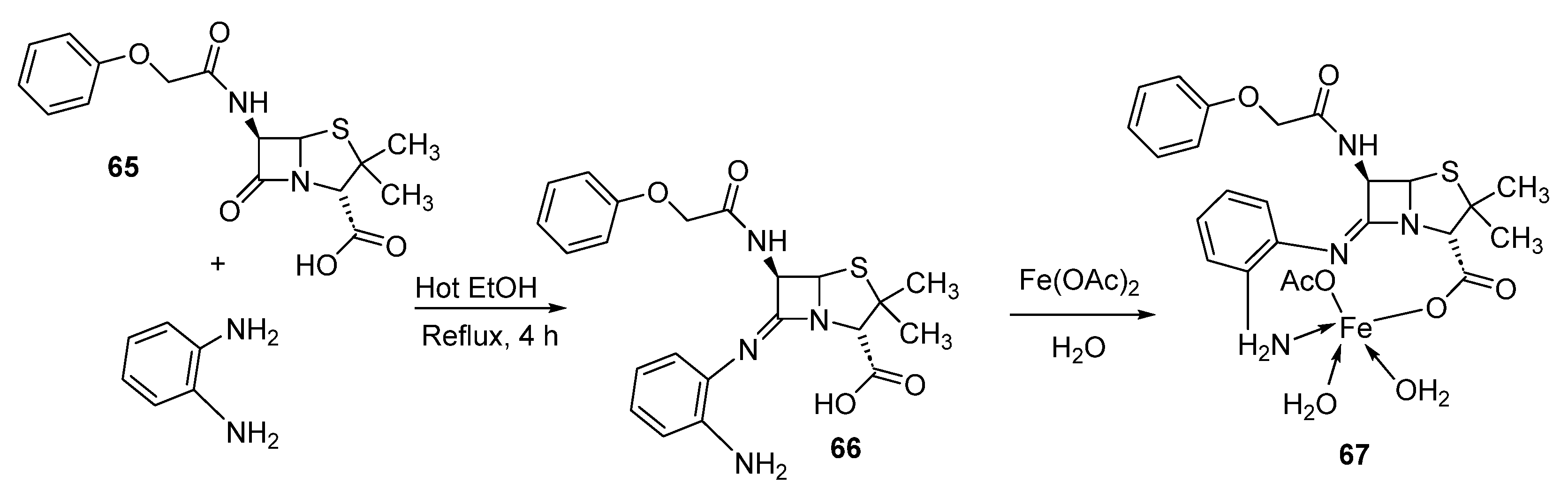
- Anacona, J.R.; Ruiz, K.; Loroño, M.; Celis, F. Antibacterial activity of transition metal complexes containing a tridentate NNO phenoxymethylpenicillin-based Schiff base. An anti-MRSA iron (II) complex. Appl. Organomet. Chem. 2019, 33, e4744. [Google Scholar] [CrossRef]
- Mumtaz, A.; Mahmud, T.; Elsegood, M.; Weaver, G.W. Synthesis, Characterization and in vitro Biological Evaluation of a New Schiff Base Derived from Drug and its Complexes with Transition Metal Ions. Rev. Chim. 2019, 69, 1678–1681. [Google Scholar] [CrossRef]
- Al-Wasidi, A.S.; Naglah, A.M.; Al-Omar, M.A.; Al-Obaid, A.-R.M.; Alosaimi, E.H.; El-Metwaly, N.M.; Refat, M.S.; Ahmed, A.S.; El-Deen, I.M.; Soliman, A.H.; et al. Manganese (II), ferric (III), cobalt (II) and copper (II) thiosemicarbazone Schiff base complexes: Synthesis, spectroscopic, molecular docking and biological discussions. Mater. Express 2020, 10, 290–300. [Google Scholar] [CrossRef]
- Kumar, K.S.; Aravindakshan, K. Synthesis, Characterization Antimicrobial and Antioxidant Studies of Complexes of Fe (III), Ni (II) and Cu (II) with Novel Schiff Base Ligand (E)-Ethyl 3-((2-Aminoethyl) Imino) Butanoate. J. Pharm. Chem. Biol. Sci. 2017, 5, 177–186. [Google Scholar]
- Mukhtar, H.; Sani, U.M.; Shettima, U.A. Synthesis, Physico-chemical and Antimicrobial Studies on Metal (II) Complexes with Schiff Base Derived from Salicylaldehyde and 2,4-Dinitrophenylhydrazine. Int. Res. J. Pure Appl. Chem. 2019, 19, 1–8. [Google Scholar] [CrossRef]
- Knittl, E.T.; Abou-Hussein, A.A.; Linert, W. Syntheses, characterization, and biological activity of novel mono- and binuclear transition metal complexes with a hydrazone Schiff base derived from a coumarin derivative and oxalyldihydrazine. Monatshefte Für Chem.-Chem. Mon. 2018, 149, 431–443. [Google Scholar] [CrossRef]
- Abdullah, A.A.A. Biomedical applications of selective metal complexes of indole, benzimidazole, benzothiazole and benzoxazole: A review (From 2015 to 2022). Saudi Pharm J. 2023, 31, 101698. [Google Scholar] [CrossRef]
- Alosaimi, A.M.; Mannoubi, I.E.l.; Zabin, S.A. In Vitro Antimicrobial and In Vivo Molluscicidal Potentialities of Fe(III), Co(II) and Ni(II) Complexes Incorporating Symmetrical Tetradentate Schiff Bases (N2O2). Orient. J. Chem. 2020, 36, 373. [Google Scholar] [CrossRef]
- Priteshkumar, M.T.; Rajesh, J.P.; Ranjan, K.G.; Sunil, H.C.; Ankurkumar, J.K.; Yati, H.V.; Parth, T.; Anjali, B.T.; Jatin, D.P. Synthesis, Spectral Characterization, Thermal Investigation, Computational Studies, Molecular Docking, and In Vitro Biological Activities of a New Schiff Base Derived from 2-Chloro Benzaldehyde and 3,3′-Dimethyl-[1,1′-biphenyl]-4,4′-diamine. ACS Omega 2023, 8, 33069–33082. [Google Scholar] [CrossRef]
