ADAM33′s Role in Asthma Pathogenesis: An Overview
Abstract
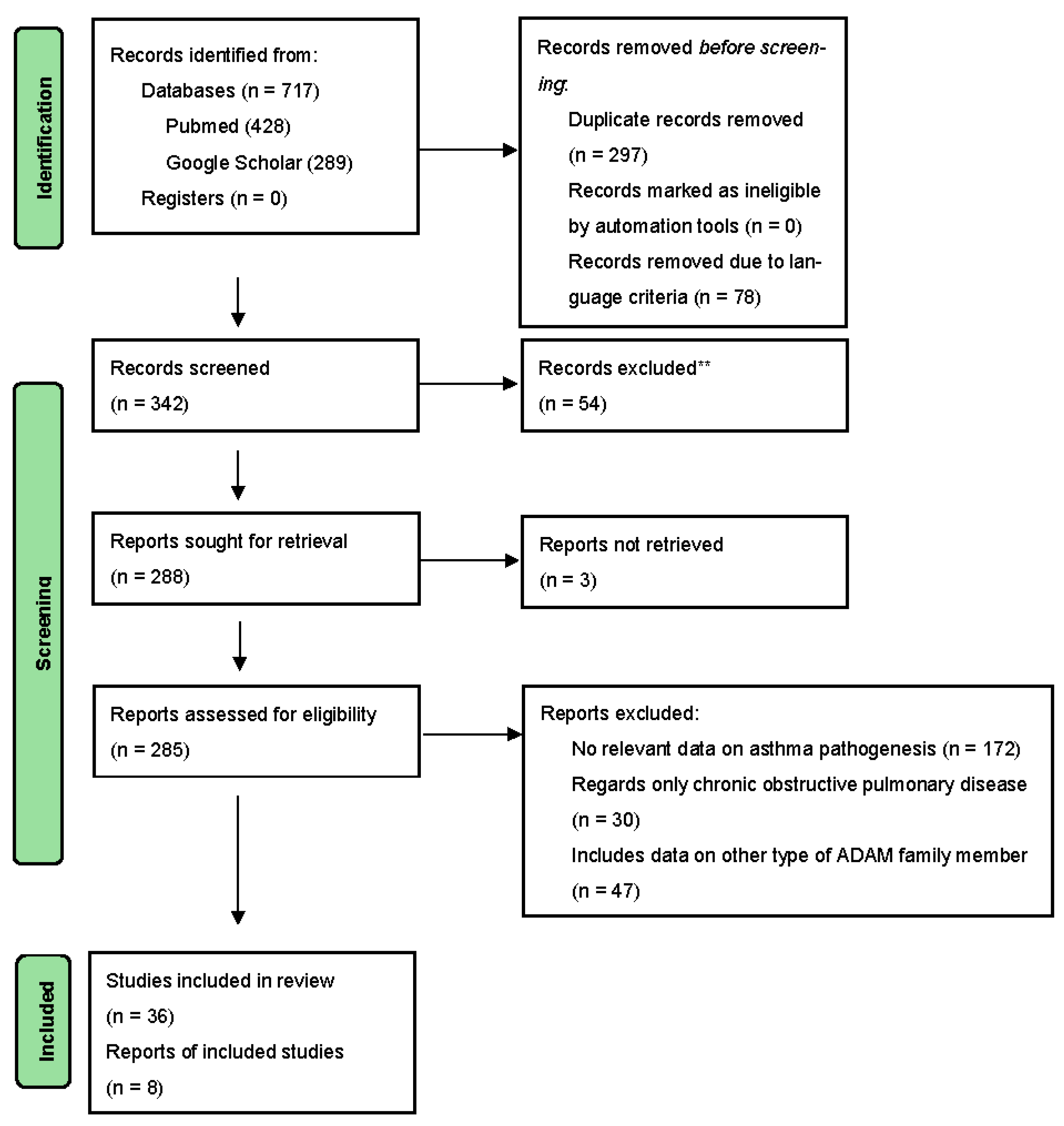
:1. Introduction
2. Methodology
2.1. ADAM Molecules in General
2.2. Structure
2.3. Function
2.4. SNPs
2.5. ADAM33 Haplotypes’ Association with Asthma
3. Environment
4. Proposed Pathogenesis
4.1. Conventional Treatment Responsiveness
4.2. Gene Therapy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Global Asthma Report 2022. Int. J. Tuberc. Lung Dis. 2022, 26, 1–104. [CrossRef]
- Ruan, Z.; Shi, Z.; Zhang, G.; Kou, J.; Ding, H. Asthma susceptible genes in children. Medicine 2020, 99, e23051. [Google Scholar] [CrossRef]
- Moheimani, F.; Hsu, A.C.-Y.; Reid, A.T.; Williams, T.; Kicic, A.; Stick, S.M.; eHansbro, P.M.; Wark, P.A.; Knight, D.A. The genetic and epigenetic landscapes of the epithelium in asthma. Respir Res. 2016, 17, 119. [Google Scholar] [CrossRef]
- Dreymueller, D.; Uhlig, S.; Ludwig, A. ADAM-family metalloproteinases in lung inflammation: Potential therapeutic targets. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L325–L343. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.R.; Kelly, J.F.C.; Howarth, P.H.; Wilson, D.I.; Holgate, S.T.; Davies, D.E.; Whitsett, J.A.; Haitchi, H.M. Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life. JCI Insight 2016, 1, e87632. [Google Scholar] [CrossRef]
- Human Gene ADAM33 (ENST00000356518.7) from GENCODE V43. Available online: https://genome.ucsc.edu/cgi-bin/hgGene?db=hg38&hgg_type=knownGene&hgg_gene=ADAM33 (accessed on 15 February 2023).
- Tripathi, P.; Awasthi, S.; Gao, P. ADAM Metallopeptidase Domain 33 (ADAM33): A Promising Target for Asthma. Mediat. Inflamm. 2014, 2014, 572025. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T.; Davies, D.E.; Powell, R.M.; Holloway, J.W. ADAM33: A newly identified protease involved in airway remodelling. Pulm. Pharmacol. Ther. 2006, 19, 3–11. [Google Scholar] [CrossRef]
- Li, H.F.; Yan, L.P.; Wang, K.; Li, X.T.; Liu, H.X.; Tan, W. Association between ADAM33 polymorphisms and asthma risk: A systematic review and meta-analysis. Respir. Res. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Howard, T.D.; Postma, D.S.; Jongepier, H.; Moore, W.C.; Koppelman, G.H.; Zheng, S.Q.; Xu, J.F.; Bleecker, E.R.; Meyers, D.A. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J. Allergy Clin. Immunol. 2003, 112, 717–722. [Google Scholar] [CrossRef]
- Holgate, S.T.; Davies, D.E.; Rorke, S.; Cakebread, J.; Murphy, G.; Powell, R.M.; Holloway, J.W. ADAM 33 and Its Association with Airway Remodeling and Hyperresponsiveness in Asthma. Clin. Rev. Allergy Immunol. 2004, 27, 023–034. [Google Scholar] [CrossRef]
- Holgate, S.T. Mechanisms of Asthma and Implications for Its Prevention and Treatment: A Personal Journey. Allergy Asthma Immunol. Res. 2013, 5, 343. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wang, L.; Su, X.; Hu, X.F. Association between V4 polymorphism in the ADAM33 gene and asthma risk: A meta-analysis. Genet. Mol. Res. 2015, 14, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Van Eerdewegh, P.; Little, R.D.; Dupuis, J.; Del Mastro, R.G.; Falls, K.; Simon, J.; Torrey, D.; Pandit, S.; McKenny, J.; Braunschweiger, K.; et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 2002, 418, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.M.; Wicks, J.; Holloway, J.W.; Holgate, S.T.; Davies, D.E. The Splicing and Fate of ADAM33 Transcripts in Primary Human Airways Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2004, 31, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, S.D.; Owen, C.A. ADAM-33 Surfaces as an Asthma Gene. N. Engl. J. Med. 2002, 347, 936–938. [Google Scholar] [CrossRef]
- Hur, G.Y.; Broide, D.H. Genes and Pathways Regulating Decline in Lung Function and Airway Remodeling in Asthma. Allergy Asthma Immunol Res. 2019, 11, 604. [Google Scholar] [CrossRef] [PubMed]
- Baurakiades, E.; Costa, V.H.; Raboni, S.M.; de Almeida, V.R.T.; Larsen, K.S.K.; Kohler, J.N.; Gozzo, P.D.C.; Klassetn, G.; Manica, G.C.; de Noronha, L. The roles of ADAM33, ADAM28, IL-13 and IL-4 in the development of lung injuries in children with lethal non-pandemic acute infectious pneumonia. J. Clin. Virol. 2014, 61, 585–589. [Google Scholar] [CrossRef]
- Yang, Y.; Haitchi, H.M.; Cakebread, J.; Sammut, D.; Harvey, A.; Powell, R.M.; Holloway, J.W.; Howarth, P.; Holgate, S.T.; Dalvies, D.E. Epigenetic mechanisms silence a disintegrin and metalloprotease 33 expression in bronchial epithelial cells. J. Allergy Clin. Immunol. 2008, 121, 1393–1399.e14. [Google Scholar] [CrossRef]
- Sunadome, H.; Matsumoto, H.; Petrova, G.; Kanemitsu, Y.; Tohda, Y.; Horiguchi, T.; Kita, K.; Kuwabara, K.; Tomii, K.; Fujimura, N.; et al. IL4Rα and ADAM33 as genetic markers in asthma exacerbations and type-2 inflammatory endotype. Clin. Exp. Allergy 2017, 47, 998–1006. [Google Scholar] [CrossRef]
- Andrews, A.; Nasir, T.; Bucchieri, F.; Holloway, J.; Holgate, S.; Davies, D. IL-13 receptor α 2: A regulator of IL-13 and IL-4 signal transduction in primary human fibroblasts. J. Allergy Clin. Immunol. 2006, 118, 858–865. [Google Scholar] [CrossRef]