- Naureen, B.; Miana, G.A.; Shahid, K.; Asghar, M.; Tanveer, S.; Sarwar, A. Iron (III) and zinc (II) monodentate Schiff base metal complexes: Synthesis, characterisation and biological activities. J. Mol. Struct. 2021, 1231, 129946. [Google Scholar] [CrossRef]
- Singh, N.P.; Kumar, K.; Kumar, A.; Agarwal, U. Synthesis, characterization and antimicrobial activity of Mn(II),Fe(II), Ni(II),Co(II) and Zn(II) complexes of schiff base derived from 2,2-Dimethylpropane 1, 3-Diamine and 5-Chloro isatin. Rasayan J. Chem. 2020, 13, 215–221. [Google Scholar] [CrossRef]
- Borase, J.N.; Mahale, R.G.; Rajput, S.S.; Shirsath, D.S. Design, synthesis and biological evaluation of heterocyclic methyl substituted pyridine Schiff base transition metal complexes. SN Appl. Sci. 2021, 3, 197. [Google Scholar] [CrossRef]
- Deshmukh, S.Y.; Padole, N.S.; Wadekar, M.P.; Chaudhari, M.A. Synthesis, Spectroscopic and Antimicrobial Studies of Cu (II) and Fe (III) Complexes of Heterocyclic Schiff Base Ligand. J. Chem. Pharm. Res. 2021, 13, 1–5. [Google Scholar]
- Sutha, S.; Perumal, S.; Liviu, M.; Jeyaprakash, D.; Sundaram, A.N. Synthesis, structural elucidation, biological, antioxidant and nuclease activities of some 5-Fluorouracil–amino acid mixed ligand complexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 134, 333–344. [Google Scholar] [CrossRef]
- Savcı, A.; Buldurun, K.; Kırkpantur, G. A new Schiff base containing 5-FU and its metal Complexes: Synthesis, Characterization, and biological activities. Inorg. Chem. Commun. 2021, 134, 109060. [Google Scholar] [CrossRef]
- Anacona, J.R.; Rodriguez, H. Metalloantibiotics: Synthesis and antibacterial activity of cefepime metal complexes. J. Coord. Chem. 2009, 62, 2212–2219. [Google Scholar] [CrossRef]
- Kumar, G.; Devi, S.; Kumar, D. Synthesis of Schiff base 24-membered trivalent transition metal derivatives with their anti-inflammation and antimicrobial evaluation. J. Mol. Struct. 2016, 1108, 680–688. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Sadeek, S.A.; Camele, I.; Mohamed, A.A. Biological and Spectroscopic Investigations of New Tenoxicam and 1. 10-Phenthroline Metal Complexes. Molecules 2020, 25, 1027. [Google Scholar] [CrossRef] [PubMed]
- Sakr, S.H.; Elshafie, H.S.; Camele, I.; Sadeek, S.A. Synthesis, spectroscopic, and biological studies of mixed ligand complexes of emifloxacin and glycine with Zn (II), Sn (II), and Ce (III). Molecules 2018, 23, 1182. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Sadeek, S.A.; Camele, I.; Mohamed, A.A. Biochemical Characterization of New Gemifloxacin Schiff Base (GMFX-o-phdn) Metal Complexes and Evaluation of Their Antimicrobial Activity against Some Phyto- or Human Pathogens. Int. J. Mol. Sci. 2022, 23, 2110. [Google Scholar] [CrossRef]