- Ghaffar, O.; Hamid, Q.; Renzi, P.M.; Allakhverdi, Z.; Molet, S.; Hogg, J.C.; Stephanie, A.S.; Luster, A.D.; Lamkhioued, B. Constitutive and Cytokine-Stimulated Expression of Eotaxin by Human Airway Smooth Muscle Cells. Am. J. Respir. Crit. Care Med. 1999, 159, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Jin, M.; Cai, Y.; Bai, C.; Shen, Y.; Yuan, Z.; Hu, Y.; Holgate, S. The effects of Th2 cytokines on the expression of ADAM33 in allergen-induced chronic airway inflammation. Respir. Physiol. Neurobiol. 2009, 168, 289–294. [Google Scholar] [CrossRef]
- Song, G.G.; Kim, J.H.; Lee, Y.H. Association between ADAM33 S2 and ST+4 polymorphisms and susceptibility to asthma: A meta-analysis. Gene 2013, 524, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Bora, E.; Arikan-Ayyildiz, Z.; Firinci, F.; Çankaya, T.; Giray-Bozkaya, Ö.; Uzuner, N.; Ülgetnalp, A. ADAM33 Gene Polymorphisms Are Not Associated with Asthma in Turkish Children. Pediatr. Allergy Immunol. Pulmonol. 2012, 25, 97–100. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, S.W.; Chang, H.K.; Kim, H.Y.; Rhim, T.; Lee, J.H.; Jang, A.S.; Koh, E.S.; Park, C.S. A Disintegrin and Metalloproteinase 33 Protein in Patients with Asthma. Am. J. Respir. Crit. Care Med. 2006, 173, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.C.; Mogas, A.K.; Olivenstein, R.; Fiset, P.O.; Chakir, J.; Bourbeau, J.; Pierre, E.; Lemiere, C.; Martin, J.; Hamid, Q. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J. Allergy Clin. Immunol. 2007, 119, 863–871. [Google Scholar] [CrossRef]
- Tripathi, P.; Awasthi, S.; Husain, N.; Prasad, R.; Mishra, V. Increased expression of ADAM33 protein in asthmatic patients as compared to non-asthmatic controls. Indian J. Med. Res. 2013, 137, 507–514. [Google Scholar]
- Awasthi, S.; Tripathi, P.; Prasad, R. Environmental risk factors for asthma in Lucknow: A case–control study. Clin. Epidemiol. Glob. Health 2013, 1, 115–123. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, M.; Qian, X.; Li, C.; Yue, L.; Liu, Y.; Shi, S. Interaction between early-life pet exposure and methylation pattern of ADAM33 on allergic rhinitis among children aged 3–6 years in China. Allergy Asthma Clin. Immunol. 2021, 17, 44. [Google Scholar] [CrossRef]
- Available online: https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr20%3A3667975%2D3682010&hgsid=1923650600_JkYLKkUGeuFlQAdhC2wncsgr3D3f (accessed on 15 February 2023).
- Vishweswaraiah, S.; Ramachandra, N.; Jayaraj, B.; Holla, A.; Chakraborty, S.; Agrawal, A.; Mahesh, P.A. Haplotype analysis of ADAM33 polymorphisms in asthma: A pilot study. Indian J. Med. Res. 2019, 150, 272. [Google Scholar]
- Liu, Y.; Wang, Z.-H.; Zhen, W.; Lu, S.-J.; Liu, Z.; Zou, L.-Y.; Xu, J.-J. Association between Genetic Polymorphisms in the ADAM33 Gene and Asthma Risk: A Meta-Analysis. DNA Cell Biol. 2014, 33, 793–801. [Google Scholar] [CrossRef]
- Deng, R.; Zhao, F.; Zhong, X. T1 polymorphism in a disintegrin and metalloproteinase 33 (ADAM33) gene may contribute to the risk of childhood asthma in Asians. Inflamm. Res. 2017, 66, 413–424. [Google Scholar] [CrossRef]
- Sun, F.J.; Zou, L.Y.; Tong, D.M.; Lu, X.Y.; Li, J.; Deng, C.B. Association between ADAM metallopeptidase domain 33 gene polymorphism and risk of childhood asthma: A meta-analysis. Braz. J. Med. Biol. Res. 2017, 50, e6148. [Google Scholar] [CrossRef]
- Zihlif, M.; Zihlif, N.; Obeidat, N.M.; Mahafza, T.; Froukh, T.; Ghanim, M.T.; AI-Akhras, F.M.; Naffa, R. Association between ADAM33 Polymorphisms and Susceptibility with Adult and Childhood Asthma among Jordanians. Genet Test Mol Biomark. 2014, 18, 767–774. [Google Scholar] [CrossRef]
- Liang, S.; Wei, X.; Gong, C.; Wei, J.; Chen, Z.; Deng, J. A disintegrin and metalloprotease 33 (ADAM33) gene polymorphisms and the risk of asthma: A meta-analysis. Hum. Immunol. 2013, 74, 648–657. [Google Scholar] [CrossRef]
- Chiang, C.H.; Lin, M.W.; Chung, M.Y.; Yang, U.C. The association between the IL-4, ADRβ2 and ADAM 33 gene polymorphisms and asthma in the Taiwanese populationstar. J. Chin. Med. Assoc. 2012, 75, 635–643. [Google Scholar] [CrossRef]
- Sultana, S.; Banerjee, P.; Ganai, I.; Laha, A.; Sultana, N.; Biswas, H.; Saha, N.C.; Moitra, S.; Poddetr, S. Polymorphism in ADAM33 gene associated with asthmatics in West Bengal, India—An investigation by in-silico analysis. World Allergy Organ. J. 2023, 16, 100834. [Google Scholar] [CrossRef]
- Zayed, K.S.; Kudhair, B.K.; Aziz, D.Z.; Lafta, I.J. The G allele of the ADAM33 T1 polymorphism (rs2280091) is a risk factor associated with asthma severity among the Iraqi Arab population. Hum. Gene 2023, 36, 201181. [Google Scholar] [CrossRef]
- Gupta, K.; Mishra, A.; Ali Khan, A.; Deb Nath, S. Correlation of antioxidants and IgE levels and its comparison with ADAM33 gene polymorphism (V4) in asthma patients of Jammu and Kashmir. J. Cardiovasc. Dis. Res. 2023, 14, 1100–1104. [Google Scholar]
- Vishweswaraiah, S.; Ramachandra, N.B.; Joshi, N.; Parthasarathi, A.; Kaleem Ullah, M.; Siddaiah, J.B.; Holla, A.D.; Chakraborty, S.; Agrawal, A.; Mahesh, P.A. Association between ADAM33 Single-Nucleotide Polymorphisms and Treatment Response to Inhaled Corticosteroids and a Long-Acting Beta-Agonist in Asthma. Diagnostics 2023, 13, 405. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.C.; Nickerson, D.A. Definition and Clinical Importance of Haplotypes. Annu. Rev. Med. 2005, 56, 303–320. [Google Scholar] [CrossRef]
- Stram, D.O.; Seshan, V.E. Multi-SNP Haplotype Analysis Methods for Association Analysis. In Statistical Human Genetics; Humana Press: New York, NY, USA, 2012; pp. 423–452. [Google Scholar]
- Vergara, C.I.; Acevedo, N.; Jiménez, S.; Martínez, B.; Mercado, D.; Gusmão, L.; Barnes, K.C.; Caraballo, L. A Six-SNP Haplotype of ADAM33 Is Associated with Asthma in a Population of Cartagena, Colombia. Int. Arch. Allergy Immunol. 2010, 152, 32–40. [Google Scholar] [CrossRef]