- Heaton, V.J.; Ambler, J.E.; Fisher, L.M. Potent Antipneumococcal Activity of Gemifloxacin Is Associated with Dual Targeting of Gyrase and Topoisomerase IV, an In Vivo Target Preference for Gyrase, and Enhanced Stabilization of Cleavable Complexes In Vitro. Antimicrob. Agents Chemother. 2000, 44, 3112–3117. [Google Scholar] [CrossRef][Green Version]
- Sultana, N.; Naz, A.; Arayne, M.S.; Mesaik, M.A. Synthesis, characterization, antibacterial, antifungal and immunomodulating activities of gatifloxacin–metal complexes. J. Mol. Struct. 2010, 969, 17–24. [Google Scholar] [CrossRef]
- Ahmed, Y.M.; Omar, M.M.; Mohamed, G.G. Synthesis, spectroscopic characterization, and thermal studies of novel Schiff base complexes: Theoretical simulation studies on coronavirus (COVID-19) using molecular docking. J. Iran. Chem. Soc. 2022, 19, 901–919. [Google Scholar] [CrossRef]
- Sivaprakash, G.P.; Tharmaraj, M.; Jothibasu, A. Arun, antimicrobial analysis of schiff base ligands pyrazole and diketone metal complex against pathogenic organisms. Int. J. Adv. Res. 2017, 5, 2656–2663. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.A.; Dar, O.A.; Gull, P.; Wani, M.Y.; Hashmi, A.A. Heterocyclic Schiff base transition metal complexes in antimicrobial and anticancer chemotherapy. MedChemComm 2018, 9, 409. [Google Scholar] [CrossRef] [PubMed]
- Gehad, G.; Omar, M.M.; Yasmin, M.A. Metal complexes of Tridentate Schiff base: Synthesis, Characterization, Biological Activity and Molecular Docking Studies with COVID-19 Protein Receptor. Z. Anorg. Allg. Chem. 2021, 647, 2201–2218. [Google Scholar]
- Hidayati, N.F.; Purwaningrum, W. Synthesis and characterization schiff base and complexes with Copper (II) and Iron (II) and their application as antibacterial agents. J. Phys. Conf. Ser. 2019, 1282, 012074. [Google Scholar] [CrossRef]
- Kitouni, S.; Chafai, N.; Chafaa, S.; Houas, N.; Ghedjati, S.; Djenane, M. Antioxidant activity of new synthesized imine and its corresponding α-aminophosphonic acid: Experimental and theoretical evaluation. J. Mol. Struct. 2023, 1281, 135083. [Google Scholar] [CrossRef]
- El-Lateef, H.M.A.; El-Dabea, T.; Khalaf, M.M.; Abu-Dief, A.M. Recent Overview of Potent Antioxidant Activity of Coordination Compounds. Antioxidants 2023, 12, 213. [Google Scholar] [CrossRef]