- Kedda, M.A.; Duffy, D.L.; Bradley, B.; O’Hehir, R.E.; Thompson, P.J. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur. J. Hum. Genet. 2006, 14, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Hirota, T.; Hasegawa, K.; Obara, K.; Matsuda, A.; Akahoshi, M.; Nakashima, K.; Shirakawa, T.; Doi, S.; Fujita, K.; Suzuki, Y.; et al. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin. Exp. Allergy 2006, 36, 884–891. [Google Scholar] [CrossRef]
- Wang, J.; Wen, J.; Si-Ma-Yi, M.H.R.G.L.; He, Y.B.; Tu-Er-Xun, K.L.B.N.; Xia, Y. Association of ADAM33 gene polymorphisms with asthma in the Uygur population of China. Biomed Rep. 2013, 1, 447–453. [Google Scholar] [CrossRef]
- Raby, B.A.; Silverman, E.K.; Kwiatkowski, D.J.; Lange, C.; Lazarus, R.; Weiss, S.T. ADAM33 polymorphisms and phenotype associations in childhood asthma. J. Allergy Clin. Immunol. 2004, 113, 1071–1078. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, H.-S.; Park, S.W.; Jang, A.S.; Uh, S.T.; Rhim, T. ADAM33 polymorphism: Association with bronchial hyper-responsiveness in Korean asthmatics. Clin. Exp. Allergy 2004, 34, 860–865. [Google Scholar] [CrossRef]
- Bukvic, B.K.; Blekic, M.; Simpson, A.; Marinho, S.; Curtin, J.A.; Hankinson, J.; Aberle, N.; Custovic, A. Asthma severity, polymorphisms in 20p13 and their interaction with tobacco smoke exposure. Pediatr. Allergy Immunol. 2013, 24, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Awasthi, S.; Ganesh, S. Association of ADAM33 Gene Polymorphisms with Reduction of Lung Function as Measured by Peak Expiratory Flow Rate Among Healthy Male Smokers and Non-Smokers. IUP J. 2 Genet. Evol. 2011, 4, 18–26. [Google Scholar]
- Reijmerink, N.E.; Kerkhof, M.; Koppelman, G.H.; Gerritsen, J.; de Jongste, J.C.; Smit, H.A.; Brunekreef, B.; Postma, D.S. Smoke exposure interacts with ADAM33 polymorphisms in the development of lung function and hyperresponsiveness. Allergy 2009, 64, 898–904. [Google Scholar] [CrossRef]
- Khoramipour, M.; Jalali, A.; Abbasi, B.; Hadi Abbasian, M. Evaluation of the association between clinical parameters and ADAM33 and ORMDL3 asthma gene single-nucleotide polymorphisms with the severity of COVID-19. Int. Immunopharmacol. 2023, 123, 110707. [Google Scholar] [CrossRef]
- Vishweswaraiah, S.; Veerappa, A.M.; Mahesh, P.A.; Jayaraju, B.S.; Krishnarao, C.S.; Ramachandra, N.B. Molecular interaction network and pathway studies of ADAM33 potentially relevant to asthma. Ann. Allergy Asthma Immunol. 2014, 113, 418–424.e1. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Han, W.; Zhou, Z.S. ADAM33 polymorphisms are associated with asthma and a distinctive palm dermatoglyphic pattern. Mol. Med. Rep. 2013, 8, 1795–1800. [Google Scholar] [CrossRef] [PubMed]
- Zihlif, M.; Imraish, A.; Al-Rawashdeh, B.; Qteish, A.; Husami, R.; Husami, R. The Association of IgE Levels with ADAM33 Genetic Polymorphisms among Asthmatic Patients. J. Pers. Med. 2021, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.M.; Hamilton, L.M.; Holgate, S.T.; Davies, D.E.; Holloway, J.W. ADAM33: A novel therapeutic target for asthma. Expert Opin. Ther. Targets 2003, 7, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Long, J.; Chen, J.; Jiang, X.; Zhu, J.; Jin, Y.; Lin, F.; Zhong, J.; Xu, R.; Mao, L.; et al. Overexpression of soluble ADAM33 promotes a hypercontractile phenotype of the airway smooth muscle cell in rat. Exp. Cell Res. 2016, 349, 109–118. [Google Scholar] [CrossRef]
- Pendergraff, H.M.; Krishnamurthy, P.M.; Debacker, A.J.; Moazami, M.P.; Sharma, V.K.; Niitsoo, L. Locked Nucleic Acid Gapmers and Conjugates Potently Silence ADAM33, an Asthma-Associated Metalloprotease with Nuclear-Localized mRNA. Mol. Ther. Nucleic Acids 2017, 8, 158–168. [Google Scholar] [CrossRef]
- Shin, M.; Chan, I.L.; Cao, Y.; Gruntman, A.M.; Lee, J.; Sousa, J. Intratracheally administered LNA gapmer antisense oligonucleotides induce robust gene silencing in mouse lung fibroblasts. Nucleic Acids Res. 2022, 50, 8418–8430. [Google Scholar] [CrossRef]

| SNP | Population | Characteristics |
|---|---|---|
| F + 1 | Asian in general [9], children in general [9], Jordanian [34], Northern Indian [34], German [34], Icelandic [34], and Chinese [34] | Associated with lung function deterioration in early life [34] |
| Q1, | Asian in general [9], Chinese Han [9], adults in general [9], and Caucasian children [9] | Homozygous for minor alleles of SNP Q-1 (CC), rapid decline in FEV1 of 9.6 mL/year [34] |
| T2, | Asian in general [9,35], children in general [9], West Bengal, and India [38] | Associated with the type 2 endotype of asthma [20] |
| S + 1 | South India [41] | |
| S1 | West Bengal, India [38] | Interaction between prenatal exposure to cigarette smoke in relation to the development of BHR [43] Associated with respiratory impedance at the age of 8 [43] |
| S2 | Jordanian, British, Europeans, Black Americans, White Americans, Hispanic Americans, and Thai [34,35] | Interaction between prenatal exposure to cigarette smoke in relation to the development of BHR [43] associated with predicted FEV1% [43] |
| ST + 4 | Children in general [9] | |
| ST + 7 | US White and Dutch White [10] | |
| T1 | Asian children [32], West Bengal, India [38], and Iraqi Arab population [14,39] | An allele associated with higher eosinophil count, increased airway hyperresponsiveness [36], increased inflammatory cell counts, and a decline in lung function in patients with COPD [32] Site corresponds to the domain involved in intracellular signaling [32] |
| V4 | Caucasian in general [9], adults in general [9], Jordanian children [34], United States/United Kingdom combined children [34], United Kingdom children [34], Dutch White children [34], Indian children [34], Dutch in general [10], and South India [40] | Associated with an increased risk of asthma with no significant differences in population selectivity [13] Located in the 3′UTR; changes may affect transcription in relationship to closely located T1 [34,35] Linked to decreased lung function and impaired improvement in lung function after three months of ICS + LABA [40] Heterozygotes with reduced FEV1 among smokers have been observed [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sleziak, J.; Gawor, A.; Błażejewska, M.; Antosz, K.; Gomułka, K. ADAM33′s Role in Asthma Pathogenesis: An Overview. Int. J. Mol. Sci. 2024, 25, 2318. https://doi.org/10.3390/ijms25042318
Sleziak J, Gawor A, Błażejewska M, Antosz K, Gomułka K. ADAM33′s Role in Asthma Pathogenesis: An Overview. International Journal of Molecular Sciences. 2024; 25(4):2318. https://doi.org/10.3390/ijms25042318
Chicago/Turabian StyleSleziak, Jakub, Antoni Gawor, Marta Błażejewska, Katarzyna Antosz, and Krzysztof Gomułka. 2024. "ADAM33′s Role in Asthma Pathogenesis: An Overview" International Journal of Molecular Sciences 25, no. 4: 2318. https://doi.org/10.3390/ijms25042318