- Nevin, T.; Memet, Ş. Synthesis and Spectral Studies of Novel Co(II), Ni(II), Cu(II), Cd(II), and Fe(II).Metal Complexes with N-[5′-Amino-2,2′-bis(1,3,4-thiadiazole)-5-yl]-2-hydroxybenzaldehyde Imine (HL). Spectrosc. Lett. 2009, 5, 258–267. [Google Scholar] [CrossRef]
- Ercan, B. Kinetic Properties of Peroxidase Enzyme from Chard (Beta vulgaris Subspecies cicla) Leaves. Int. J. Food Prop. 2013, 16, 1293–1303. [Google Scholar] [CrossRef]
- Turan, N.; Buldurun, K. Synthesis, characterization and antioxidant activity of Schiff base and its metal complexes with Fe(II), Mn(II), Zn(II), and Ru(II) ions: Catalytic activity of ruthenium(II) complex. Eur. J. Chem. 2018, 9, 22–29. [Google Scholar] [CrossRef]
- Said, M.A.; Al-unizi, A.; Al-Mamary, M.; Alzahrani, S.; Lentz, D. Easy coordinate geometry indexes, τ4 and τ5 and HSA study for unsymmetrical Pd(II), Fe(II), Zn(II), Mn(II), Cu(II) and VO(IV) complexes of a tetradentate ligand: Synthesis, characterization, properties, and antioxidant activities. Inorg. Chim. Acta 2020, 505, 119434. [Google Scholar] [CrossRef]
- Preeti, S.; Preeti, Y.; Kushneet, K.S.; Anurag, T.; Shilpika, B.M. Advancement in the synthesis of metal complexes with special emphasis on Schiff base ligands and their important biological aspects. Results Chem. 2024, 7, 101222. [Google Scholar] [CrossRef]
- Elkanzi, N.A.A.; Ali, A.M.; Hrichi1, H.; Abdou, A. New mononuclear Fe(III), Co(II), Ni(II), Cu(II), and Zn(II) complexes incorporating 4-{[(2 hydroxyphenyl) imino]methyl}phenyl-4-methylbenzenesulfonate (HL): Synthesis, characterization, theoretical, anti-inflammatory, and molecular docking investigation. Appl. Organomet. Chem. 2022, 36, e6665. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Targeting ferroptosis in pancreatic cancer: A double-edged sword. Trends Cancer 2021, 7, 891–901. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anane, J.; Owusu, E.; Rivera, G.; Bandyopadhyay, D. Iron–Imine Cocktail in Drug Development: A Contemporary Update. Int. J. Mol. Sci. 2024, 25, 2263. https://doi.org/10.3390/ijms25042263
Anane J, Owusu E, Rivera G, Bandyopadhyay D. Iron–Imine Cocktail in Drug Development: A Contemporary Update. International Journal of Molecular Sciences. 2024; 25(4):2263. https://doi.org/10.3390/ijms25042263
Chicago/Turabian StyleAnane, Judith, Esther Owusu, Gildardo Rivera, and Debasish Bandyopadhyay. 2024. "Iron–Imine Cocktail in Drug Development: A Contemporary Update" International Journal of Molecular Sciences 25, no. 4: 2263. https://doi.org/10.3390/ijms25042263
APA StyleAnane, J., Owusu, E., Rivera, G., & Bandyopadhyay, D. (2024). Iron–Imine Cocktail in Drug Development: A Contemporary Update. International Journal of Molecular Sciences, 25(4), 2263. https://doi.org/10.3390/ijms25042263








